Introduction
Lung cancer remains the most common cause of cancer
mortality worldwide (1,2). Approximately 80% of all lung cancer
patients are diagnosed with non-small-cell lung cancer (NSCLC)
(3). Currently, lung cancer therapy
is mainly based on Tumor-Node-Metastasis (TNM) disease staging and
tumor histological classification. However, despite progress in
surgical techniques, chemotherapy and radiotherapy, the 5-year
survival rate of patients with lung cancer remains low (~16%)
(4,5).
Therefore, there is a continuous need to identify specific and
sensitive biomarkers that may improve cancer patient management.
Such markers should allow prediction and prognostication of patient
survival, disease free survival or treatment response (6). Therefore, the current study aimed to
investigate potential molecular markers which may become novel
prognostic factors in NSCLC, specifically jagged 1 (Jag1), jagged 2
(Jag2), delta-like protein 1 (Dll1), delta-like protein 3 (Dll3),
delta-like protein 4 (Dll4), Notch 1 and hairy and enhancer of
split-1 (Hes1). The present study focused on the Notch signaling
pathway, which is known to have a significant role in tumorigenesis
and cancer progression (7,8). The Notch family consists of four
receptors (Notch 1–4) and five ligands (Jag1, Jag2, Dll1, Dll3 and
Dll4) (9). Notably, receptors and
ligands are typically presented on neighboring cells; therefore,
ligand binding is triggered via direct cell-cell communication
(10). Notch ligands function as
Notch signaling agonists, exerting their actions through
intercellular interactions (11).
However, in mammals, binding between Notch ligands and Notch
receptors remains a non-selective process (12). Recent data revealed that certain Notch
ligands may be highly expressed in lung cancer cells (5). Furthermore, it has long been known that
the lung constitutes the richest source of Notch ligand and
receptor mRNA (13). However, to the
best of our knowledge, to date the role of Notch ligands in cancer
pathogenesis remains to be fully elucidated. Notably, a low rate of
mutations observed in Notch ligands in cancer may make them a good
target for research on cancer therapy concepts (14). However, despite available knowledge,
there remains limited information on the level of Notch ligand
expression in NSCLC patients (15–17).
Materials and methods
Patients and tissue samples
The present study was performed on 61 pairs of tumor
and matched unaffected lung tissue specimens obtained from patients
with various stages of NSCLC, aged from 39.8 to 78.1 years (mean,
62.5 years; standard deviation, 8.4 years) who underwent a curative
surgery between March 2003 and October 2009 at the Department of
Thoracic Surgery, Bialystok Medical University Hospital (Poland).
Detailed patient characteristics are presented in Table I.
 | Table I.Patient characteristics. |
Table I.
Patient characteristics.
| Characteristic | n | % |
|---|
| Gender |
|
|
| Male | 46 | 75.4 |
|
Female | 15 | 24.6 |
| Age at diagnosis,
years |
|
|
| Median
(range) | 61.6
(39.8–78.1) |
|
| Mean | 62.5 |
|
| Smoking history |
|
|
|
Smoker | 54 | 88.5 |
|
Non-smoker | 7 | 11.5 |
| Histology |
|
|
|
Squamous cell carcinoma | 25 | 41.0 |
|
Adenocarcinoma | 28 | 45.9 |
| Large
cell carcinoma | 8 | 13.1 |
| Tumor size, T |
|
|
|
T1a | 6 | 9.8 |
|
T1b | 7 | 11.5 |
|
T2a | 26 | 42.6 |
|
T2b | 10 | 16.4 |
| T3 | 12 | 19.7 |
| Lymph node status,
N |
|
|
| N0 | 45 | 73.8 |
| N1 | 16 | 26.2 |
| Tumor stage |
|
|
| IA
(I) | 11 | 18.0 |
| IB
(II) | 19 | 31.2 |
| IIA
(III) | 11 | 18.0 |
| IIB
(IV) | 20 | 32.8 |
| Follow-up period,
months |
|
|
|
Median | 49 |
|
|
Range | 5–86 |
|
| Status |
|
|
|
Alive/censored | 33 | 54.1 |
|
Succumbed to lung cancer | 27 | 44.3 |
| Other
cause of mortality | 1 | 1.6 |
| Relapse-free
survival time, months |
|
|
|
Median | 47 |
|
|
Range | 3–86 |
|
The samples were collected upon obtaining informed
consent from the patients at the time of surgery, and the present
study was approved by the Ethics Committee of the Medical
University of Bialystok (Bialystok, Poland). Tissue samples were
processed immediately following surgical removal. Tumor tissue and
unaffected lung tissue specimens from the same lobe or lung of the
patient were snap-frozen in liquid nitrogen, followed by storage at
−80°C. Prior to processing, the sections of frozen tissue specimens
were stained with hematoxylin and eosin and evaluated by
pathologists to confirm the suitability of tumor cell content. Only
the tumor samples that contained at least 50% tumor cells following
microscopic observation using the Leica DM 2000 LED light
microscope (Leica Microsystems GmbH, Wetzlar, Germany), as well as
unaffected lung tissue samples without malignant cells, were used
for further analysis.
RNA extraction and quality
control
Total RNA was isolated from fresh-frozen tissue
specimens using the mirVana miRNA isolation kit (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
protocol. The 100-µl resulting RNA extracts were stored at −80°C
prior to further processing. Quantity and quality of RNA assessment
was performed using a UV/VIS spectrophotometer NanoDrop 2000c
(Thermo Fisher Scientific, Inc.). The level of integrity required
for quantitation (RNA integrity number >7) was determined for
the extracted total RNA using the Agilent RNA 6000 Nano kit on a
Bioanalyzer 2100 (Agilent Technologies, Inc., Santa Clara, CA,
USA). A total of 500 ng of the RNA was reverse transcribed into
cDNA in a reaction with High Capacity RNA-to-cDNA Master mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol.
Quantitative polymerase chain reaction
(qPCR)
mRNA expression levels of Jag1, Jag2,
Dll1, Dll3, Dll4, Notch1 and
Hes1 were evaluated in the tumor and unaffected lung tissues
using comparative qPCR. The TaqMan probes (Hs01070032_m1
Jag1, Hs00171432_m1 Jag2, Hs00194509_m1 Dll1,
Hs01085096_m1 Dll3, Hs00184092_m1 Dll4, Hs00172878_m1
Hes1, Hs01062014_m1 Notch1) and the TaqMan Assay kit
(all from Applied Biosystems; Thermo Fisher Scientific, Inc.) were
used to perform PCR. The expression of the above-mentioned genes
[by the change-in-cycling-threshold ΔCq method (18,19)] were
calculated and normalized to ribosomal 18SRNA gene
expression (Hs99999901_s1 18SRNA). The following
thermocycling conditions were used: 50°C for 2 min; 95°C for 10
min; 40 cycles of 95°C for 15 sec and 60°C for 60 sec. Each sample
was analyzed in triplicate. All reactions were performed using the
ABI PRISM® 7900HT Sequence Detection system (Thermo
Fisher Scientific, Inc.).
Statistical analysis
For statistical analysis, GraphPad Prism software
(version 5.01; GraphPad Software, Inc., La Jolla, CA, USA) was
used. The normality of distribution was analyzed using the
Shapiro-Wilk W test. Based on the results, the following tests were
used: i) Student's t-test, for normally distributed variables; ii)
Mann-Whitney U test, for variables whose distributions differed
from normal in at least one of the compared groups. Survival curves
were created with the Kaplan-Meier method, and the log-rank test
was used to determine differences between survival proportions. Cox
proportional hazards model was applied to assess the prognostic
strength of high or low Notch ligand expression levels. P<0.05
was considered to indicate a statistically significant
difference.
Results
A total of 61 NSCLC and adjacent noncancerous lung
tissue samples were analyzed by reverse transcription-qPCR.
Initially, it was observed that Notch receptors and ligands were
expressed in NSCLC and noncancerous lung tissue at detectable
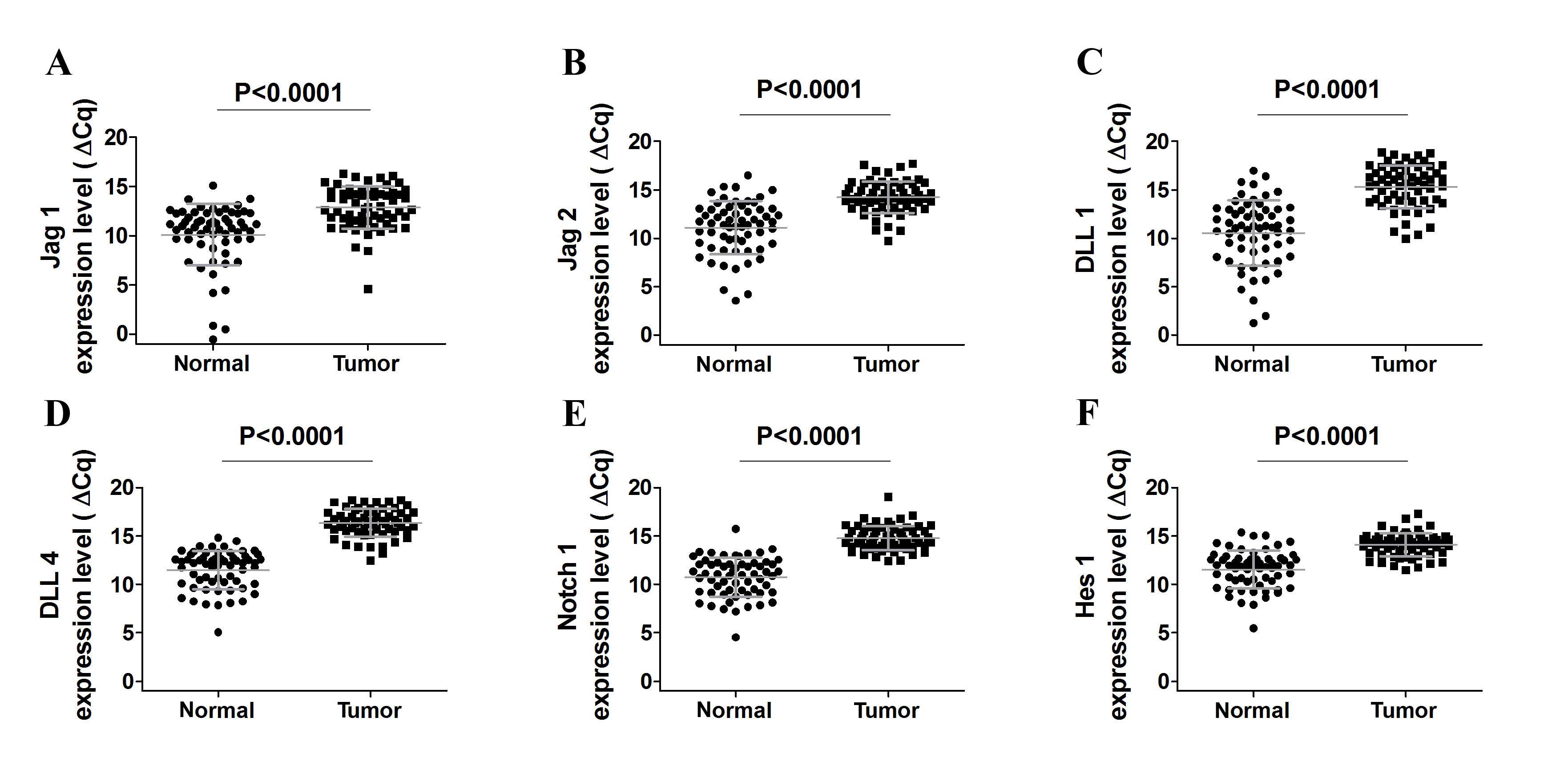
levels (Fig. 1A-E). Notably, Jag1,
Jag2, DLL1 and DLL4 expression levels were lower in
cancer when compared to the noncancerous lung tissue (all
P<0.0001; Fig. 1A-F), which was
indicated by increased ΔCq values. Furthermore, DLL3
expression was consistently low and was observed only in 21/61
tumor tissues, and consequently, no significant differences in gene
expression in tumor to non-tumor lung tissue were observed (data
not shown). In 4/25 squamous cell carcinoma tumor samples,
detectable levels of DLL3 expression were observed.
DLL3 was expressed in 14/28 adenocarcinoma samples and in
3/8 large cell carcinoma samples (Table
II).
 | Table II.Delta-like protein 3 expression in
tumors. |
Table II.
Delta-like protein 3 expression in
tumors.
| Histology | Lack of DLL3
expression, n (%) | Presence of
DLL3 expression, n (%) | Total, n (%) |
|---|
| Adenocarcinoma | 14 (50) | 14 (50) | 28 (100) |
| Large cell
carcinoma |
5 (62.5) |
3 (37.5) | 8
(100) |
| Squamous cell
carcinoma | 21 (84) | 4
(16) | 25 (100) |
| Total | 40 (65.6) | 21 (43.4) | 61 (100) |
Subsequently, the present study analyzed the
expression of Hes1, a basic helix-loop-helix transcriptional
repressor that is a downstream target of Notch signaling. Notably,
Notch 1 and Hes1 expression was decreased in tumor
samples compared to corresponding noncancerous lung tissue (both
P<0.0001) and these observations were consistent with the
results of the Notch ligands gene expression investigation
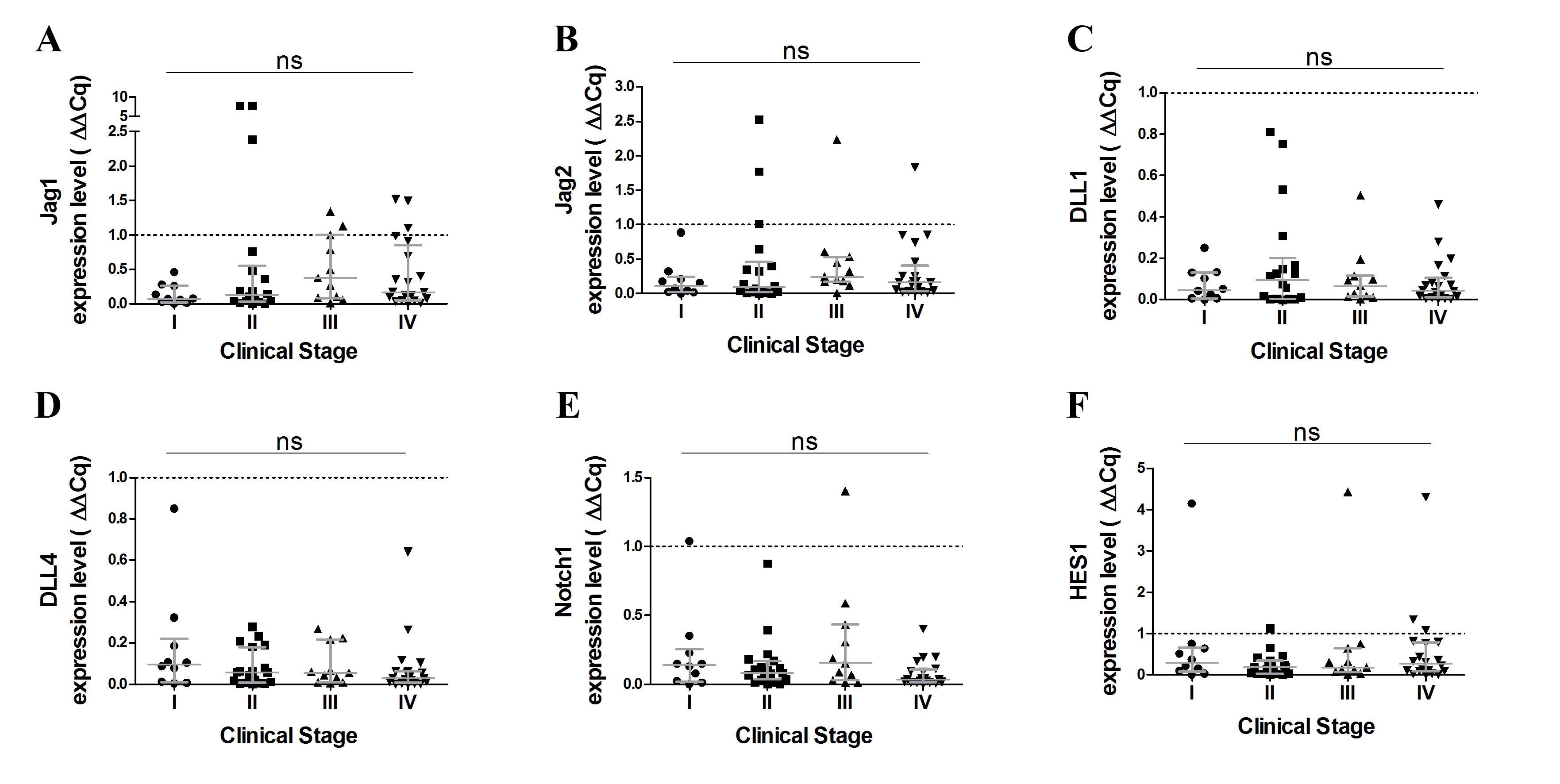
(Fig. 1E). Furthermore, no
significant differences were observed in terms of Notch receptors
and ligand expression levels at various stages of the disease
(Fig. 2).
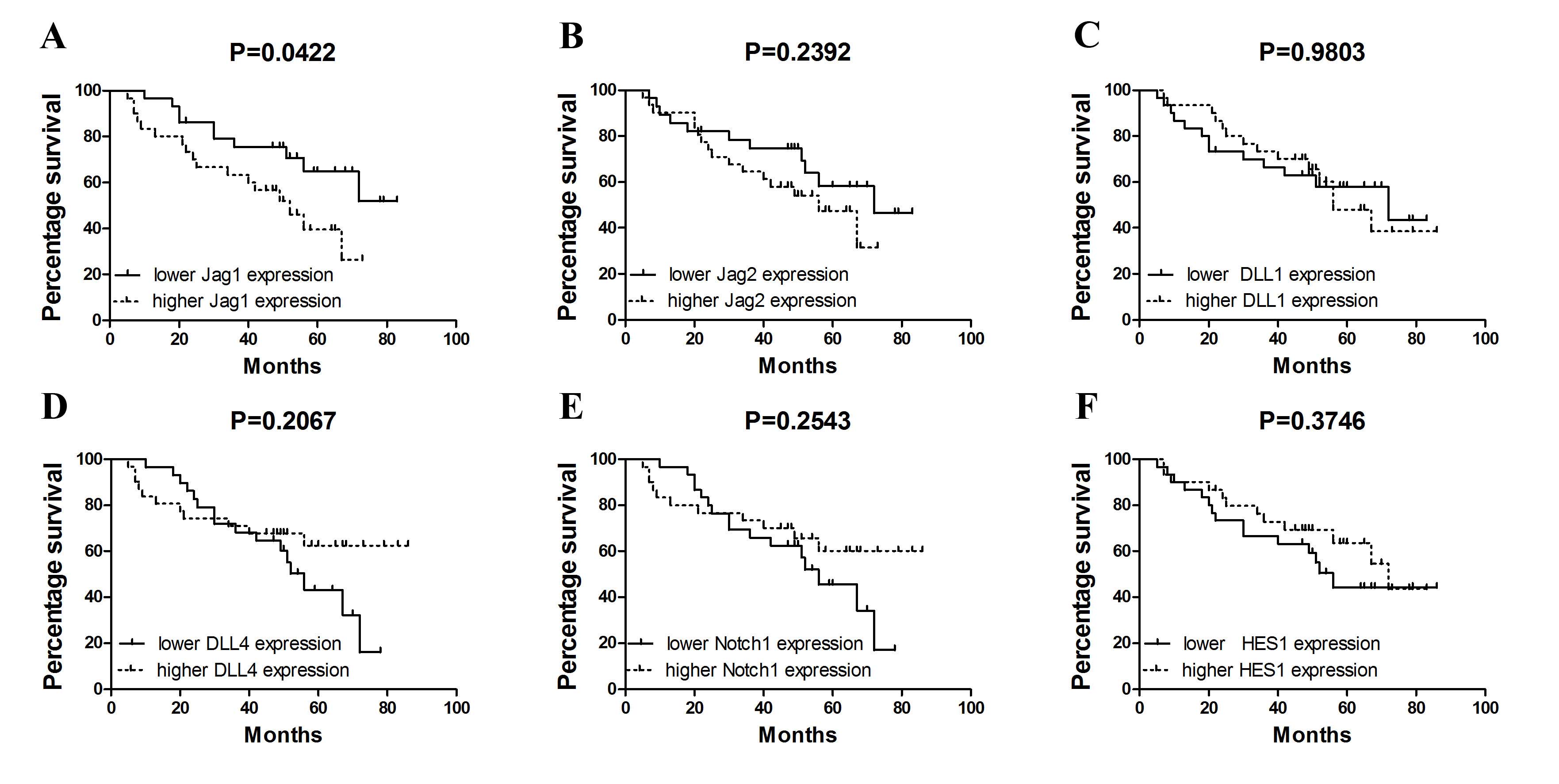
Furthermore, the present study analyzed whether
quantitative assessment of Notch ligand expression in NSCLC may
improve patient prognostication. Having used Cox regression
analysis, it was observed that increased expression of Jag1
(above the median value observed in all NSCLC patients, 0.1478 for
Jag1, 0.1641 for Jag2, 0.0590 for Dll1, 0.0545
for Dll4, 0.0791 for Notch1 and 0.2022 for
HES1) may serve as an indicator of poor overall survival
(hazard ratio, 2.220; 95% confidence interval, 1.005–4.905;
P=0.048) in NSCLC. Notably, by the use of Kaplan-Meier estimates it
was demonstrated that increased Jag1 expression in tumor
tissue is associated with shortened overall survival (P=0.0422;
Fig. 3A). By contrast, no
statistically significant associations between survival and ligand
expression for other genes were identified (Jag2, P=0.2392; DLL1,
P=0.9803; DLL4, P=0.2067; Notch1, P=0.2543; HES1, P=0.3746;
Fig. 3B-F).
Discussion
In the present study, the expression of Notch
ligands, Notch 1 and its target gene Hes1, were
analyzed in NSCLC tissues. It was observed that Notch ligands,
including Jag1, Jag2, Dll1 and Dll4, were expressed
in tumor and unaffected lung tissue samples in patients with NSCLC.
By contrast, Dll3 expression was detected in 21/61 tumor
tissue samples. The results presented in the current study reveal a
potentially novel role of Notch ligands in NSCLC, as it was
demonstrated that the Notch signaling pathway is significantly
deregulated in NSCLC patients. The levels of Notch ligands were
consistently lower in tumor tissues, when compared to adjacent
noncancerous lung tissues from the same patient. Thus, the present
study aimed to evaluate the potential role of the analyses of
tumor-associated alterations of Notch signaling in prognostication
of NSCLC patients. The identification of novel prognostic markers
capable of predicting overall survival and clinical consequences of
applied therapies is required in NSCLC. The present study
demonstrated that increased expression of Jag1 may be an
indicator of poor overall survival. This finding is notable as
tumor tissues were characterized by reduced Notch signaling levels
compared to noncancerous tissues, but increased expression of
Jag1 indicated a less favorable clinical outcome. Thus, the
results of the present study add another piece of evidence to the
complex role of Notch signaling in the pathogenesis of NSCLC. In
certain ways, the present findings do not support the results of a
recent meta-analysis performed by Yuan et al (20) who demonstrated that increased
expression of Notch 1 was more frequently accompanied by
lymph node metastasis and more advanced TNM stage. Consistently,
Yuan et al (20) observed that
patients with Notch 1 or Notch 3 overexpression
presented with significantly poorer overall survival. Similarly, in
another recent study, Notch 1 overexpression was observed to
increase the metastatic potential of NSCLC cells (21). To a certain extent, these findings
were contrasted by Nguyen et al (22) who discovered that the expression of
Notch 1 receptors in NSCLC tissues was negatively associated with
stage and nodal status, but not tumor size. Notably, the expression
of activated form of Notch 1, N1-ICD (intracellular domain)
was low and neither significantly associated with stage nor nodal
status (22). To date, it has been
postulated that the effects of Notch signaling contributing to
maintaining a balance between cell proliferation and apoptosis may
be either oncogenic or tumor-suppressive depending on the cancer
type (7). In the present study, it
may be hypothesized that these effects differ even within a single
cancer type. Similarly, Notch-associated signals have dual negative
and positive (oncogenic and tumor-suppressive) roles in the course
of hematological malignancies (23).
In the present study, reduced expression levels of Notch ligands
and receptors in tumor tissues were consistently observed compared
to control samples. By contrast, increased expression levels of
Jag1 were observed to be positively associated with a less
favorable clinical outcome.
The results of the present study warrant additional
studies on whether downregulation of Notch ligands and receptors is
induced by tumors in order to suppress endogenous anti-tumor
mechanisms, or if it represented a mechanism of self-defense
against the developing tumor. The idea of downregulation of Notch
signaling is novel for lung cancer but it is not unexpected in the
light of studies performed on other types of cancer, including
endometrial cancer. Jonusiene et al (24) investigated the expression of Notch
receptors (Notch 1, Notch 2, Notch 3 and
Notch 4), ligands (Jag1, Jag2 and Dll1)
and target gene Hes1 in endometrial cancer and adjacent
non-tumor endometrial tissue from endometrial cancer patients. The
mRNA levels of Notch receptors and ligands were reduced in
endometrial cancer compared with adjacent non-tumor tissue
(24). Furthermore, in contrast to
the results of the present study, the expression of Notch1,
Notch4 and Dll1 in IB stage adenocarcinoma was
significantly reduced compared with the expression of these
molecules at the IA stage (16).
Considered along with the results of the present study, these
findings suggest that Notch-mediated signaling may support
tumor-suppressing mechanisms in certain types of cancer, including
lung or endometrial cancer.
In conclusion, the present study reported that Notch
1 and Notch ligands are downregulated in tumors when compared to
noncancerous lung tissue in NSCLC patients. Furthermore, the
present study demonstrated that quantitative measurement of
Jag1 expression may improve prognostication of NSCLC patient
survival. In contrast to the work of previous authors, the present
study hypothesizes that the role of Notch signaling in the
pathogenesis of NSCLC cannot be simply linked to either
upregulation or downregulation of its ligands and receptors in
tumor tissue. The identification of alternative factors influencing
Notch-related signaling pathways in the settings of NSCLC remains
warranted.
Acknowledgements
The present study was supported by the Budget for
Science between the years 2013 and 2015, project no., IP2012 033872
(Iuventus Plus), to Joanna Pancewicz-Wojtkiewicz.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Greenhalgh J, Dwan K, Boland A, Bates V,
Vecchio F, Dundar Y, Jain P and Green JA: First-line treatment of
advanced epidermal growth factor receptor (EGFR) mutation positive
non-squamous non-small cell lung cancer. Cochrane Database Syst Rev
CD010383. 2016. View Article : Google Scholar
|
|
4
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kosakowska EA, Stec R, Charkiewicz R,
Skoczek M and Chyczewski L: Molecular differences in the KRAS gene
mutation between a primary tumor and related metastatic sites-case
report and a literature review. Folia Histochem Cytobiol.
48:597–602. 2010.PubMed/NCBI
|
|
6
|
Ludwig JA and Weinstein JN: Biomarkers in
cancer staging, prognosis and treatment selection. Nat Rev Cancer.
5:845–856. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Radtke F and Raj K: The role of Notch in
tumorigenesis: Oncogene or tumour suppressor? Nat Rev Cancer.
3:756–767. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sethi N and Kang Y: Notch signalling in
cancer progression and bone metastasis. Br J Cancer. 105:1805–1810.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pancewicz J and Nicot C: Current views on
the role of Notch signaling and the pathogenesis of human leukemia.
BMC Cancer. 11:5022011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Aster JC, Pear WS and Blacklow SC: The
Varied Roles of Notch in Cancer. Annual Review of Pathology:
Mechanisms of Disease. 12:null. 2017. View Article : Google Scholar
|
|
11
|
D'Souza B, Miyamoto A and Weinmaster G:
The many facets of Notch ligands. Oncogene. 27:5148–5167. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shimizu K, Chiba S, Kumano K, Hosoya N,
Takahashi T, Kanda Y, Hamada Y, Yazaki Y and Hirai H: Mouse jagged1
physically interacts with notch2 and other notch receptors.
Assessment by quantitative methods. J Biol Chem. 274:32961–32969.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu K, Moghal N and Egan SE: Notch
signaling in lung development and disease. Adv Exp Med Biol.
727:89–98. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li D, Masiero M, Banham AH and Harris AL:
The notch ligand JAGGED1 as a target for anti-tumor therapy. Front
Oncol. 4:2542014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nasarre P, Potiron V, Drabkin H and Roche
J: Guidance molecules in lung cancer. Cell Adh Migr. 4:130–145.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jin MM, Ye YZ, Qian ZD and Zhang YB: Notch
signaling molecules as prognostic biomarkers for non-small cell
lung cancer. Oncol Lett. 10:3252–3260. 2015.PubMed/NCBI
|
|
17
|
Choi K, Ahn YH, Gibbons DL, Tran HT,
Creighton CJ, Girard L, Minna JD, Qin FX and Kurie JM: Distinct
biological roles for the notch ligands Jagged-1 and Jagged-2. J
Biol Chem. 284:17766–17774. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Endoh H, Tomida S, Yatabe Y, Konishi H,
Osada H, Tajima K, Kuwano H, Takahashi T and Mitsudomi T:
Prognostic model of pulmonary adenocarcinoma by expression
profiling of eight genes as determined by quantitative real-time
reverse transcriptase polymerase chain reaction. J Clin Oncol.
22:811–819. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bolkun L, Grubczak K, Schneider G, Zembko
P, Radzikowska U, Singh P, Kloczko J, Ratafczak MZ, Moniuszko M and
Eljaszewicz A: Involvement of BAFF and APRIL in resistance to
apoptosis of acute myeloid leukemia. J Cancer. 7:1979–1983. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yuan X, Wu H, Xu H, et al: Meta-analysis
reveals the correlation of Notch signaling with non-small cell lung
cancer progression and prognosis. Sci Rep. 5:103382015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li C, Song G, Zhang S, Wang E and Cui Z:
Wnt3a increases the metastatic potential of non-small cell lung
cancer cells in vitro in part via its upregulation of Notch3. Oncol
Rep. 33:1207–1214. 2015.PubMed/NCBI
|
|
22
|
Nguyen D, Rubinstein L, Takebe N, Miele L,
Tomaszewski JE, Ivy P, Doroshow JH and Yang SX: Notch1 phenotype
and clinical stage progression in non-small cell lung cancer. J
Hematol Oncol. 8:92015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Leong KG and Karsan A: Recent insights
into the role of Notch signaling in tumorigenesis. Blood.
107:2223–2233. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jonusiene V, Sasnauskiene A, Lachej N,
Kanopiene D, Dabkeviciene D, Sasnauskiene S, Kazbariene B and
Didziapetriene J: Down-regulated expression of Notch signaling
molecules in human endometrial cancer. Med Oncol. 30:4382013.
View Article : Google Scholar : PubMed/NCBI
|