Introduction
Colorectal cancer (CRC) is one of the most commonly
diagnosed cancers, and is a common cause of cancer-associated
mortalities in developed countries (1,2). In China,
CRC is the fifth most common cancer type and the fourth most common
cause of cancer-associated mortality (3). CRC prognosis is dependent on tumor stage
at presentation (4). The 5-year
overall survival ranges from 93% for stage I patients to 8% for
stage IV patients (5). Currently, the
only curative treatment is surgical resection, while a modest
number of patients that survive benefit from chemotherapy (6). Despite numerous efforts having been made
in the past few decades, CRC survival rates have not been
significantly improved (7). As a
result, novel biomarkers that are of clinical value are thus
urgently required to facilitate early detection and allow
personalized treatment strategies for patients at high risk of CRC,
thus improving the compliance rates (8). A family of small regulatory RNAs termed
microRNAs (miRNAs or miRs) has emerged with important functions in
development, cell differentiation, and regulation of cell cycle and
apoptosis (9–11).
miRNAs are a class of short 22 nucleotide-long
non-coding RNAs that repress protein translation through binding to
their target messenger RNAs (mRNAs) (12,13).
Bioinformatics and cloning studies have estimated that miRNAs may
regulate 30% of all human genes and control hundreds of gene
targets (14). In the past decade, a
particular important role by miRNAs in tumor pathogenesis has
surfaced (15). Indeed, globally
abnormal miRNA expression patterns can classify human cancers,
invasion, progression and response to therapy (16). It has been well-documented that
certain specific miRNAs contribute to cell transformation,
proliferation and tumorigenesis by serving as targets of genomic
lesions that are associated with activation of oncogenes and
inactivation of tumor suppressor genes in cancer cells, including
amplification, deletion and epigenetic silencing (17). In addition, miRNAs can provide
functional links downstream of classic oncogenic and
tumor-suppressor signaling pathways (18). Furthermore, numerous studies have
demonstrated the pro- and anti-tumorigenic activities of specific
miRNAs both in vitro and in vivo (19,20).
miRNA expression profiling in stage III CRC
identified >11 miRNAs, including miR-21 and miR-141, that were
significantly upregulated (21). It
has been well documented that miR-21 functions as an oncogene and
modulates tumorigenesis through the regulation of genes such as
B-cell lymphoma-2 (22). miR-141 has
been also implicated in tumorigenesis of several types of cancer,
including nasopharyngeal cancer (23). An early study revealed that
upregulation of miR-141, which was closely associated with late
stage and poor survival in CRC, was only observed in CRC patients'
blood, but not in cancer tissues, suggesting that such an increase
may be derived from inflammatory responses in these patients
(1). However, a more recent study
demonstrated that miR-141 was significantly elevated in CRC tissue
samples at stage III, compared with non-cancerous (NC) adjacent
colorectal mucosa (21).
Experimentally validated target genes of miR-141 include
mitogen-activated protein kinase kinase 4 (MAP2K4) (23). MAP2K4 is a tumor suppressor gene that
can phosphorylate c-Jun N-terminal kinase or p38 with dual
specificity, resulting in the activation of the stress-activated
protein kinase pathway, which has been associated with apoptosis
and neoplastic transformation (24,25).
In the present study, the hypothesis that miR-141
promotes cell proliferation of colon cancer by inhibiting MAP2K4
was tested. The results demonstrated that miR-141 was significantly
upregulated in CRC cancer tissues, but barely detected in NC
adjacent colorectal mucosa. By contrast, the levels of MAP2K were
significantly elevated in normal tissues, but it was present at
less detectable levels in CRC. In addition, it was further
functionally confirmed that miR-141 inhibited MAP2K4 in a variety
of colon cancer lines, thus promoting the proliferation of cancer
cells. Our results suggest that miR-141 can be a therapeutic target
for the treatment of CRC.
Materials and methods
Cell culture
The human colon carcinoma cell lines HT29, T84 and
LS174 (ATCC, Manassas, VA, USA) were maintained in RPMI-1640 medium
supplemented with 10% fetal bovine serum (ATCC). The cells were
grown in a humidified incubator at 37°C with 5% CO2.
Bioinformatics analysis
To investigate whether miR-141 targets MAP2K4, a
bioinformatics analysis was performed using TargetScan (www.targetscan.org) and microRNA.org
(http://www.microrna.org/microrna/home.do). miR-141 was
shown to target sites 75–81 and 192–199 of the 3′-untranslated
region (UTR) of MAP2K4 mRNA.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNAs were extracted with TRIzol (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA). Dried RNA
pellets were re-suspended in appropriate volumes of
diethylpyrocarbonate-trated H2O (Sigma-Aldrich; Merck
Millipore, Darmstadt, Germany). Complementary DNA synthesis was
achieved with miR-141-specific, U6 small nuclear RNA
(snRNA)-specific or oligo-dT primers using SuperScript II Reverse
Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.). qPCR
was performed using an ABI 7300 Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The cycling conditions
were as follows: 95°C for 1 min, followed by 40 cycles at 55°C for
30 sec and 70°C for 30 sec. miR-141 or MAP2K4 mRNA levels were
determined relative to U6 or GAPDH expression, respectively, using
a SYBR Green PCR kit (Qiagen GmbH, Hilden, Germany). Fold-change in
expression was determined by the quantification cycle (Cq) method
using the formula 2-ΔΔCq (26). The primers used were as follows: U6
snRNA forward, 5′-TGCGGGTGCTCGCTTCGGCAGC-3′ and reverse,
5′-GGGTCCGAGGTGCACTGGATACGACAAAATATGG-3′; miR-141 forward,
5′-ATCGCCAGGATAAATTGACGCA-3′ and reverse,
5′-CCGCCTTAACACTGTCTGGTA-3′; MAP2K4 forward,
5′-GATGAATCCAAAAGGCCAAA-3′ and reverse,
5′-TCAATCGACATACATGGGAGAG-3′; and GAPDH forward,
5′-CTCCCGCTTCGCTCTCTG-3′ and reverse, 5′-CTGGCGACGCAAAAGAAG-3′.
Western blotting
Whole cell lysates were re-suspended in 1X SDS
loading buffer, boiled at 95°C for 5 min and centrifuged AT 3,000 ×
g for 5 min at 4°C. Supernatants were resolved on 10% SDS-PAGE and
transferred onto polyvinylidene difluoride membranes (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Membranes were blocked in
5% nonfat milk in PBS with Tween-20 (PBST) [10 mM phosphate buffer
(pH 7.2), 150 mM NaCl and 0.1% Tween-20] for 60 min, washed three
times with PBST and incubated with anti-MAP2K4 antibody (1:500;
cat. no. sc-376838; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA) at 4°C overnight. The blots were probed with anti-β-actin
antibody (1:500; cat. no. A-5316; Sigma-Aldrich; Merck Millipore)
as a loading control. Membranes were washed three times with PBST,
incubated with horseradish peroxidase (HRP)-conjugated secondary
antibody (1:5,000; cat. no. 115-035-003; Jackson ImmumoResearch,
West Grove, PA, USA) and developed with Immun-Star HRP Substrate
(Bio-Rad Laboratories, Inc.).
Immunohistochemistry
Briefly, the tissue samples were fixed in buffered
formalin for 24 h, dehydrated in 70% ethanol, paraffin-embedded and
sectioned (5-µm). The slides were incubated with proteinase K
(Roche Diagnostics, Indianapolis, IN, USA) for 20 min to help
uncover the hidden antigens. Following incubation with the primary
antibody against MAP2K4 at 1:100 dilution overnight at 4°C, the
sections were washed with TBS containing Tween-20 (TBST) prior to
being incubated with the HRP-conjugated secondary antibody diluted
in TBS. Sections were washed again with TBST, and the secondary
antibody was detected using the VECTASTAIN® Elite ABC
HRP kit (Thermo Fisher Scientific, Inc.) for 30 min. Two
independent pathologists evaluated the intensity of cytoplasmic
staining of MAP2K4 on a scale of 0 to 4 (0 corresponds to the
absence of staining and 4 is the highest degree of staining). The
range of the scale is 2–3 for NC and 0–1 for colonic adenocarcinoma
(CAC).
Cell proliferation assay
MTT assay was performed to estimate the effect of
miR-141 on human CAC cell proliferation, as previously described
(27). Cells were seeded into 96-well
plates (5,000 cells/well in 200 µl medium) and incubated for 24 h
at 37°C, 5% CO2. Pancreatic ductal adenocarcinoma cells
were transfected with miR-141 mimics (miR-141) or miR-141
antagomirs (Ant-mIR-141) (IDT DNA, Coralville, IA, USA) using
Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). Cells cultured
in complete medium were used as a blank control. At the end of
culturing, 20 µl of a 5 mg/ml MTT solution (Sigma-Aldrich; Merck
Millipore) was added per well, and the cells were incubated for
another 4 h at 37°C. Supernatants were then removed, and formazan
crystals were dissolved in 150 µl dimethyl sulfoxide
(Sigma-Aldrich; Merck Millipore). Finally, the optical density was
determined at 490 nm using Victor 3 Multi-Label Microplate Reader
(PerkinElmer, Inc., Waltham, MA, USA). In each assay, five parallel
wells were analyzed, and the results were collected as the mean of
≥3 independent experiments.
Luciferase assay
cDNA strands corresponding to the 3′-UTR of MAP2K4
mRNA containing the wild-type (WT) or mutant seed sequence (the
distal one) of miR-141 were synthesized and the complementary
strands were annealed at 75°C for 2 min, cooled to room temperature
for 5 min and cloned into the pGL4.32 vector (Promega Corporation,
Madison, WI, USA) immediately downstream of the firefly luciferase
(FL) gene. The resulting plasmids were sequence-validated prior to
further analysis. HT29 cells were transfected with the plasmid for
48 h using Lipofectamine 2000, and luciferase assays were performed
using the luciferase assay system (Promega Corporation) and the
Victor 3V plate reader (PerkinElmer, Inc., Waltham, MA, USA),
according to the manufacturer's protocol.
Statistical analysis
Data are expressed as the mean ± standard deviation.
Student's t-test was used to compare the differences between two
groups. Statistical analyses were performed using Microsoft Excel
2013 (Microsoft Corporation, Redmond, WA, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-141 is upregulated, but MAP2K4 is
downregulated, in CRC
The levels of miR-141 and MAP2K4 were determined in
CRC tissue specimens. Total RNAs were extracted from a total of 12
colon cancer and NC adjacent tissue samples for assessment of
miR-141 and MAP2K4 mRNAs levels by qPCR. The same tissues were
formaldehyde-fixed, paraffin-embedded and subjected to
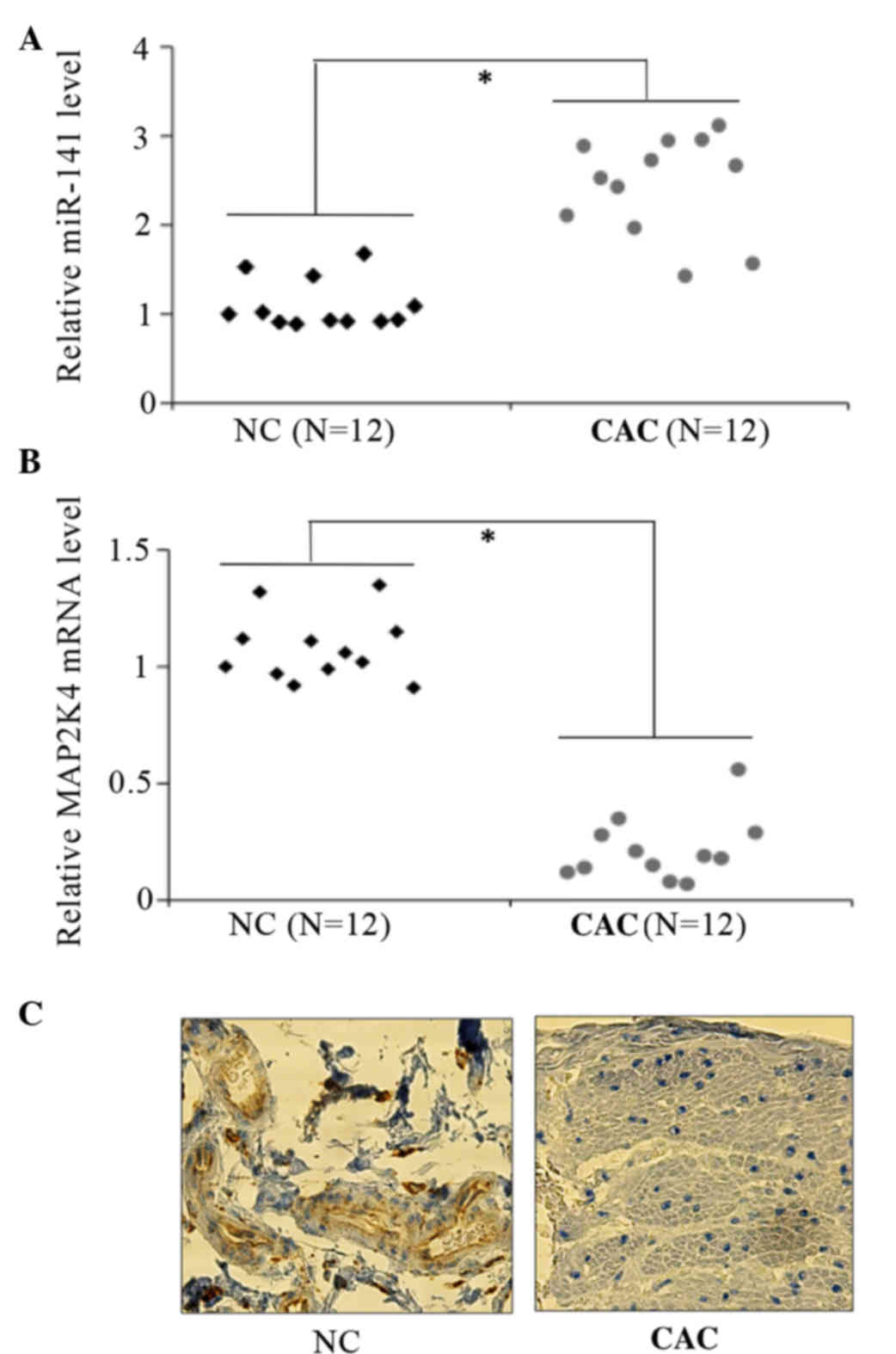
immunohistochemical staining. It was observed that miR-14 was
significantly upregulated in CRC cancer tissues (Fig. 1A). By contrast, MAP2K4 mRNA levels
decreased by an average of >60% (Fig.
1B), compared with the NC adjacent colon mucosa. In addition,
positive cytoplasmic staining for MAP2K4 was observed in the normal
colon, but not in cancer tissues (Fig.
1C), with the use of immunohistochemistry. These results
suggest that MAP2K4 is a target of miR-141 in CRC.
miR-141 promotes cell proliferation in
vitro
To investigate the effect of miR-141 on CRC cancer
cell growth, human CAC cell lines, including HT29, T84 and LS174,
were transiently transfected with miR-141 mimics or miR-141
antagomirs (Ant-miR-141) and their respective non-specific
controls. The RT-qPCR analysis revealed that the levels of miR-141
increased by 3–4-fold with transfection of miR-141 mimics in all
three CAC cell lines (Fig. 2A). By
contrast, transfection of miR-141 antagomirs led to a significant
decrease in miR-141 levels (Fig. 2B),
consistent with a previous finding (28).
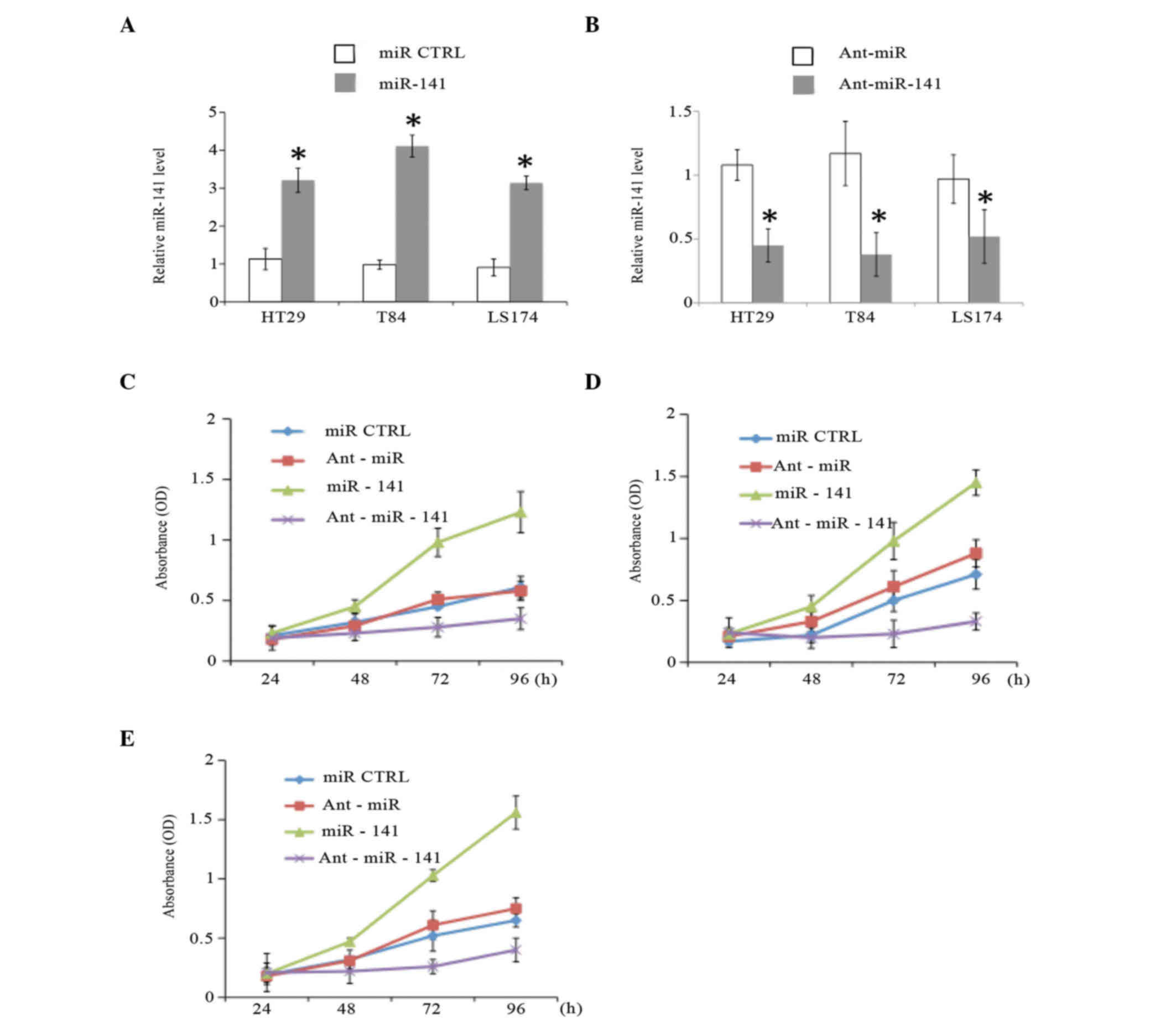 | Figure 2.miR-141 promotes the proliferation of
CAC cells in vitro. (A and B) The expression levels of miR-141 were
determined at 48 h post-transfection in CAC cell lines (HT29, T84
and or LS174) transfected with (A) miR-141 mimics or (B)
Ant-miR-141 and their respective controls (miR CTRL or Ant-miR,
respectively; 40 nM) by reverse transcription-quantitative
polymerase chain reaction. The proliferation of CAC cell lines (C)
HT29, (D) T84 and (E) LS174, which were transiently transfected
with miR-141 mimics, Ant-miR-141 or their respective controls, was
analyzed by MTT proliferation assay. *P<0.01 (n=3).miR,
microRNA; CTRL, control; Ant, antagomir; OD, optical density.; CAC,
colonic adenocarcinoma. |
The MTT proliferation assay was
performed in cancer cell lines
The results indicated that miR-141 mimics promoted,
while miR-141 antagomirs inhibited, cancer cell growth (P<0.01)
in all three CAC cell lines (Fig.
2C-E). The cell cycle distribution was also analyzed in HT29
cells, and it was noticed that a larger proportion of cells
transfected with miR-141 antagomirs accumulated in the G1 phase,
whereas the S-phase population significantly decreased (P<0.01;
data not shown). These results suggested that miR-141 acts as an
oncogene to promote cancer cell proliferation in the pathogenesis
of CRC.
MAP2K4 constitutes a direct target of
miR-141 in CRC
MAP2K4 is a putative tumor-suppressor gene and an
experimentally observed target gene for miR-141 (25). To investigate whether miR-141 targets
MAP2K4 in CRC, a bioinformatics analysis was performed with the use
of TargetScan and miRanda, and multiple miR-141 target sites in
MAP2K4 were identified. Next, it was validated whether miR-141
directly targets MAP2K4 mRNA in CAC cells. An increase in
intracellular concentration of miR-141 inhibited, while elevated
miR-141 antagomirs remarkably increased, the protein levels of
MAP2K4 in all three human CAC cell lines (Fig. 3A).
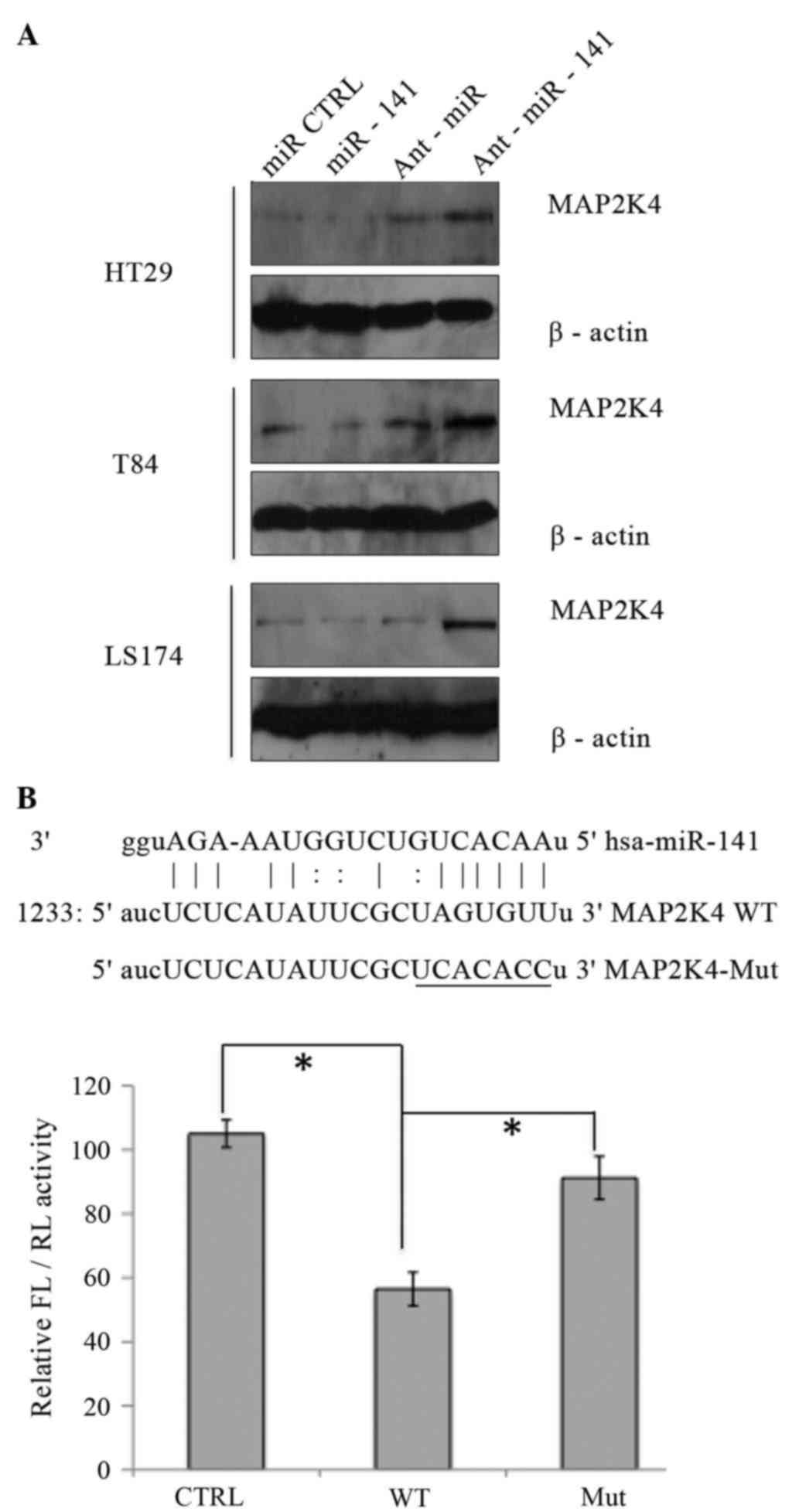 | Figure 3.MAP2K4 is one of the miR-141 targets.
(A) The MAP2K4 expression levels in colonic adenocarcinoma cell
lines upon transfection with miR-141 mimics, Ant-miR-141 and their
respective control (miR CTRL or Ant-miR, respectively; 40 nM) were
determined by western blotting. (B) Top panel: Predicted target
site of miR-141 in the 3′-UTR region of MAP2K4 messenger RNA.
Bottom panel: The luciferase activity in the presence of both WT or
Mut MAP2K4 3′-UTR and miR-141 mimics was compared with that of the
control. *P<0.01 (n=3). miR, microRNA; CTRL, control; Ant,
antagomir; MAP2K4, mitogen-activated protein kinase kinase 4; UTR,
untranslated region; FL, firefly luciferase; RL, Renilla
luciferase; WT, wild type; Mut, mutant; hsa, Homo sapiens. |
In order to further confirm the direct interaction
between miR-141 and MAP2K4, a miR-141 target site was introduced at
the 3′-UTR of MAP2K4 mRNA, downstream of the FL reporter gene, the
regulation of which could be mediated by miR-141. In addition, a
mutated version of this target site was used as a negative control.
HT29 cells were transfected with the control FL plasmid, the FL
plasmid with the miR-141 target site, or the FL plasmid with a
mutation of the miR-141 target site. In addition, the
Renilla luciferase reporter plasmid was co-transfected as an
internal reference. As shown in Fig.
3B, a significant decrease in FL activity was observed in cells
transfected with the FL reporter plasmid with WT miR-141 target
site. By contrast, a de-repression in FL activity was observed in
cells transfected with miR-141-mutant FL reporter plasmid.
Taken together, these results indicate that MAP2K4
is a direct target of miR-141 in CRC cells.
miR-141 supports CRC cell growth by
inhibiting MAP2K4
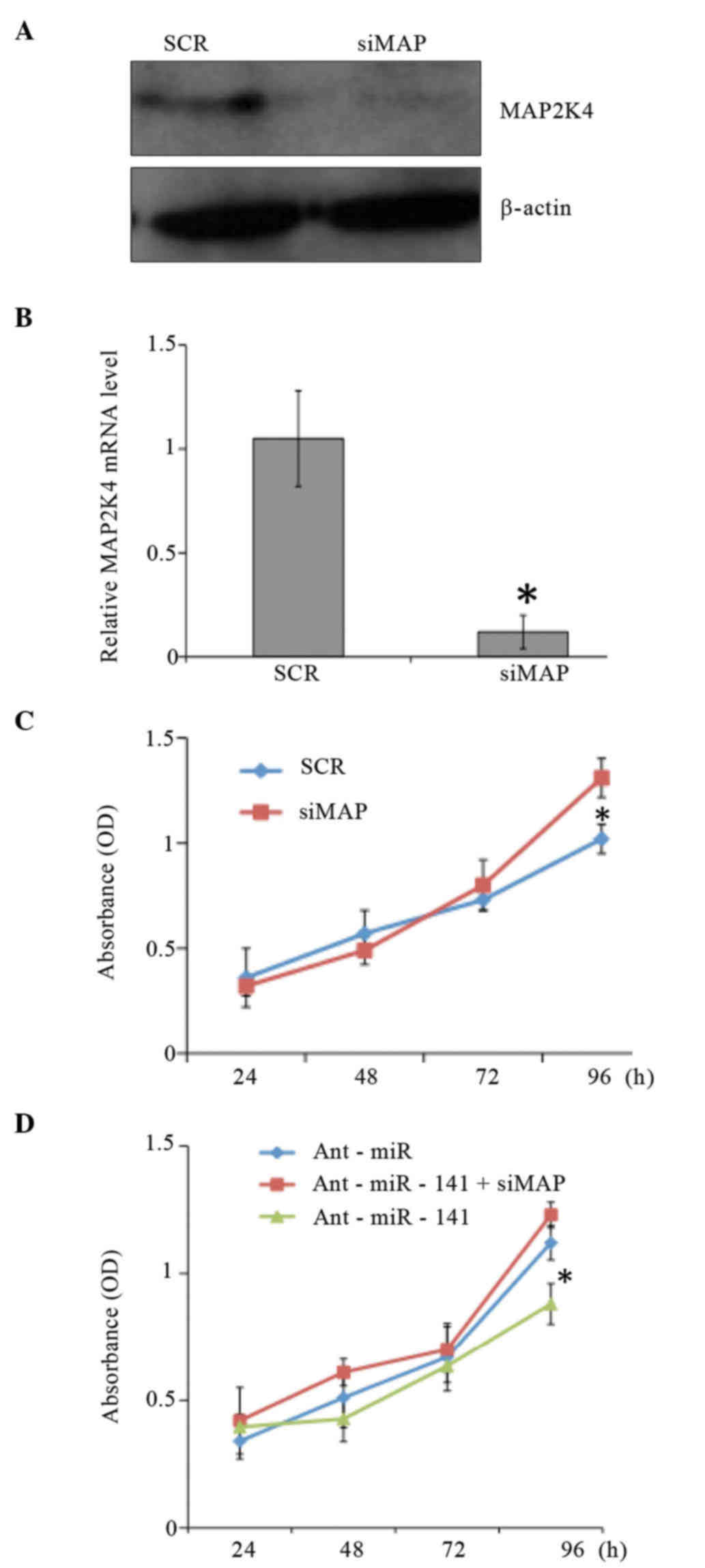
To elucidate whether the growth-promoting effect of
miR-141 is mediated by the repression of MAP2K4 in CRC cells, HT29
cells were transfected with small interfering RNA against MAP2K4.
Then, cell growth was examined by MTT assay. As shown in Fig. 4, the MTT assay results demonstrated
that gene silencing of MAP2K4 led to the proliferation of HT29
cells. Furthermore, the inhibitory effects of miR-141 antagomirs on
cell growth were reversed when MAP2K4 was downregulated.
Collectively, these results suggest that miR-141 is
involved in the proliferation of CRC cells by directly inhibiting
MAP2K4.
Discussion
A comprehensive detailing of the molecular
mechanisms underlying CRC initiation and progression will
facilitate the identification of novel biomarkers for early
diagnosis and therapy of CRC, thus improving the outcome of
patients with CRC. Over the past decade, miRNAs emerged as a new
class of gene regulators involved in a variety of cancers (17,18). The
present study has demonstrated for the first time that miR-141
plays a critical role in colonic tumorigenesis by inhibiting the
tumor-suppressor gene MAK2K4 to promote cancer proliferation. Our
results support that miR-141 can be used for early diagnosis and as
a potential drug target in the treatment of CRC.
miRNAs are an abundant class of endogenous small RNA
molecules of 20–25 nucleotides in length (10,29). As
important regulators in animals and plants, miRNAs regulate gene
expression by either repressing protein translation or
destabilizing their target mRNA levels (30–32).
miRNAs can function as oncogenes or as tumor-suppressor genes
(18). Numerous miRNAs are directly
involved in the pathogenesis of human cancers, including lung,
breast, brain, liver and colon cancer, as well as leukemia, by
regulating cell proliferation and apoptosis, processes that are
important in cancer formation (17).
Overexpressed miRNAs in cancers, such as mir-17–92, may function as
oncogenes and may promote cancer development by negatively
regulating tumor-suppressor genes and/or genes that control cell
differentiation or apoptosis (33).
Underexpressed miRNAs in cancers, such as let-7, function as
tumor-suppressor genes, and may inhibit cancers by regulating
oncogenes and/or genes that control cell differentiation or
apoptosis (34). miRNA expression
profiles may become useful biomarkers for cancer diagnostics. In
addition, miRNA therapy could be a powerful tool for cancer
prevention and therapeutics.
With regard to the regulation of miR-141, multiple
lines of evidence support that epigenetic mechanisms are involved
in the regulation of miR-141 expression in both normal and cancer
cells (35). There is an inverse
correlation between the expression and DNA methylation states of
miR-141 in normal human and mouse cell lines, as well as in human
breast and prostate cancer cell lines (35,36). In
addition, different histone codes exist between miR-141 expressing
and non-expressing cells that accurately represent the expression
and DNA methylation states (35).
Additionally, the epigenetic modifier 5-aza-2′-deoxycytidine
relieves the repression of miR-141 in cancer cell lines (35). Collectively, these findings indicate
that miR-141 is an evolutionarily conserved epigenetically labile
miRNA cluster (35). Since de
novo DNA (cytosine-5-)-methyltransferase 3 beta (DNMT3B) is
frequently repressed in human CRC cell lines and primary CRC tumors
by aberrant DNA hypermethylation of its distal promoter (37), we hypothesize that upregulation of
miR-141 in CRC may result from a reduction in DNMT3B, and that
DNMT3B is likely to be responsible for the hypermethylation of
miR-141, which usually occurs in healthy colon tissues. However,
this hypothesis deserves further investigation.
Our understanding of the biological functions of
miRNAs relies on the identification of their specific target genes.
The prediction of the majority of miRNA-target sequence matching is
based on imperfect complementarity of the miRNA with the 3′-UTRs of
its target mRNAs (38). The
requirement for complementarity of the 5′ seed region could result
in hundreds of possible targets (39). In the present study, MAP2K4 was
experimentally validated as a direct target of miR-141 in CRC
cells, adding information to previously reported cell types
(23).
Numerous studies have demonstrated that MAP2K4 is a
putative tumor-suppressor gene (24,25,40). By
systematically characterizing the biochemical properties of a large
panel of cancer-associated MAP2K4 mutations, Ahn et al
examined the consequences of MAP2K4 inactivation in genetically
engineered mouse models of human lung cancer driven by mutant KRAS
alone or in combination with mutant tumor protein p53 (TP53), and
profiled transcriptional changes induced by MAP2K4 depletion in
KRAS/TP53-mutant lung adenocarcinoma cells (24). The findings from that study revealed
that MAP2K4 functions as a tumor suppressor in lung adenocarcinoma
and inhibits tumor cell invasion by decreasing peroxisome
proliferator-activated receptor γ2 expression (24). In addition, MAP2K4 inactivation
increased the multiplicity and accelerated the growth of incipient
lung neoplasias, and promoted the invasion and metastasis of
KRAS/TP53-mutant lung adenocarcinoma cells, leading to the
conclusion that MAP2K4 exerts its tumor-suppressor activity at both
the early and late stages of lung tumorigenesis (24). Given the inverse correlation of
miR-141 and MAP2K4 in CRC identified in the present study, we
speculate that the signaling pathway of miR-141-MAP2K4 is likely to
be associated with the prognosis of CRC. Targeting this signaling
pathway may represent a novel approach for the treatment of
CRC.
In summary, the present study reports that miR-141
is frequently upregulated in CRC and is associated with cancer cell
proliferation of CRC. It further demonstrates that miR-141 promotes
cell growth of CRC cell by repressing the tumor-suppressor gene
MAP2K4. We propose that the strategy of targeting miR-141 may
provide an effective therapeutic approach for CRC patients who have
exhausted other modes of treatment.
References
|
1
|
Cheng H, Zhang L, Cogdell DE, Zheng H,
Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR and Zhang W:
Circulating plasma MiR-141 is a novel biomarker for metastatic
colon cancer and predicts poor prognosis. PLoS One. 6:e177452011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pox CP: Controversies in colorectal cancer
screening. Digestion. 89:274–281. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang YL, Zhang ZS, Wu BP and Zhou DY:
Early diagnosis for colorectal cancer in China. World J
Gastroenterol. 8:21–25. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
American Cancer Society, . What are the
survival rates for colorectal cancer by stage? http://www.cancer.org/Cancer/ColonandRectumCancer/DetailedGuide/colorectal-cancer-survival-ratesAccessed.
October 15–2015.
|
|
5
|
O'Connell JB, Maggard MA and Ko CY: Colon
cancer survival rates with the new American Joint Committee on
Cancer sixth edition staging. J Natl Cancer Inst. 96:1420–1425.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bijan MD and Azadeh S: An overview of
colorectal cancer survival rates and prognosis in Asia. World J
Gastrointest Oncol. 4:71–75. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Takuji T, Mayu T, Takahiro T and Rikako I:
Biomarkers for Colorectal Cancer. Int J Mol Sci. 11:3209–3225.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP and Chen CZ: Micromanagers of
gene expression: The potentially widespread influence of metazoan
microRNAs. Nat Rev Genet. 5:396–400. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sonja H and Damjan G: MicroRNAs as Novel
Biomarkers in Colorectal Cancer. Front Genet. 3:1802012.PubMed/NCBI
|
|
16
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: microRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Iorio MV and Croce CM: MicroRNA
involvement in human cancer. Carcinogenesis. 33:1126–1133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vega A Brunet, Pericay C, Moya I, Ferrer
A, Dotor E, Pisa A, Casalots À, Serra-Aracil X, Oliva JC, Ruiz A
and Saigí E: MicroRNA expression profile in stage III colorectal
cancer: Circulating miR-18a and miR-29a as promising biomarkers.
Oncol Rep. 30:320–326. 2013.PubMed/NCBI
|
|
22
|
Si ML, Zhu S, Wu H, Lu Z, Wu F and Mo YY:
miR-21-mediated tumor growth. Oncogene. 26:2799–2803. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang L, Deng T, Li X, Liu H, Zhou H, Ma
J, Wu M, Zhou M, Shen S, Li X, et al: microRNA-141 is involved in a
nasopharyngeal carcinoma-related genes network. Carcinogenesis.
31:559–566. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ahn YH, Yang Y, Gibbons DL, Creighton CJ,
Yang F, Wistuba II, Lin W, Thilaganathan N, Alvarez CA, Roybal J,
et al: Map2k4 functions as a tumor suppressor in lung
adenocarcinoma and inhibits tumor cell invasion by decreasing
peroxisome proliferator-activated receptor γ2 expression. Mol Cell
Biol. 31:4270–4285. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Davis SJ, Choong DY, Ramakrishna M, Ryland
GL, Campbell IG and Gorringe KL: Analysis of the mitogen-activated
protein kinase kinase 4 (MAP2K4) tumor suppressor gene in ovarian
cancer. BMC Cancer. 11:1732011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He D, Miao H, Xu Y, Xiong L, Wang Y, Xiang
H, Zhang H and Zhang Z: MiR-371-5p facilitates pancreatic cancer
cell proliferation and decreases patient survival. PLoS One.
9:e1129302014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Krutzfeldt J, Rajewsky N, Braich R, Rajeev
KG, Tuschl T, Manoharan M and Stoffel M: Silencing of microRNAs in
vivo with ‘antagomirs’. Nature. 438:685–689. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ambros V: microRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang B, Love TM, Call ME, Doench JG and
Novina CD: Recapitulation of short RNA-directed translational gene
silencing in vitro. Mol Cell. 22:553–560. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wakiyama M, Takimoto K, Ohara O and
Yokoyama S: Let-7 microRNA-mediated mRNA deadenylation and
translational repression in a mammalian cell-free system. Genes
Dev. 21:1857–1862. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Djuranovic S, Nahvi A and Green R:
miRNA-mediated gene silencing by translational repression followed
by mRNA deadenylation and decay. Science. 336:237–240. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Olive V, Bennett MJ, Walker JC, Ma C,
Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ and He L: miR-19
is a key oncogenic component of mir-17–92. Genes Dev. 23:2839–2849.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee YS and Dutta A: The tumor suppressor
microRNA let-7 represses the HMGA2 oncogene. Genes Dev.
21:1025–1030. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vrba L, Jensen TJ, Garbe JC, Heimark RL,
Cress AE, Dickinson S, Stampfer MR and Futscher BW: Role for DNA
methylation in the regulation of miR-200c and miR-141 expression in
normal and cancer cells. PLoS One. 5:e86972010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Batista L, Bourachot B, Mateescu B, Reyal
F and Mechta-Grigoriou F: Regulation of miR-200c/141 expression by
intergenic DNA-looping and transcriptional read-through. Nat
Commun. 7:89592016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huidobro C, Urdinguio RG, Rodriguez RM,
Mangas C, Calvanese V, Martínez-Camblor P, Ferrero C, Parra-Blanco
A, Rodrigo L, Obaya AJ, et al: A DNA methylation signature
associated with aberrant promoter DNA hypermethylation of DNMT3B in
human colorectal cancer. Eur J Cancer. 48:2270–2281. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Felekkis K, Touvana E, Stefanou Ch and
Deltas C: microRNAs: A newly described class of encoded molecules
that play a role in health and disease. Hippokratia. 14:236–240.
2010.PubMed/NCBI
|
|
39
|
Hibio N, Hino K, Shimizu E, Nagata Y and
Ui-Tei K: Stability of miRNA 5′terminal and seed regions is
correlated with experimentally observed miRNA-mediated silencing
efficacy. Sci Rep. 2:9962012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Teng DH, Perry WL III, Hogan JK, Baumgard
M, Bell R, Berry S, Davis T, Frank D, Frye C, Hattier T, et al:
Human mitogen-activated protein kinase kinase 4 as a candidate
tumor suppressor. Cancer Res. 57:4177–4182. 1997.PubMed/NCBI
|