Introduction
Gastric cancer is the second most common cancer in
the world, accounting for one million cancer-associated mortalities
annually and representing a major worldwide public health problem
(1). With advances in early
detection, standardized surgical treatment and improved
perioperative care, the outcomes of patients with gastric cancer
have notably improved (2). However,
diagnosis of gastric cancer at early stages remains a challenge in
clinical settings due to asymptomatic or nonspecific symptoms in
early lesions, as well as lack of effective biomarkers. Gastric
cancer development and progression involves multiple factors
(3). Helicobacter pylori
infection is a major risk factor in gastric cancer development
(4). Tobacco smoke, alcohol
consumption and dietary factors, including smoked foods, salt-rich
foods, red meat and pickled vegetables are also associated with
increased gastric cancer incidence (5). These factors potentially involve chronic
inflammation and gastric carcinogenesis (5). Over the past decades, efforts have been
made to elucidate the underlying mechanisms and to identify novel
diagnostic biomarkers and therapeutic targets for gastric cancer.
However, gastric cancer mortality remains high (6). Therefore, additional investigation and
attention is required to identify novel biomarkers for early
diagnosis, prognosis and treatment response prediction of gastric
cancer and/or to identify novel targets to control gastric
cancer.
Nusse and Varmus identified the first member of the
Wnt gene family in 1982 (7), and it
is now well accepted that these highly conserved Wnt secreted
proteins are critical mediators of cell-to-cell signaling, cell
fate and pattern formation during embryonic development. The Wnt
family of proteins includes at least 19 secreted-type glycoproteins
with 22–24 conserved cysteine residues (8). This canonical Wnt pathway regulates
target gene expression in the nucleus to control cell proliferation
(9), and the non-canonical pathways
regulate numerous other aspects of cell biology, including cell
motility and morphology (10).
Altered expression of Wnt gene family proteins is associated with
human carcinogenesis (7). In gastric
cancer, Wnt signaling may promote self-renewal of gastric cancer
stem cells (CSCs); therefore, targeting of Wnt signaling may lead
to a clinical gastric cancer therapy (11). Wnt10B, localized on human chromosome
12q13 (12), was implicated in cancer
development and progression by regulating nuclear factor-κB and
Notch pathways in osteosarcoma cells (13). However, Wnt10B is a bi-functional
protein; one function involves β-catenin/transcription factor
activation to promote tumor progression and the second function is
associated with downregulating cell growth through a
β-catenin-independent mechanism (14). In the present study, the association
between Wnt10B mRNA and protein levels in human gastric cancer
tissues and clinicopathological data from patients was first
assessed. Wnt10B expression in regulation of gastric cancer cell
proliferation and migration capacity in vitro was then
investigated.
Materials and methods
Gastric cancer tissue samples
The primary gastric cancer tissues and their matched
adjacent noncancerous tissues (collected >5–10 cm away from the
primary lesion) were obtained from 25 patients with gastric cancer
who underwent surgical resection in Nanjing Medical
University-Affiliated Wuxi Second Hospital (Jiangsu, China) between
September 2014 and January 2015. There were 17 males and 8 females
with a median age of 61 years (range, 29–75 years). All patients
were histologically diagnosed with gastric adenocarcinoma and had
not received any treatment prior to surgery. The present study was
approved by the Ethics Committee of Nanjing Medical
University-Affiliated Wuxi Second Hospital and informed consent was
obtained and documented from all patients prior to involvement in
the present study. Cancer tissues and adjacent noncancerous tissues
were obtained and immediately stored at −80°C. For the present
study, tissues were confirmed by pathological examination and used
for reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) and western blotting.
Cell line and culture
The human gastric cancer SGC-7901 cell line was
obtained from the Institute of Biochemistry and Cell Biology
(Chinese Academy of Sciences, Shanghai, China), and maintained in
Dulbecco's modified Eagle's medium (DMEM) with low-glucose (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.)
at 37°C in a humidified incubator with 95% air and 5%
CO2.
Lentivirus and Wnt10B knockdown in
SGC-7901 cells
A lentiviral expression vector carrying Wnt10B short
hairpin RNA (shRNA) and Lenti-green fluorescent protein (GFP)-shRNA
as a negative control vector were obtained from Sigma-Aldrich (EMD
Millipore, Billerica, MA, USA). The Wnt10B shRNA lentiviral vector
was generated by ligation of Wnt10B siRNA sequences from the
Tet-pLKO-puro vector (Sigma-Aldrich; EMD Millipore). Wnt10B shRNA
oligonucleotide sequences were
5′-CCGGCGGGCTCTAAGCAATGAGATTCTCGAGAATCTCATTGCTTAGAGCCCGTTTTTG-3′
and
5′-AATTCAAAAACGGGCTCTAAGCAATGAGATTCTCGAGAATCTCATTGCTTAGAGCCCG-3′,
while the sequences of the negative control shRNA were
5′-CCGGGCAAGCTGACCCTGAAGTTCATCTCGAGATGAACTTCAGGGTCACGTTGCTTTTTG-3′
and
5′-AATTCAAAAAGCAAGCTGACCCTGAAGTTCATCTCGAGATGAACTTCAGGGTCACGTTGC-3′.
The recombinant lentivirus was produced by co-transfecting
pLKO-GFP-shRNA or pLKO-Wnt10B-shRNA into HEK293T cells (Cell Bank
of Type Culture Collection of the Chinese Academy of Sciences,
(Beijing, China) that were previously stably transfected with
packaging PU1562 and PU1563 plasmids using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. The
virus-containing supernatant was harvested at 48 and 72 h following
transfection. Once the lentiviral titer was established, gastric
cancer SGC-7901 cells were infected with multiplicity of infection
of the lentivirus and selected with 1 µg/ml of puromycin
(Invitrogen; Thermo Fisher Scientific, Inc.) for 15 days to obtain
stable cell sublines. To induce shRNA expression, the cells were
treated with 80 µg/ml doxycycline (Sigma-Aldrich: EMD Millipore)
and the efficiency of Wnt10B knockdown was assessed by RT-qPCR and
western blot analysis.
RNA isolation and RT-qPCR
Total RNA was isolated from tissues and cells using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and 2
µl aliquots of RNA samples were reversely transcribed into cDNA
using a HiScript 1st strand cDNA synthesis kit (Vazyme, Piscataway,
NJ, USA) according to the manufacturer's protocol. The qPCR
amplifications were performed using the QuantiTect SYBR-Green PCR
kit (Toyobo Co., Ltd., Osaka, Japan) according to the
manufacturer's protocol. Thermal cycle parameters were as follows:
95°C for 5 min; 40 cycles at 95°C for 30 sec, 60°C for 30 sec, and
72°C for 30 sec; and 65–95°C drawing dissociation curve. β-actin
was used as an internal control. The expression of each gene was
defined from the threshold cycle (Cq) and the melting temperatures
were recorded. Using the 2−ΔΔCq method (15), relative changes in mRNA expression
were analyzed. The sequences of specific primers are presented in
Table I.
 | Table I.Primer sequences for quantitative
polymerase chain reaction amplification. |
Table I.
Primer sequences for quantitative
polymerase chain reaction amplification.
| Gene | Primer sequences | Amplicon size,
bp | Tm, °C |
|---|
| Wnt10B |
| 395 | 66 |
|
Forward |
5′-GCAATGGCAGCGCTCAACTC-3′ |
|
|
|
Reverse |
5′-GGCGCCAGGTGGTAACTGAA-3′ |
|
|
| β-actin |
| 265 | 56 |
|
Forward |
5′-CACGAAACTACCTTCAACTCC-3′ |
|
|
|
Reverse |
5′-CATACTCCTGCTTGCTGATC-3′ |
|
|
| Oct4 |
| 285 | 60 |
|
Forward |
5′-TTGAGGCTCTGCAGCTTAG-3′ |
|
|
|
Reverse |
5′-GCCGGTTACAGAACCACAC-3′ |
|
|
| Nanog |
| 292 | 60 |
|
Forward |
5′-CCTGATTCTTCCACCAGTCC-3′ |
|
|
|
Reverse |
5′-TGCTATTCTTCGGCCAGTTG-3′ |
|
|
Protein extraction and western blot
analysis
Tissues and cells were homogenized and lysed in
radioimmunoprecipitation assay buffer supplemented with proteinase
inhibitors (1 mM nphenylmethanesulfonyl fluoride; Vazyme,
Piscataway, NJ, USA). The protein concentration of each sample was
determined by the bicinchoninic acid protein assay kit (CWBIO,
Beijing, China). Equal amounts of protein were subjected to
SDS-PAGE on 12% SDS-PAGE gels and then transferred to
polyvinylidene difluoride membranes. For western blot analysis, the
membrane was blocked in 5% (w/v) skim milk for 1 h at 37°C and
incubated with a primary antibody for 12 h at 4°C, followed by a
horseradish peroxidase-conjugated anti-rabbit secondary antibody
(#AP188P; EMD Millipore) for 1 h at 37°C. Protein bands were
detected by the enhanced chemiluminescence detection system (GE
Healthcare Life Sciences, Chalfont, UK). The primary antibodies
used were: Anti-Wnt10B (dilution, 1:200; sc-25524; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA); GAPDH (dilution, 1:2,000;
#CW0100A; Kangcheng Pharmaceutical Co. Ltd., Guangzhou, China);
Ki67 (dilution, 1:1,000; #9449; Cell Signaling Technology, Inc.,
Danvers, MA, USA); β-catenin (dilution, 1:1,000; #8480; Cell
Signaling Technology, Inc.); cyclin D1 (dilution, 1:500; #P24385;
Bioworld Technology, Inc., St. Louis Park, MN, USA); N-cadherin
(dilution, 1:1,000; #13116; Cell Signaling Technology, Inc.);
E-cadherin (dilution, 1:1,000; #14472; Cell Signaling Technology,
Inc.); octamer-binding transcription factor 4 (Oct4; dilution,
1:200; #sc-365509; Santa Cruz Biotechnology, Inc.); and Nanog
antibody (dilution, 1:200; #25045; Signalway Antibody, Danvers, MA,
USA).
Immunofluorescence
Tissue samples or cells were fixed in 4%
paraformaldehyde for 20 min, permeabilized for 3 min with 0.1%
Triton X-100, and blocked with 5% bovine serum albumin for 20 min.
The sections were then incubated with indicated aforementioned
primary antibodies overnight at 4°C, followed by incubation with
Cy3-labeled anti-mice IgG secondary antibody at a dilution of 1:800
(#CW0145; Kangcheng Pharmaceutical Co. Ltd.), at 37°C for 45 min.
The nuclei were then counterstained with DAPI (Sigma-Aldrich, EMD
Millipore). The sections were reviewed under a fluorescent
microscope (Nikon, Tokyo, Japan) and images were sequentially
captured for evaluation.
Cell counting assay
Cells were seeded at a density of 1×104
cells per well onto 24-well plates and grown for up to 4 days. The
cells were cultured at 37°C in humidified air containing 5%
CO2. At the end of each experiment, the cells were
collected and counted in triplicate for each group at the indicated
time points (0, 1, 3 and 4 days). The data were summarized as a
percentage of the control.
Tumor cell wound healing assay
Cells were seeded at a density of 5×105
cells per well onto 6-well plates and incubated for ~24 h at 37°C
to reach >95% confluency of the monolayer. Wounds were then
created using a sterile plastic micropipette tip on the cell
monolayer and the plates were washed with PBS twice and 37°C
cultured for an additional 24 h. Images of the cells were captured
at 0 and 24 h to measure the wound healing for five randomly
selected fields. The width of wound healing at the original
scratches was measured and calculated using NIH Image version 1.62
program (http://rsb.info.nih.gov/nih-image/). The data are
expressed as the mean ± standard deviation and presented as a
percentage of the control.
Tumor cell Transwell migration
assay
SGC-7901 cells were seeded at a density of
1×105 per well into the upper chamber of a 6.5 mm
Transwell insert (#3422; Corning Incorporated, Corning, NY, USA).
DMEM supplemented with 10% FBS was added into the bottom chamber
and the cells were incubated at 37°C for 16 h. At the end of the
experiment, cells remaining on the membrane surface were removed
with cotton swabs and cells that had migrated through the membrane
(pore size, 8 mm) were fixed with 4% paraformaldehyde, stained for
15 min at room temperature with 1% crystal violet (Sigma-Aldrich;
EMD Millipore), and images were captured by light microscope
(Nikon, Tokyo, Japan).
Statistical analysis
Differences in the expression of Wnt10B mRNA in
paired tumor and normal tissues was analyzed by the Mann-Whitney U
test with GraphPad Prism 5.0 software (GraphPad Software, La Jolla,
CA, USA). Association between Wnt10B expression and
clinicopathological factors was estimated by Fisher's exact test.
Statistically significant correlation between two continuous
variables was analyzed by the Spearman's rho test. The statistical
association between two groups was analyzed by Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Upregulated Wnt10B mRNA is associated
with lymph node metastasis of gastric cancer
To assess the potential role of Wnt10B in gastric
cancer, RT-qPCR analysis of Wnt10B was performed in 25 pairs of
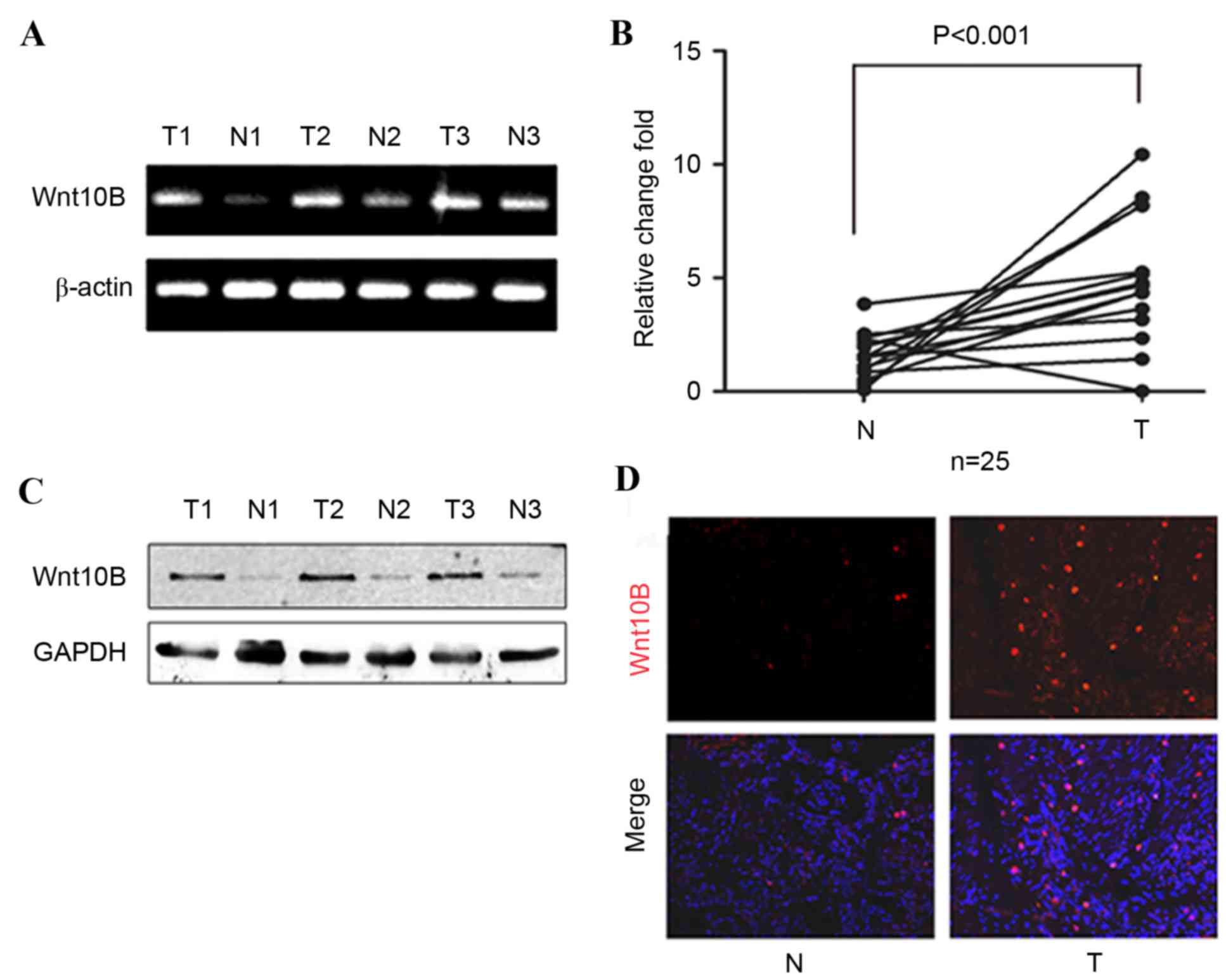
gastric cancer and adjacent normal tissues. Levels of Wnt10B mRNA
in gastric cancer tissues were significantly higher compared with
the paired adjacent normal tissues (P<0.001; Fig. 1A and B). Western blotting and
immunofluorescence were then performed to analyze the expression of
Wnt10B protein in these samples, and it was identified that levels
of Wnt10B protein were also increased in gastric cancer tissues
(Fig. 1C and D).
It was then determined whether associations exist
between Wnt10B mRNA level and clinicopathological characteristics
of patients. High levels of Wnt10B were revealed to be associated
with lymph node metastasis (P=0.022; Table II), but no associations were observed
with other clinicopathological characteristics, including age,
gender, tumor size, histological grade or clinical stages.
 | Table II.Association of Wnt10B expression with
clinicopathological factors from patients with gastric cancer. |
Table II.
Association of Wnt10B expression with
clinicopathological factors from patients with gastric cancer.
| Factor | Number (%) | High Wnt10B, n | Low Wnt10B, n | P-value |
|---|
| Age |
|
|
| 0.63 |
| ≤60
years | 10 (40) | 8 | 2 |
|
| >60
years | 15 (60) | 10 | 5 |
|
| Gender |
|
|
| 0.60 |
|
Male | 17 (68) | 12 | 5 |
|
|
Female | 8
(32) | 6 | 2 |
|
| Size |
|
|
| 0.63 |
| ≤5
cm | 19 (76) | 10 | 9 |
|
| >5
cm | 6
(24) | 3 | 3 |
|
| Stage |
|
|
| 0.60 |
|
I/II | 18 (72) | 11 | 7 |
|
|
III/IV | 7
(28) | 4 | 3 |
|
| Histological
grade |
|
|
| 0.45 |
|
W/M | 7
(28) | 3 | 4 |
|
|
Poorly/signet | 18 (72) | 10 | 8 |
|
| Tumor grade |
|
|
| 0.54 |
|
T1/T2 | 19 (76) | 11 | 8 |
|
|
T3/T4 | 6
(24) | 4 | 2 |
|
| Lymph node
metastasis |
|
|
| 0.022 |
|
Present | 20 (80) | 20 | 0 |
|
|
Absent | 5
(20) | 2 | 3 |
|
Wnt10B knockdown inhibits gastric
cancer cell proliferation and migration in vitro
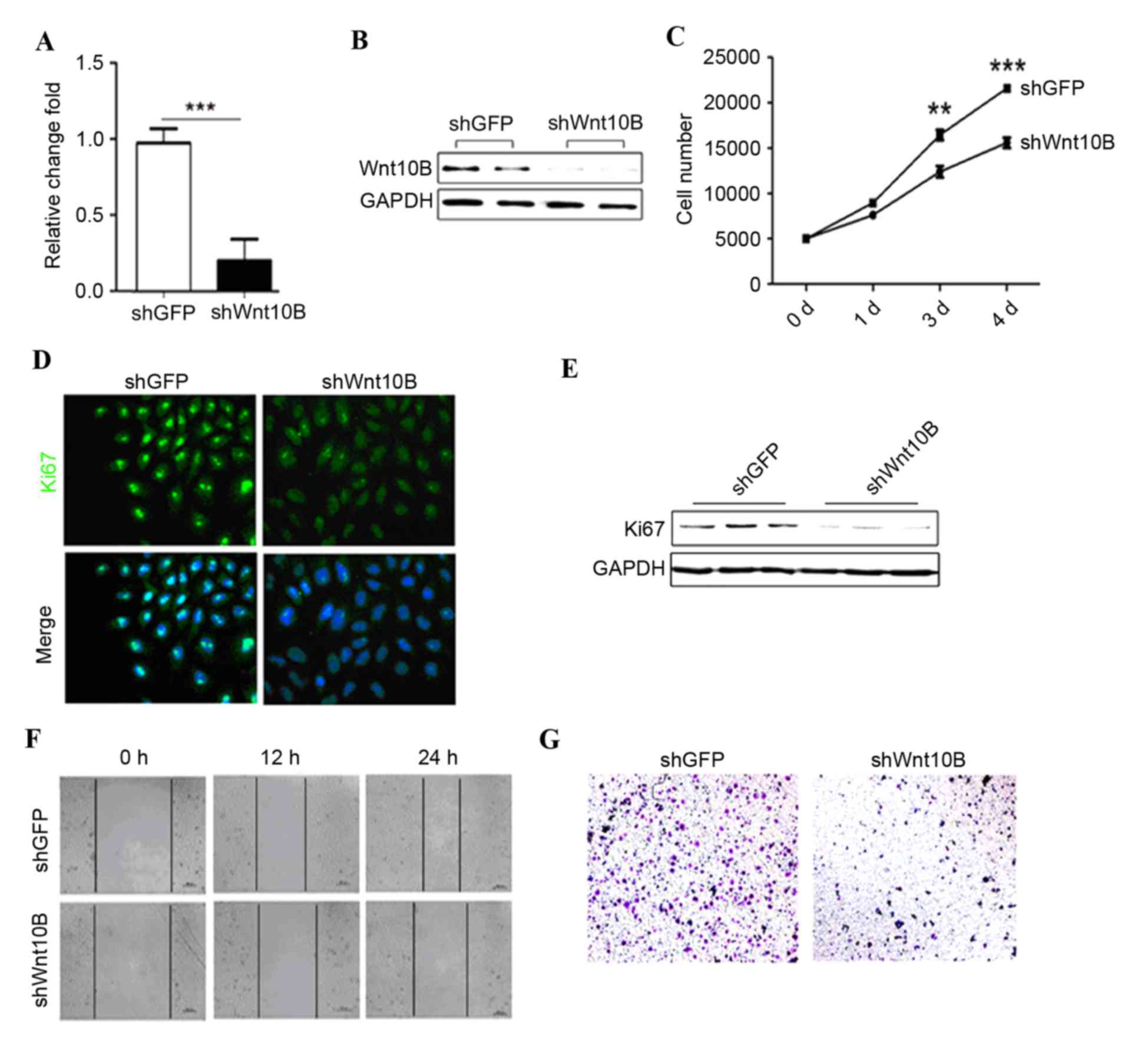
Wnt10B expression was knocked down in gastric cancer
SGC-7901 cells using lentivirus carrying Wnt10B shRNA. Wnt10B shRNA
significantly reduced levels of Wnt10B mRNA and protein in the
Wnt10B shRNA-infected (shWnt10B) SGC-7901 cells compared with the
negative control shRNA (P<0.001; Fig.
2A and B). In addition, knockdown of Wnt10B expression
inhibited gastric cancer proliferation compared with the control
shRNA-infected gastric cancer cells (Fig.
2C). Ki67 immunofluorescent-staining and western blot analysis
data confirmed that Wnt10B shRNA reduced tumor cell proliferation
(Fig. 2D and E).
The role of Wnt10B in regulation of gastric cancer
cell motility was assessed using wound healing and Transwell
migration assays. Wnt10B knockdown markedly reduced gastric cancer
cell wound healing and migration capacity compared with the
controls (Fig. 2F and G).
Knockdown of Wnt10B expression
inhibits the Wnt/β-catenin signaling pathway and cancer cell
stemness
A number of previous studies demonstrated that
activation of the Wnt/β-catenin signaling pathway was able to
induce epithelial-mesenchymal transition (EMT) of cells and acquire
cell stemness (12,16–19). Thus,
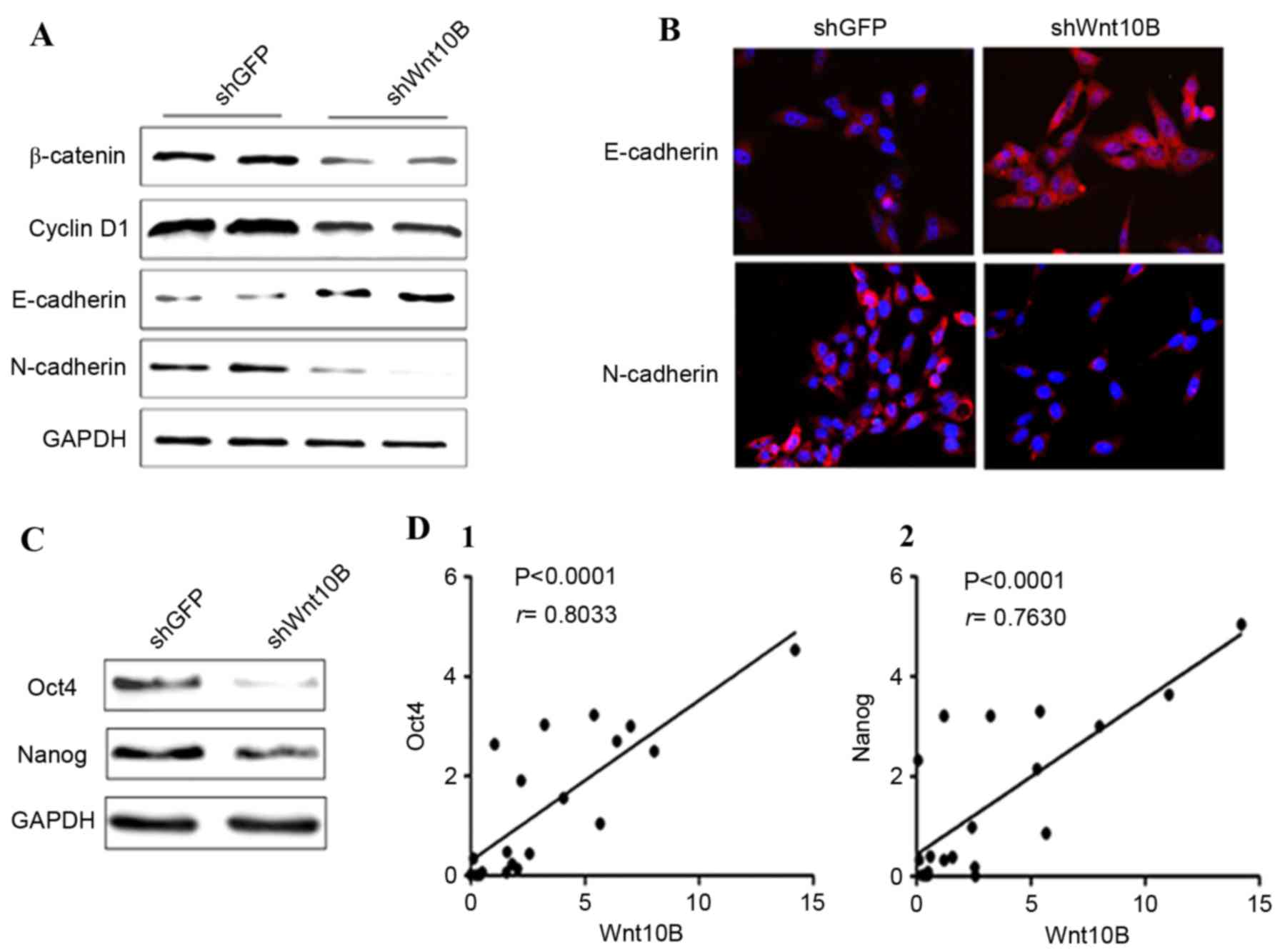
the present study confirmed the effects of Wnt10B knockdown on
gastric cancer cells, and it was identified that Wnt10B knockdown
reduced expression of β-catenin and the targeting gene cyclin D1 in
SGC-7901 cells (Fig. 3A). The
expression of the epithelial cell marker E-cadherin was
upregulated, whereas expression of the mesenchymal cell marker
N-cadherin was downregulated (Fig. 3A and
B), indicating a reversed EMT phenomenon in gastric cancer
cells. Subsequently, expression of the stem cell markers Oct4 and
Nanog was detected in the Wnt10B knocked down cells. Expression of
Oct4 and Nanog proteins was decreased in Wnt10B knocked down cells
compared with the control cells (Fig.
3C). Therefore, it was speculated that Wnt10B may increase the
number of stem cells in gastric cancer tissue. Thus, the Wnt10B
mRNA and stem cell markers Oct4 and Nanog mRNA were detected in
gastric cancer tissues by real-time RT-PCR, and the association
between Wnt10B, Oct4 and Nanog was analyzed. The results
demonstrated that there was a positive correlation between Wnt10B
and Oct4 (r=0.8033), and Wnt10B and Nanog (r=0.7630) in gastric
cancer tissues (Fig. 3D).
Discussion
One hallmark of tumor biology is activation of the
canonical Wnt pathway and upregulated stability and nuclear
localization of β-catenin to promote expression of targeted genes
during human tumorigenesis or development (20–22).
Upregulation of Wnt/β-catenin signaling can enhance EMT in
different types of cancer cells, including prostate, hepatocellular
and gastric cancer cells (16–19), as
well as proliferation and formation of CSCs (11,23). The
present study first analyzed the association of Wnt10B expression
in human gastric cancer tissues with clinicopathological data from
patients, and then investigated the role of Wnt10B in the
regulation of gastric cancer cell proliferation and migration, and
the underlying molecular events in vitro. The present data
demonstrated that Wnt10B mRNA and protein were upregulated in
gastric cancer tissues compared with the paired normal tissues. The
upregulated Wnt10B expression was associated with gastric cancer
metastasizing to lymph nodes. In addition, knockdown of Wnt10B
expression reduced gastric cancer cell proliferation and migration
and expression of Ki67 protein. Knockdown of Wnt10B expression also
inhibited tumor cell EMT by upregulating E-cadherin and
downregulating N-cadherin and tumor cell stemness, via
downregulation of Oct4 and Nanog expression. The data from the
present study indicated that knockdown of Wnt10B expression may
have a role in suppressing gastric cancer progression. Additional
studies are required to investigate whether targeting Wnt10B
expression or activity may be a novel strategy to control gastric
cancer.
Wnt10B, a member of the Wnt family of proteins
(24), is conserved among diverse
species and serves a crucial role in normal tissue development
(25,26). A previous study revealed that Wnt10B
activates canonical β-catenin signaling to promote development of
breast cancer (27). It was also
suggested that Wnt10B is expressed differently in benign and
malignant diseases, and has the potential to be a tumor biomarker
in molecular diagnosis of human cancer (28). In the present study, the usefulness of
detection of Wnt10B expression to differentiate normal tissues from
tumor tissues was confirmed. In addition, the present data also
revealed that Wnt10B expression is associated with gastric cancer
lymph node metastasis, indicating that Wnt10B is not only altered
early in gastric cancer development, but is also associated with
gastric cancer progression. Additional studies with a larger sample
size are required to confirm the present data.
Furthermore, the present data demonstrated that
knockdown of Wnt10B expression inhibits proliferation and migration
of gastric cancer cells. It is widely accepted that cancer
metastasis is one of the malignant properties of cancer and the
main cause of cancer-associated mortality (29). Wnt10B-associated tumor cell migration
and wound healing may promote gastric cancer progression.
Phenotypically, tumor cell EMT is an important process in cancer
development and metastasis (30).
When cancer cells acquire EMT phenotypes, they invade the
surrounding tissue and eventually metastasize to distant sites
(31). The present study showed that
knockdown of Wnt10B inhibits migration of gastric cancer cells via
suppression of tumor cell EMT. This is evident by enhanced
expression of the epithelial marker E-cadherin, but reduced
expression of mesenchymal marker N-cadherin in gastric cancer
cells.
In addition, CSCs have been identified in a variety
of human cancers, such as breast, brain, prostate, melanoma, colon,
liver, pancreatic and head and neck cancer, and are able to
initiate tumorigenesis and cause tumor metastasis (32). A previous study demonstrated that
activation of the Wnt/β-catenin pathway in cancer cells induces
their stem cell characteristics (33). The stem cell markers Oct4,
sex-determining region Y-box 2, Nanog and Krüppel-like factor 4 are
important target genes of the Wnt/β-catenin pathway (34). In the present study, it was identified
that Wnt10B level was positively associated with stem cell markers
in gastric cancer tissues, supporting that Wnt10B may be involved
in the generation and maintenance of CSCs in gastric cancer. These
data were also confirmed in Wnt10B knockdown cells, demonstrating
that reduced Wnt10B expression also downregulated expression of
Oct4 and Nanog in gastric cancer cells in vitro. Further
studies may confirm whether Wnt10B knockdown inhibits β-catenin
translocation into cell nuclei, as shown by previous studies in
other cancers (35–38).
The present study provides proof-of-principle,
demonstrating that alteration in Wnt10B expression is associated
with gastric cancer. Future studies may confirm the effects of
Wnt10B in gastric cancer to provide a novel diagnostic and
prognostic biomarker for gastric cancer and a therapeutic strategy
to control gastric cancer. In addition, the mechanisms responsible
for the aberrant expression of Wnt10B in gastric cancer will also
be investigated. Furthermore, the authors will perform animal
experiments to verify the effects of Wnt10B knockdown in
suppression of gastric cancer metastasis.
Acknowledgements
The present study was supported in part by a grant
from the National Natural Science Foundation of China (grant no.
81301503).
References
|
1
|
Tan YK and Fielding JW: Early diagnosis of
early gastric cancer. Eur J Gastroenterol Hepatol. 18:821–829.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ding YB, Xia TS, Wu JD, Chen GY, Wang S
and Xia JG: Surgical outcomes for gastric cancer of a single
institute in southeast China. Am J Surg. 203:217–221. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee YY and Derakhshan MH: Environmental
and lifestyle risk factors of gastric cancer. Arch Iran Med.
16:358–365. 2013.PubMed/NCBI
|
|
4
|
González CA, Sala N and Rokkas T: Gastric
cancer: Epidemiologic aspects. Helicobacter. 18 Suppl 1:S34–S38.
2013. View Article : Google Scholar
|
|
5
|
World Cancer Report 2014. World Health
Organization. Chapter 5.4. 2014
|
|
6
|
Wang M, Gu H, Qian H, Zhu W, Zhao C, Zhang
X, Tao Y, Zhang L and Xu W: miR-17-5p/20a are important markers for
gastric cancer and murine double minute 2 participates in their
functional regulation. Eur J Cancer. 49:2010–2021. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nusse R and Varmus HE: Many tumors induced
by the mouse mammary tumor virus contain a provirus integrated in
the same region of the host genome. Cell. 31:99–109. 1982.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen H, Wang Y and Xue F: Expression and
the clinical significance of Wnt10a and Wnt10b in endometrial
cancer are associated with the Wnt/β-catenin pathway. Oncol Rep.
29:507–514. 2013.PubMed/NCBI
|
|
9
|
Cadigan KM and Liu YI: Wnt signaling:
Complexity at the surface. J Cell Sci. 119:395–402. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kohn AD and Moon RT: Wnt and calcium
signaling: Beta-catenin independent pathways. Cell Calcium.
38:439–446. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mao J, Fan S, Ma W, Fan P, Wang B, Zhang
J, Wang H, Tang B, Zhang Q, Yu X, et al: Roles of Wnt/β-catenin
signaling in the gastric cancer stem cells proliferation and
salinomycin treatment. Cell Death Dis. 5:e10392014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Katoh M: WNT FGF gene clusters (Review).
Int J Oncol. 21:1269–1273. 2002.PubMed/NCBI
|
|
13
|
Mödder UI, Oursler MJ, Khosla S and Monroe
DG: Wnt10b activates the Wnt, notch, and NFκB pathways in U2OS
osteosarcoma cells. J Cell Biochem. 112:1392–1402. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yoshikawa H, Matsubara K, Zhou X, Okamura
S, Kubo T, Murase Y, Shikauchi Y, Esteller M, Herman JG, Wei Wang X
and Harris CC: WNT10B functional dualism:
Beta-catenin/Tcf-dependent growth promotion or independent
suppression with deregulated expression in cancer. Mol Biol Cell.
18:4292–4303. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu H, Yin J, Wang H, Jiang G, Deng M,
Zhang G, Bu X, Cai S, Du J and He Z: FOXO3a modulates WNT/β-catenin
signaling and suppresses epithelial-to-mesenchymal transition in
prostate cancer cells. Cell Signal. 27:510–518. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jiang L, Yang YD, Fu L, Xu W, Liu D, Liang
Q, Zhang X, Xu L, Guan XY, Wu B, et al: CLDN3 inhibits cancer
aggressiveness via Wnt-EMT signaling and is a potential prognostic
biomarker for hepatocellular carcinoma. Oncotarget. 5:7663–7676.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cong N, Du P, Zhang A, Shen F, Su J, Pu P,
Wang T, Zjang J, Kang C and Zhang Q: Downregulated microRNA-200a
promotes EMT and tumor growth through the wnt/β-catenin pathway by
targeting the E-cadherin repressors ZEB1/ZEB2 in gastric
adenocarcinoma. Oncol Rep. 29:1579–1587. 2013.PubMed/NCBI
|
|
19
|
Voon DC, Wang H, Koo JK, Nguyen TA, Hor
YT, Chu YS, Ito K, Fukamachi H, Chan SL, Thiery JP and Ito Y: Runx3
protects gastric epithelial cells against epithelial-mesenchymal
transition-induced cellular plasticity and tumorigenicity. Stem
Cell. 30:2088–2099. 2012. View Article : Google Scholar
|
|
20
|
Brown AM: Canonical Wnt signaling:
High-throughput RNAi widens the path. Genome Biol. 6:2312005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li C, Song G, Zhang S, Wang E and Cui Z:
Wnt3a increases the metastatic potential of non-small cell lung
cancer cells in vitro in part via its upregulation of Notch3. Oncol
Rep. 33:1207–1214. 2015.PubMed/NCBI
|
|
22
|
Qi L, Sun B, Liu Z, Cheng R, Li Y and Zhao
X: Wnt3a expression is associated with epithelial-mesenchymal
transition and promotes colon cancer progression. J Exp Clin Cancer
Res. 33:1072014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shojima K, Sato A, Hanaki H, Tsujimoto I,
Nakamura M, Hattori K, Sato Y, Dohi K, Hirata M, Yamamoto H and
Kikuchi A: Wnt5a promotes cancer cell invasion and proliferation by
receptor-mediated endocytosis-dependent and -independent
mechanisms, respectively. Sci Rep. 5:80422015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee FS, Lane TF, Kuo A, Shackleford GM and
Leder P: Insertional mutagenesis identities a member of the Wnt
gene family as a candidate oncogene in the mammary epithelium of
int-2/Fgf-3 transgenic mice. Proc Natl Acad Sci USA. 92:2268–2272.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nusse R and Varmus HE: Wnt genes. Cell.
69:1073–1087. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Moon RT, Brown JD and Torres M: WNTs
modulate cell fate and behavior during vertebrate development.
Trends Genet. 13:157–162. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wend P, Runke S, Wend K, Anchondo B,
Yesayan M, Jardon M, Hardie N, Loddenkemper C, Ulasov I, Lesniak
MS, et al: WNT10B/β-catenin signalling induces HMGA2 and
proliferation in metastatic triple-negative breast cancer. EMBO Mol
Med. 5:264–279. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bui TD, Rankin J, Smith K, Huguet EL,
Ruben S, Strachan T, Harris AL and Lindsay S: A novel human gene,
WNT10B, maps to 12q13 and is expressed in human breast carcinomas.
Oncogene. 14:1249–1253. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guan X: Cancer metastases: Challenges and
opportunities. Acta Pharm Sin B. 5:402–418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ye X and Weinberg RA:
Epithelial-mesenchymal plasticity: A central regulator of cancer
progression. Trends Cell Biol. 25:675–686. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lee JM, Dedhar S, Kalluri R and Thompson
EW: The epithelial-mesenchymal transition: New insights in
signaling, development, and disease. J Cell Biol. 172:973–981.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Takaishi S, Okumura T and Wang TC: Gastric
cancer stem cells. J Clin Oncol. 26:2876–2882. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Arend RC, Londoño-Joshi AI, Straughn JM Jr
and Buchsbaum DJ: The Wnt/β-catenin pathway in ovarian cancer: A
review. Gynecol Oncol. 131:772–779. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Holland JD, Klaus A, Garratt AN and
Birchmeier W: Wnt signaling in stem and cancer stem cells. Curr
Opin Cell Biol. 25:254–264. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bao Z, Wang Y, Yang L, Wang L, Zhu L, Ban
N, Fan S, Chen W, Sun J, Shen C and Cui G: Nucleostemin promotes
the proliferation of human glioma via Wnt/β-Catenin pathway.
Neuropathology. 36:237–249. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wangpu X, Yang X, Zhao J, Lu J, Guan S, Lu
J, Kovacevic Z, Liu W, Mi L, Jin R, et al: The metastasis
suppressor, NDRG1, inhibits ‘stemness’ of colorectal cancer via
down-regulation of nuclear β-catenin and CD44. Oncotarget.
6:33893–33911. 2015.PubMed/NCBI
|
|
37
|
Guo Q and Qin W: DKK3 blocked
translocation of β-catenin/EMT induced by hypoxia and improved
gemcitabine therapeutic effect in pancreatic cancer Bxpc-3 cell. J
Cell Mol Med. 19:2832–2841. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Burkhalter RJ, Westfall SD, Liu Y and
Stack MS: Lysophosphatidic acid initiates epithelial to mesenchymal
transition and induces β-catenin-mediated transcription in
epithelial ovarian carcinoma. J Biol Chem. 290:22143–22154. 2015.
View Article : Google Scholar : PubMed/NCBI
|