Introduction
Neuroblastoma is a common, malignant childhood tumor
of the sympathetic nervous system that originates from primordial
neural crest cells. Neuroblastoma is affected by age, localization
and the degree of differentiation, and it exhibits marked
variations in clinical presentation and biological characteristics
(1,2).
Certain cases of neuroblastoma spontaneously regress to benign
neuroblastoma, whereas others are difficult to cure and have poor
prognoses (3–6). Despite significant improvements in the
treatment of early-stage neuroblastoma and infantile neuroblastoma
over past decades, the mechanism of neuroblastoma tumorigenesis
remains unclear.
Transcriptional co-activator with PDZ-binding motif
(TAZ) is a transcriptional co-activator which has been reported to
bind to a variety of transcription factors, including thyroid
transcription factor-1, T-Box 5 and paired box 3 (7). TAZ functions as a major effector of the
Hippo pathway and is involved in tissue homeostasis regulation,
regeneration and tumorigenesis. TAZ has been reported to be a
positive regulator of cell proliferation and tumorigenesis of
breast cancer (8). A previous study
also proposed that TAZ is involved in cell proliferation and in the
tumorigenesis of neuroblastoma (9),
however, the potential molecular mechanisms have not been
sufficiently elucidated.
In the present study, TAZ was demonstrated to be
associated with neuroblastoma cell proliferation and tumorigenesis,
and insight was gained into the underlying mechanisms, which may
promote the development of potential neuroblastoma treatments.
Materials and methods
Cell culture
The human neuroblastoma cell line BE(2)-C was obtained from the American Type
Culture Collection (ATCC; Manassas, VA, USA) and was cultured in
DMEM/F12 (a 1:1 mixture of Dulbecco's modified Eagle's medium
(DMEM) and Ham's nutrient mixture F12) (Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(FBS; Thermo Fisher Scientific, Inc.). Additional neuroblastoma
cell lines, SK-N-AS (cat. no. CRL-2137; ATCC), SK-N-DZ (cat. no.
CRL-2149; ATCC) and SK-N-F1 (cat. no. CRL-2141; ATCC) were grown in
DMEM (Thermo Fisher Scientific, Inc.) supplemented with 10% FBS.
All cells were cultured at 37°C in a 5% CO2 humidified
incubator and the medium was replaced every two days.
Lentiviral production and
infection
The lentiviral constructs pCDH-CMV-EF1-TAZ and
pCDH-CMV-EF1-cop green fluorescent protein (GFP) (System
Biosciences, Inc., Palo Alto, CA, USA) were used for overexpression
assays. For the downregulation of TAZ, the lentiviral constructs
pLK0.1-puro-GFP-specified small interfering RNA (GFPsi) and
pLK0.1-puro-TAZ-specified small interfering RNA (TAZsi; TAZsi#1:
CCGGCGGACTTCATTCAAGAGGAA TCTCGAGATTCCTCTTGAATGAAGTCCGTTTTTG,
TAZsi#2: CCG GCTGTACGAGCTCATCGAGAAGCTCGA
GCTTCTCGATGAGCTCGTACAGTTTTTG) were mixed with 0.5 µg pLP1, pLP2 and
pLP/VSVG (Invitrogen; Thermo Fisher Scientific, Inc.) plasmids and
transfected into 293FT packaging cells (Invitrogen; Thermo Fisher
Scientific, Inc.) using Lipofectamine 2000 reagent (Thermo Fisher
Scientific, Inc.). Virus-containing supernatants were harvested and
titered, passed through a 0.45 µm filter and used to infect target
cells. Subsequent to the final round of infection, target cells
were grown in the presence of 2 µg/ml puromycin (Thermo Fisher
Scientific, Inc.), and drug-resistant cells were harvested. Target
cells were infected with two rounds in total, for 24 h at a
time.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Cells were harvested and lysed with TRIzol (Thermo
Fisher Scientific, Inc.) for total RNA purification. RNA was
reverse transcribed into cDNA using SuperScript II reverse
transcriptase (Thermo Fisher Scientific, Inc.). The buffer, dNTPs
and oligo (dT) were purchased from Thermos Fisher Scientific, Inc.
TAZ mRNA transcripts were measured using RT2
SYBR-Green/fluorescein PCR master mix (Takara Biotechnology Co.,
Ltd., Dalian, China). RT-qPCR reactions were performed in
triplicate using an iQ5 Real-Time PCR system (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). Individual values were normalized against
those of a glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
control. The GAPDH RT-qPCR primer sequences were as follows:
5′-GTCTCCTCTGACTTCAACAGCG-3′ (sense) and
5′-ACCACCCTGTTGCTGTAGCCAA-3′ (antisense). The TAZ RT-qPCR primer
sequences were as follows: 5′-GGGTTAGGGTGCTACAGTGTCC-3′ (sense) and
5′-GGGTCTGTTGGGGATTGATG-3′ (antisense). The conditions for PCR were
95°C for 30 sec followed by 42 cycles of 95°C for 10 sec and 60°C
for 30 sec. Relative mRNA expression levels were determined using
the 2−ΔΔCq method (10).
Western blot analysis
Cells were harvested and washed once with ice-cold
PBS. Proteins were extracted from cell pellets using SDS Lysis
Buffer (Beyotime Institute of Biotechnology, Haimen, China),
according to the manufacturer's instructions. Each protein sample
(60 µg) was separated using 10% SDS-PAGE. The proteins were then
transferred to a polyvinylidene difluoride (PVDF) membrane (Merck
KGaA, Darmstadt, Germany). Following blocking with 5% nonfat milk
in TBS with Tween-20 for 2 h, the PVDF membranes were incubated
with primary antibodies at 4°C overnight. The membranes were then
washed three times and incubated with horseradish peroxidase
(HRP)-conjugated secondary antibodies at room temperature for 2 h.
The signals were visualized by enhanced chemiluminescence (Beyotime
Institute of Biotechnology). The following primary antibodies were
used: Mouse anti-GAPDH (1:1,000; cat. no. AG019; Beyotime Institute
of Biotechnology); rabbit anti-TAZ (1:200; cat. no. sc-48805; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA); mouse
anti-cyclin-dependent kinase (CDK)2 (1:200; cat. no. sc-6248; Santa
Cruz Biotechnology, Inc.); rabbit anti-CDK4 (1:200; cat. no.
sc-601; Santa Cruz Biotechnology, Inc.); mouse anti-CDK6 (1:200;
cat. no. sc-56282; Santa Cruz Biotechnology, Inc.); rabbit
anti-cyclin D1 (1:200; cat. no. sc-753; Santa Cruz Biotechnology,
Inc.); rabbit anti-cyclin E1 (1:1,000; cat. no. ab33911; Abcam,
Cambridge, UK); rabbit anti-cyclin E2 (1:1,000; cat. no., 4132;
Cell Signaling Technology, Inc., Danvers, MA, USA); mouse
anti-cyclin A1 (1:1,000; cat. no. ab172317; Abcam); and mouse
anti-cyclin B1 (1:1,000; cat. no. 4135; Cell Signaling Technology,
Inc.). HRP-labeled goat anti-mouse immunoglobulin G (1:1,000; IgG;
cat. no. A0216; Beyotime Institute of Biotechnology) and goat
anti-rabbit IgG (1:1,000; cat. no. A0208; Beyotime Institute of
Biotechnology) were used as secondary antibodies.
Cell viablity analysis
BE(2)-C or SK-N-AS
cells were seeded onto 96-well culture plates with 800 cells/well,
and the cell viability was determined by MTT (Sigma-Aldrich; Merck
KGaA) analysis. Briefly, cells in each well were incubated with 20
µl MTT reagent in 100 µl medium (BE(2)-C in DMEM/F12, SK-N-AS in DMEM) at 37°C
for 2 h. The supernatant was then removed and 150 µl dimethyl
sulfoxide was added to dissolve the blue crystals. The absorbance
was measured at a wavelength of 560 nm using a microplate reader
(Model 550; Bio-Rad Laboratories, Inc.).
5-bromo-2-deoxyuridine (BrdU) staining
assay
Cells were grown on coverslips and incubated with 10
µg/ml BrdU (Sigma-Aldrich; Merck KGaA) at 37°C for 40 min. The
cells were then washed with PBS and fixed in 4% paraformaldehyde
(Thermo Fisher Scientific, Inc.) for 15 min at 25°C. Subsequently,
the cells were treated with 1 N HCl, blocked with 10% goat serum
(Beyotime Institute of Biotechnology) at 25°C for 1 h and incubated
with a rat primary antibody against BrdU (dilution, 1:200; cat.
no., ab6326; Abcam) at room temperature for 2 h, followed by an
Alexa Fluor 594 goat anti-rat IgG secondary antibody at room
temperature for 1 h (dilution, 1:400; Invitrogen; Thermo Fisher
Scientific, Inc.). DAPI (300 nM) was used for counterstaining. A
Nikon 80i fluorescence microscope (magnification, ×40) with
Image-Pro Plus software (version 6.0; Media Cybernetics, Inc.,
Rockville, MD, USA) was used to examine and analyze the fluorescent
signaling images. 6 fields for each sample were analyzed.
Cell cycle analysis
A total of 1×106 cells were collected by
centrifugation at 1,000 × g in 4°C for 5 min, washed twice with
ice-cold PBS, fixed with 70% ethanol and stained with 20 µg/ml
propidium iodide (Invitrogen; Thermo Fisher Scientific, Inc.). The
samples were analyzed by flow cytometry using a BD FACSVerse flow
cytometer (BD Biosciences, Franklin Lakes, NJ, USA), and the data
were analyzed with BD CellQuest Pro software version 5.1 (BD
Biosciences).
Colony formation assay
The oncogenic potential was evaluated in
vitro by determining the colony-forming ability in soft agar
cultures. For the lower layer, 1 ml of 0.6% agarose (Thermo Fisher
Scientific, Inc.) in growth medium was added to each well of a
6-well plate. For the upper layer, 2 ml of 0.3% agarose in growth
medium containing 1×103 cells was added to each well.
Following ~3 weeks of culture at 37°C, 6 fields of the colonies
were counted using a Nikon 80i light microscope.
In vivo tumorigenesis assay
For the in vivo tumor formation study,
5×106 cells per injection point were subcutaneously
injected into the lateral backsides of 6 (5 weeks old; 18–22 g)
non-obese diabetic (NOD)/severe combined immunodeficiency (SCID)
female mice (Beijing Vital River Laboratory Animal Technology Co.,
Ltd., China) and housed in an SPF room that was maintained at a
constant temperature (20–25°C) and humidity (45–55%) with a 12-h
light:dark cycle. The mice were fed a commercial diet (Beijing
Vital River Laboratory Animal Technology Co., Ltd.) and sterile
water ad libitum. GFPsi and TAZsi SK-N-AS cells were implanted in
the left and right side of each mouse, respectively. Tumor growth
was monitored by measuring the volume with calipers (volume= (π/6)
× length × width2). Following ~2 weeks, the mice were
sacrificed and the tumors were harvested and weighed.
All mice were raised under specific pathogen-free
conditions. Experimental procedures and animal welfare were
conducted according to the Guide for the Care and Use of Laboratory
Animals (Ministry of Science and Technology of China, 2006) and
were approved by the Animal Ethics Committee of Southwest
University (Chongqing, China).
Statistical analysis
All observations were confirmed by performing at
least three independent experiments. Quantitative data were
expressed as the mean ± standard deviation. Two-tailed Student's
t-tests were performed using GraphPad Prism (version 6.0; GraphPad
Software, Inc., La Jolla, CA, USA) for paired samples. P<0.05
was considered to indicate a statistically significant
difference.
Results
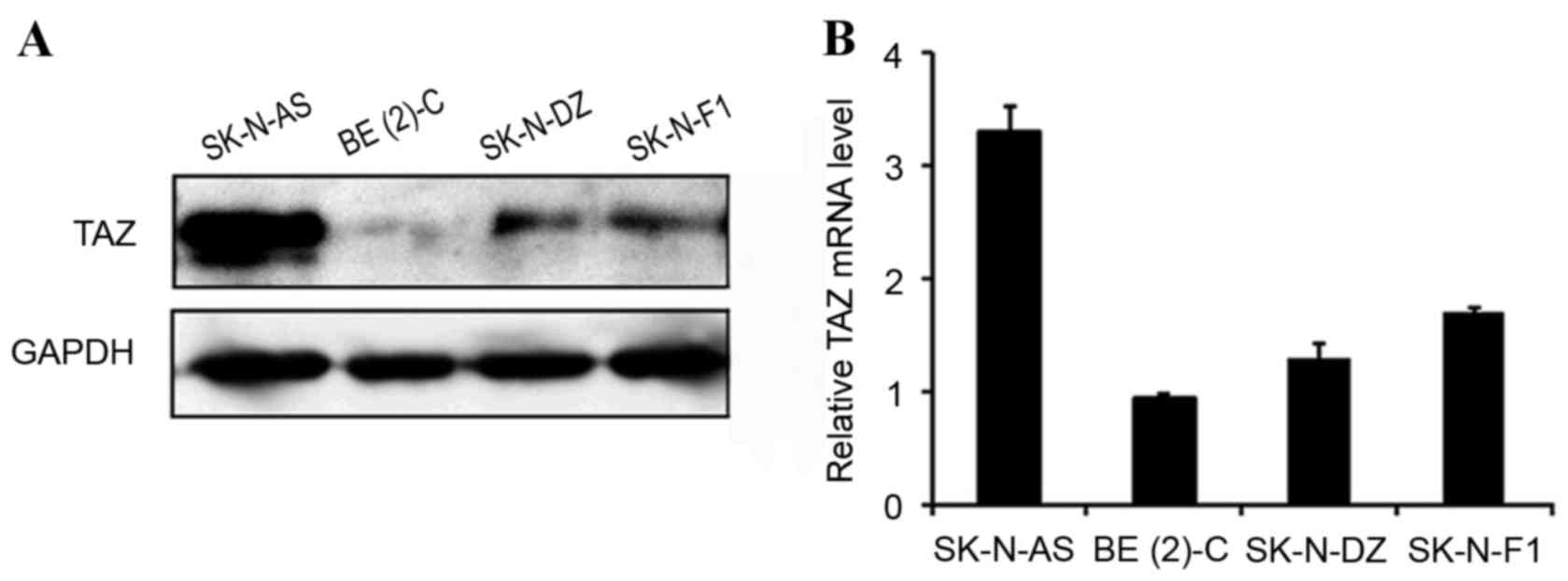
TAZ is commonly expressed in
neuroblastomas
To investigate the expression profile of TAZ in
neuroblastomas, four neuroblastoma cell lines, SK-N-AS, BE(2)-C, SK-N-DZ and SK-N-F1, were analyzed
using western blotting and RT-qPCR. TAZ was detected in all four
cell lines, among which the TAZ expression level was highest in
SK-N-AS, moderate in SK-N-DZ and SK-N-F1 and lowest in BE(2)-C (Fig 1).
Western blot analysis and RT-qPCR demonstrated consistent results,
indicating that TAZ is commonly expressed in neuroblastoma cell
lines.
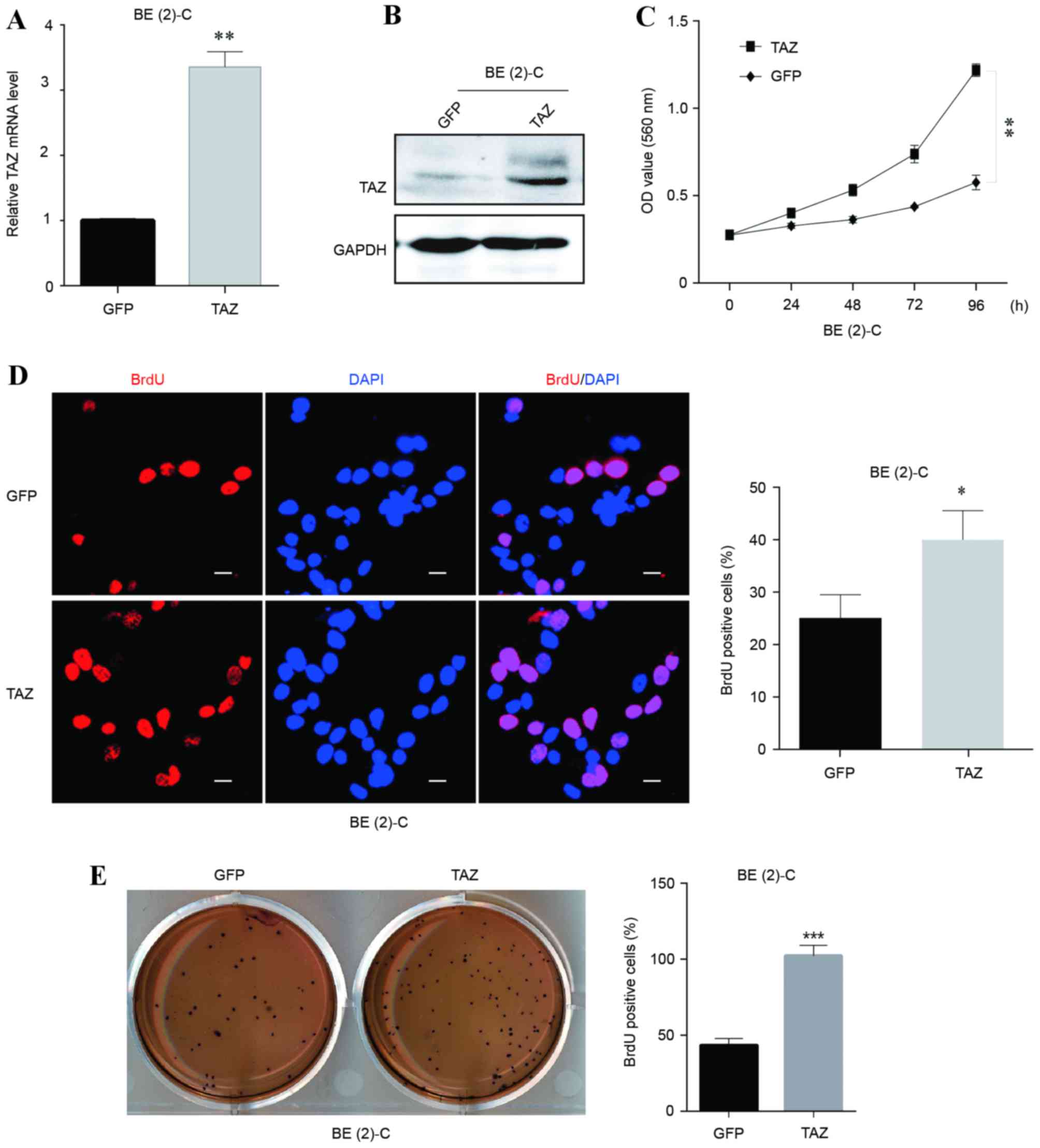
Overexpression of TAZ promotes cell
proliferation and self-renewal in neuroblastoma BE(2)-C cells
To explore the potential function of TAZ in
regulating cell growth and proliferation in neuroblastoma, TAZ was
overexpressed in the BE(2)-C cell
line. The overexpression efficiency was verified at the mRNA and
protein levels (Fig. 2A and B,
respectively). Growth curves and BrdU staining assays were employed
to examine the cell viability and proliferation abilities of
BE(2)-C cells with and without TAZ
overexpression. The results revealed that the TAZ-overexpressing
BE(2)-C cells exhibited significant
increases in viability and BrdU-positive signals (Fig. 2C and D, respectively), indicating that
the overexpression of TAZ enhanced BE(2)-C cell viability and proliferation.
Furthermore, colony formation ability was detected
using soft agar assays to evaluate the oncogenicity of cells in
vitro. TAZ-overexpressing BE(2)-C
cells formed significantly more colonies compared with the control
(Fig. 2E), indicating an increased
cell self-renewal ability and in vitro oncogenicity
following overexpression of TAZ.
Downregulation of TAZ inhibits cell
proliferation and self-renewal in neuroblastoma SK-N-AS cells
Since the SK-N-AS cell line had the highest TAZ
expression, TAZ knockdown was performed in this cell line. Two
lentiviral plasmids expressing small interfering RNA sequences
against TAZ were used, and the two succeeded in knocking down TAZ
in SK-N-AS cells (Fig. 3A and B). A
marked decrease in cell number was observed under a 20x Nikon 80i
light microscope following knockdown of TAZ (Fig. 3C). In addition, the MTT and BrdU
staining assays revealed that cell viability and proliferation were
significantly (P<0.01; P<0.001) inhibited in the TAZsi group
compared with the GFPsi group (Fig. 3D
and E, respectively).
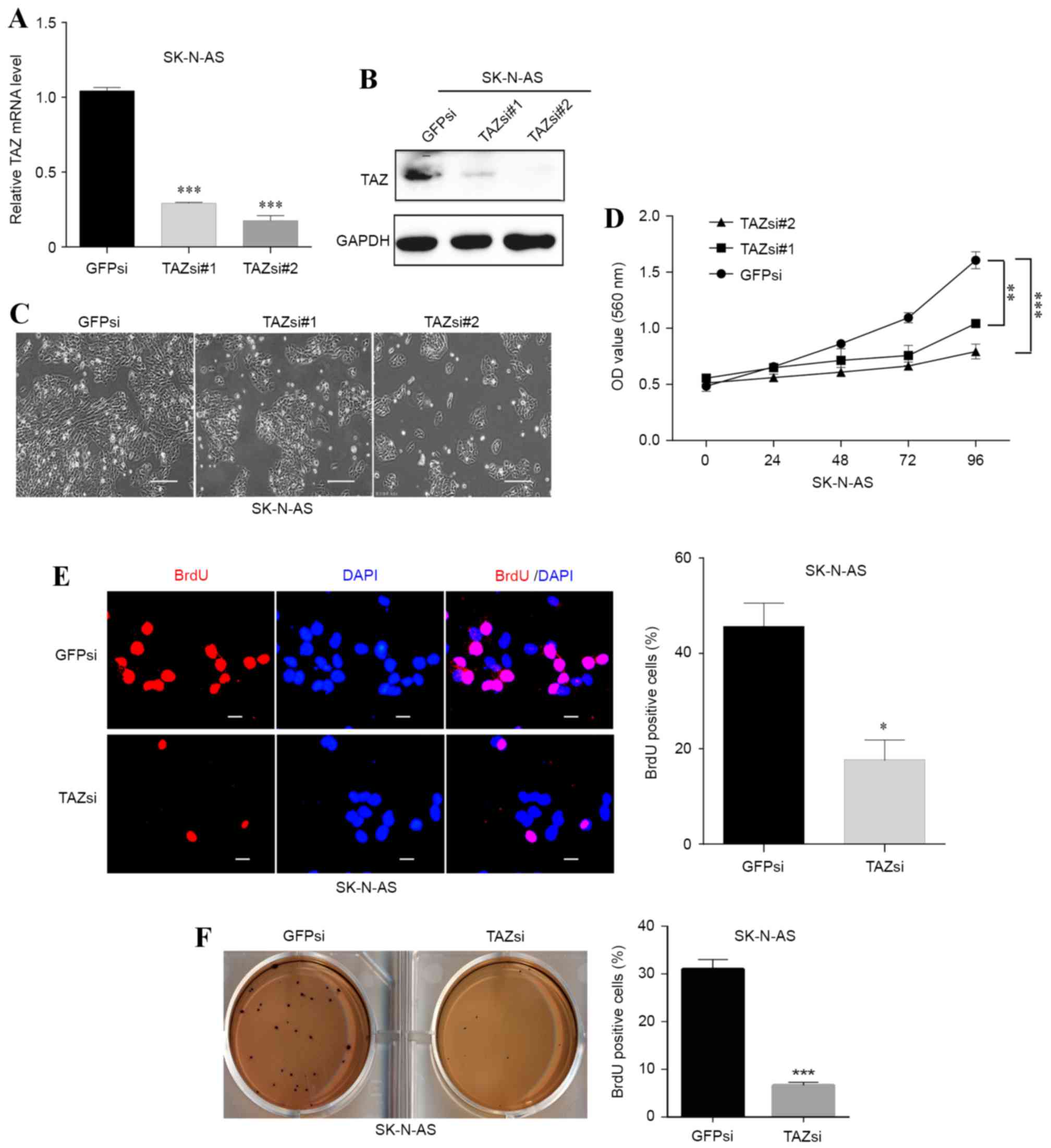 | Figure 3.Downregulation of TAZ inhibits cell
proliferation and self-renewal in SK-N-AS neuroblastoma cells. Two
small interfering RNA sequences were used to knockdown TAZ in
SK-N-AS cells. (A) The downregulation efficacy was investigated at
the mRNA level using a reverse transcription-quantitative
polymerase chain reaction and (B) at the protein level using
western blot analysis. (C) The morphology and cell number changes
following TAZ knockdown in SK-N-AS cells were examined by
microscopy (scale bars, 20 µm). (D) Following knockdown of TAZ,
SK-N-AS cell viability was monitored with the MTT assay. (E) BrdU
staining assays were conducted following TAZ downregulation in
SK-N-AS cells. Representative immunofluorescence images and the
statistical analysis of the BrdU-positive rate are depicted (scale
bars, 5 µm). (F) Soft agar assays were performed following
knockdown of TAZ in SK-N-AS cells. Representative photographs and
quantification of the colonies are depicted. For all knockdown
experiments, GFPsi was used as the control. The data were analyzed
using two-tailed Student's t-tests and are presented as the mean ±
standard deviation; n=3. *P<0.05, **P<0.01 and ***P<0.001
vs. GFPsi. TAZ, transcriptional co-activator with PDZ-binding
motif; BrdU, 5-bromo-2-deoxyuridine; GFPsi, green fluorescent
protein-specified small interfering RNA; TAZsi, transcription
co-activator with PDZ-binding motif-specified small interfering
RNA; OD, optical density; GADPH, glyceraldehyde 3-phosphate
dehydrogenase. |
The colony formation abilities of SK-N-AS cells with
or without TAZ downregulation were also examined. The results
demonstrated that the TAZsi group formed fewer colonies compared
with the control group (Fig. 3F),
indicating that the self-renewal and clonogenic abilities of the
TAZ-silenced SK-N-AS cells were weakened by knocking down TAZ.
These results indicated that TAZ was necessary for neuroblastoma
cell proliferation and self-renewal.
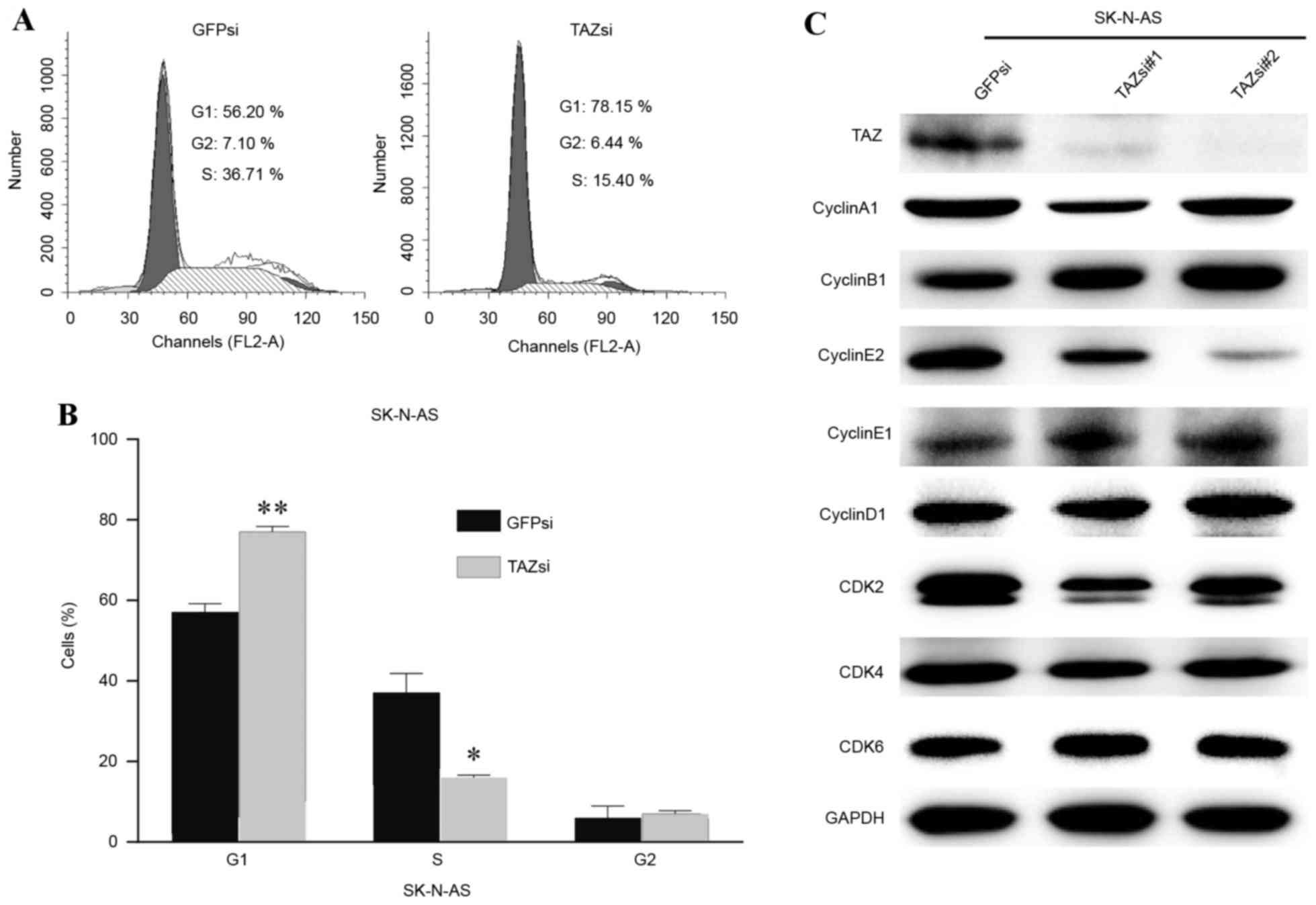
Downregulation of TAZ induces cell
cycle arrest in the G1 phase
To gain additional insights into the association
between TAZ and cell proliferation, the cell cycle status of
SK-N-AS cells was examined by flow cytometry. Following TAZ
downregulation, SK-N-AS cells exhibited a marked increase in G1
phase and a significant decrease in the S phase (Fig. 4A). Fig.
4B depicts the statistical data. Furthermore, the cell
cycle-associated protein levels of several cyclins and
cyclin-dependent kinases were examined to elucidate the molecular
mechanism underlying cell cycle arrest. Western blot analysis
results revealed that the inhibition of cyclin E2 expression was
associated with TAZ knockdown, and that there were no evident
changes in the expression of other cyclins or CDKs (Fig. 4C). Cyclin E2 is known as an essential
protein for the cell cycle G1/S transition, and its expression
peaks at the late G1 and early S phase. The present data indicated
that cell cycle arrest in the G1 phase, induced by TAZ
downregulation, may be mediated through the inhibition of cyclin E2
expression.
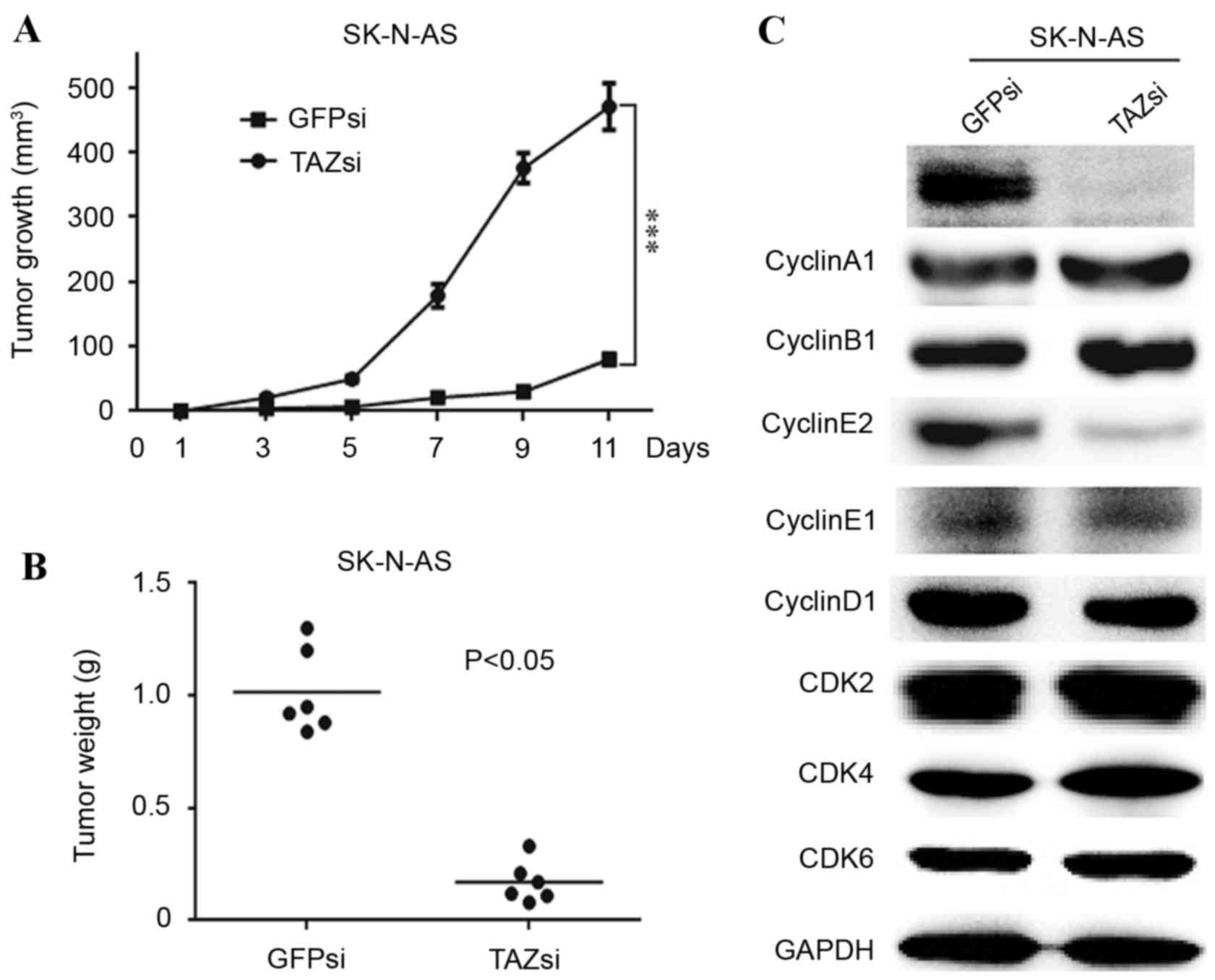
TAZ is required for tumorigenesis, and
TAZ knockdown affects cell cycle progression in vivo
As TAZ was essential for the clonogenic ability of
neuroblastoma cells in vitro (Figs. 2E and 3F), the in vivo tumorigenicity of TAZ
knockdown and control SK-N-AS cells was investigated. A
subcutaneous xenograft mouse model was used to examine the effect
of TAZ downregulation on tumorigenesis in NOD/SCID mice. The volume
and weight of the TAZsi tumors were significantly decreased
compared with the GFPsi group (Fig. 5A
and B, respectively). Furthermore, western blot analysis of the
xenograft tumors revealed that the expression patterns of the
cyclins and CDKs were similar to those of the TAZsi SK-N-AS cells.
Cyclin E2 was visibly decreased, indicating cell cycle arrest in
the TAZsi tumors (Fig. 5C).
Discussion
The transcriptional co-activator TAZ has been
reported to be associated with carcinogenesis in multiple human
malignant cancers, including breast cancer, ovarian cancer, lung
cancer and hepatocellular carcinoma (11–14). TAZ
functions as an oncogene and regulates various biological
processes, including cell proliferation, tumorigenesis, invasion,
metastasis and chemoresistance (15–22).
However, the molecular mechanism underlying TAZ-mediated regulation
of cell proliferation and tumorigenesis is unclear and requires
additional study. The results of the present study demonstrated
that the upregulation of TAZ in BE(2)-C, a neuroblastoma cell line with low TAZ
expression, promoted cell proliferation and self-renewal in
vitro. Furthermore, it was revealed that the downregulation of
TAZ in SK-N-AS (a neuroblastoma cell line with high TAZ expression)
suppressed cell proliferation and self-renewal in vitro and
tumorigenesis in vivo. These data indicated that TAZ is
essential for sustaining neuroblastoma cell proliferation and
tumorigenesis.
Cell cycle progression is associated with cell
proliferation and tumorigenesis, and cyclins and CDKs are key
proteins that regulate cell cycle progression. Data from the
present mechanistic study demonstrated that the downregulation of
TAZ in SK-N-AS cells induced cell cycle arrest in the G1 phase and
decreased the expression of cyclin E2, which is involved in the
cell cycle G1/S transition. Western blot analysis of in vivo
xenograft tumors produced the same results. The present data
provided evidence that TAZ is involved in the control of cell cycle
progression by regulating the expression of cyclin E2 in
neuroblastoma.
In conclusion, the present data revealed that TAZ is
essential for sustaining neuroblastoma cell proliferation and
tumorigenesis, and that TAZ knockdown in neuroblastoma induces cell
cycle arrest in the G1 phase, as well as cyclin E2 downregulation.
The present findings indicated that TAZ may serve as a potential
therapeutic target in neuroblastoma.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81201551 and
81502574) and the Fundamental Research Funds for the Central
Universities (grant nos. XDJK2013B020, XDJK2016C007, 2362015XK09
and XDJK2016E019).
Glossary
Abbreviations
Abbreviations:
|
BrdU
|
5-bromo-2-deoxyuridine
|
|
CDK
|
cyclin-dependent kinase
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
FBS
|
fetal bovine serum
|
|
TAZ
|
transcriptional co-activator with
PDZ-binding motif
|
References
|
1
|
Brodeur GM: Neuroblastoma: Biological
insights into a clinical enigma. Nat Rev Cancer. 3:203–216. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shimada H, Ambros IM, Dehner LP, Hata J,
Joshi VV, Roald B, Stram DO, Gerbing RB, Lukens JN, Matthay KK and
Castleberry RP: The international neuroblastoma pathology
classification (the Shimada system). Cancer. 86:364–372. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cohn SL, Pearson AD, London WB, Monclair
T, Ambros PF, Brodeur GM, Faldum A, Hero B, Iehara T, Machin D, et
al: The International neuroblastoma risk group (INRG)
classification system: An INRG Task Force report. J Clin Oncol.
27:289–297. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Modak S and Cheung NK: Neuroblastoma:
Therapeutic strategies for a clinical enigma. Cancer Treat Rev.
36:307–317. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maris JM: Recent advances in
neuroblastoma. N Engl J Med. 362:2202–2211. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cui H, Schroering A and Ding HF: p53
mediates DNA damaging drug-induced apoptosis through a
caspase-9-dependent pathway in SH-SY5Y neuroblastoma cells. Mol
Cancer Ther. 1:679–686. 2002.PubMed/NCBI
|
|
7
|
Cui CB, Cooper LF, Yang X, Karsenty G and
Aukhil I: Transcriptional coactivation of bone-specific
transcription factor Cbfa1 by TAZ. Mol Cell Biol. 23:1004–1013.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cordenonsi M, Zanconato F, Azzolin L,
Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR,
Poletti A, et al: The Hippo transducer TAZ confers cancer stem
cell-related traits on breast cancer cells. Cell. 147:759–772.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang M, Liu Y, Zou J, Yang R, Xuan F, Wang
Y, Gao N and Cui H: Transcriptional co-activator TAZ sustains
proliferation and tumorigenicity of neuroblastoma by targeting CTGF
and PDGF-β. Oncotarget. 6:9517–9530. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hiemer SE, Zhang L, Kartha VK, Packer TS,
Almershed M, Noonan V, Kukuruzinska M, Bais MV, Monti S and Varelas
X: A YAP/TAZ-regulated molecular signature is associated with oral
squamous cell carcinoma. Mol Cancer Res. 13:957–968. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hiemer SE, Szymaniak AD and Varelas X: The
transcriptional regulators TAZ and YAP direct transforming growth
factor β-induced tumorigenic phenotypes in breast cancer cells. J
Biol Chem. 289:13461–13474. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu W, Wei Y, Wu S, Wang Y, Wang Z, Sun Y,
Cheng SY and Wu J: Up-regulation of the Hippo pathway effector TAZ
renders lung adenocarcinoma cells harboring EGFR-T790M mutation
resistant to gefitinib. Cell Biosci. 5:72015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tan G, Cao X, Dai Q, Zhang B, Huang J,
Xiong S, Zhang Yy, Chen W, Yang J and Li H: A novel role for
microRNA-129-5p in inhibiting ovarian cancer cell proliferation and
survival via direct suppression of transcriptional co-activators
YAP and TAZ. Oncotarget. 6:8676–8686. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang L, Shi S, Guo Z, Zhang X, Han S, Yang
A, Wen W and Zhu Q: Overexpression of YAP and TAZ is an independent
predictor of prognosis in colorectal cancer and related to the
proliferation and metastasis of colon cancer cells. PLoS One.
8:e655392013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lei QY, Zhang H, Zhao B, Zha ZY, Bai F,
Pei XH, Zhao S, Xiong Y and Guan KL: TAZ promotes cell
proliferation and epithelial-mesenchymal transition and is
inhibited by the hippo pathway. Mol Cell Biol. 28:2426–2436. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lai D, Ho KC, Hao Y and Yang X: Taxol
resistance in breast cancer cells is mediated by the hippo pathway
component TAZ and its downstream transcriptional targets Cyr61 and
CTGF. Cancer Res. 71:2728–2738. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chan SW, Lim CJ, Loo LS, Chong YF, Huang C
and Hong W: TEADs mediate nuclear retention of TAZ to promote
oncogenic transformation. J Biol Chem. 284:14347–14358. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang H, Liu CY, Zha ZY, Zhao B, Yao J,
Zhao S, Xiong Y, Lei QY and Guan KL: TEAD transcription factors
mediate the function of TAZ in cell growth and
epithelial-mesenchymal transitio. J Biol Chem. 284:13355–13362.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao D, Zhi X, Zhou Z and Chen C: TAZ
antagonizes the WWP1-mediated KLF5 degradation and promotes breast
cell proliferation and tumorigenesis. Carcinogenesis. 33:59–67.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yuen HF, McCrudden CM, Huang YH, Tham JM,
Zhang X, Zeng Q, Zhang SD and Hong W: TAZ expression as a
prognostic indicator in colorectal cancer. PLoS One. 8:e542112013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin CW, Chang YL, Chang YC, Lin JC, Chen
CC, Pan SH, Wu CT, Chen HY, Yang SC, Hong TM and Yang PC:
MicroRNA-135b promotes lung cancer metastasis by regulating
multiple targets in the Hippo pathway and LZTS1. Nat Commun.
4:18772013. View Article : Google Scholar : PubMed/NCBI
|