Introduction
Gastric cancer (GC) is one of the most common types
of malignant cancer worldwide due to its high rates of morbidity
and mortality (1). However, an
effective treatment method for patients with GC is lacking, and
improvements to the clinical outcome remains a challenge.
Therefore, additional investigations into the underlying molecular
mechanisms for the development and progression of GC, and the
development of effective treatments for patients with GC is
essential.
Paclitaxel is a commonly used first-line
chemotherapeutic drug for treatment of numerous types of cancer
including breast (2), lung (3), colon (4),
gastric (5) and cervical cancer
(6) by regulating tubulin
polymerization, inhibiting cell cycle progression and promoting
apoptosis. However, cancer cells appear to be insensitive to
paclitaxel following long-term treatment due to the acquisition of
chemotherapeutic resistance (7).
Thus, the inhibition of cancer chemoresistance to paclitaxel may
significantly improve paclitaxel chemotherapy for various
cancers.
microRNAs (miRNAs) are a group of small noncoding
RNAs that negatively regulate target genes via either translational
repression or mRNA degradation (8).
An increasing number of studies have suggested that miRNAs perform
a crucial role in regulating cancer cells by acting as oncogenes or
tumor suppressers (9–11). Therefore, miRNAs may serve as
potential biomarkers and targets for the diagnosis and treatment of
various types of cancer (12).
miRNA-34a is an important regulator of tumor suppression, and acts
by controlling cellular proliferation, cellular differentiation and
the survival of cancer cells (13). A
previous study demonstrated that the expression of miRNA-34a in GC
was markedly downregulated, and that miRNA-34a was able to
significantly inhibit epidermal growth factor receptor
(EGFR)-signaling-dependent matrix metalloproteinase-7 (MMP-7)
activation, which suggested that miRNA-34a may serve as a tumor
suppresser to inhibit the cellular proliferation, migration and
invasion of GC cells (14). However,
the role of miRNA-34a in GC remains poorly understood.
To investigate whether miRNA-34a may enhance the
anticancer effect of paclitaxel on GC, the present study treated
human GC cancer cells with miRNA-34a combined with paclitaxel. The
results suggested that the introduction of miRNA-34a mimics and the
miRNA-34a inhibitor into GC cells is able to enhance and decrease
the chemotherapeutic efficacy of paclitaxel, respectively.
Additional studies identified that E2F transcription factor 5
(E2F5) was a potential target of miRNA-34a. The results of the
present study may increase the understanding of miRNA-34a in GC and
facilitate the development of potential chemotherapeutic against
GC.
Materials and methods
Cell culture
Human gastric cancer MKN45 and BGC823 cell lines
were purchased from the Type Culture Collection of the Chinese
Academy of Sciences (Shanghai, China). Cells were cultured in
Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.) and antibiotics (100
U ml−1 penicillin and 100 µg ml−1
streptomycin), and maintained at 37°C in a humidified cell
incubator with 5% CO2 and 95% air. Paclitaxel
(Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) was dissolved prior
to use at concentration of 0.1 mM in DMEM, stored at 4°C, and
further diluted in DMEM to obtain each final concentration.
Cell transfection
100 nM of miRNA-34a mimic, miRNA-34a inhibitor or
negative control (NC) miRNA-scrambled (SCR; Genepharma, Inc.,
Guangzhou, Guangdong, China), E2F5-specific siRNA or negative
control (NC) siRNA (Shanghai Shengong Biological Engineering
Technology & Services, Ltd., Shanghai, China) or 2 µg of
pmirGLO-E2F5 or pmirGLO-MutE2F5 reporter constructs were
transfected into MKN45 or BGC823 cells at 70% confluence using
Lipofectamine™ 2000 (Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. The siRNA sequences were as follows:
NC siRNA, CATGTCATGTGTCACATCTC; E2F5-siRNA,
CAGGACCTATCCATGTGCTGCTTAT. The miRNA sequences were not provided by
the suppliers, in order to protect commercial secrets.
3′-untranslated region (UTR)
luciferase reporter assay
Potential target genes for miRNA-34a were identified
using TargetScan (http://www.targetscan.org) software, and E2F
transcription factor 5 protein (E2F5) was identified as a target
candidate of interest.
The genomic DNA of MKN45 cells was isolated using a
QIAamp UCP DNA Micro Kit (Qiagen, Inc., Valencia, CA, USA). The
3′-UTR sequence of the human E2F5 gene containing the miRNA-34a
binding site was generated by polymerase chain reaction (PCR) with
LA Taq DNA polymerase (Takara, Dalian, China) using a Eppendorf
Mastercycler nexus Thermal Cycler (Eppendorf, Hamburg, Germany)
with conditions as follows: Initial denaturation at 95°C for 5 min,
followed by 33 cycles of 60 sec at 95°C for denaturing, 30 sec at
60°C for annealing and 60 sec at 72°C for extension, then a final
extension at 72°C for 10 min. PCR was performed with the following
primers: E2F5 UTR forward, CGG CTA GCT TCT GGA TTT CAA CTT TTC TTC;
E2F5 UTR reverse, GCG TCG ACG GAA AGT GGA ATG TCA GAA GTC (Shanghai
Shengong Biological Engineering Technology & Services, Ltd.).
The 3′-UTR was subsequently cloned into the pmirGLO-report vector
(Promega Corporation, Madison, WI, USA), which was denoted as
pmirGLO-E2F5.
The mutated (Mut) E2F5 3′-UTR miRNA-34a binding
sites oligonucleotide was generated by deletion mutagenesis with
overlapping extension PCR. Initially, two parallel PCRs were
conducted with pmirGLO-E2F5 as the template: One reaction was
performed with the previously described E2F5 UTR forward primer and
a Mut E2F5 UTR reverse primer with the sequence, ACT TTA CAA GTG
AAC GTC TGT; the other PCR was performed with a Mut forward
miRNA-34a binding site primer with the sequence
AACAGACGTTCCACTTGTAA and the previously described E2F5 UTR reverse
primer (Shanghai Shengong Biological Engineering Technology &
Services, Ltd.). A total of 10 µl equimolar mixture of the two PCR
products was used as a template with the E2F5 UTR forward and
reverse primers to obtain a Mut 3′-UTR of E2F5 oligonucleotide. The
Mut 3′-UTR oligonucleotide was subsequently cloned into the
pmirGLO-report vector, designated as pmirGLO-MutE2F5. All
constructs were sequenced by Shanghai Shengong Biological
Engineering Technology & Services, Ltd.
For the luciferase reporter assay, 3×105
MKN45 or BGC823 GC cells were seeded in each well of 24-well plates
and co-transfected with the pmirGLO-E2F5 or pmirGLO-MutE2F5
reporter constructs, and miRNA-34a mimics, miRNA-34a inhibitors or
miRNA-SCR, using Lipofectamine 2000™ (Thermo Fisher Scientific,
Inc.). Luciferase activities were determined using a
Dual-Luciferase® Reporter assay system (Promega
Corporation). Data were normalized and presented as the
firefly/Renilla luciferase ratio.
Western blot analysis
Following transfection for 24 h, cells were lysed
using cell lysis buffer (2% SDS, 6 M urea, 200 mM ammonium
bicarbonate, 0.1% protease inhibitor cocktail). Subsequent to the
quantification of cell lysate protein concentration with a
Bicinchoninic Acid Protein Assay kit (Pierce; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol, 30 µg
of cell lysate in each lane was separated with 10% SDS-PAGE and
transferred onto nitrocellulose membranes (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). The membranes were blocked with skimmed
dried milk in PBS buffer for 1 h at room temperature, and
immunoblotted using primary antibodies against E2F5 (dilution,
1:1,000; cat no. sc-1082; Santa Cruz Biotechnology, Inc.) or
β-actin (dilution, 1:5,000; cat. no. A3854; Sigma-Aldrich; Merck
KGaA) at 4°C overnight. Membranes were then incubated with the
horseradish peroxidase-conjugated secondary antibody (dilution,
1:2,000; cat no. M21002; Abmart, Shanghai, China) at room
temperature for 1 h. Immunolabeling was detected using 100 ml
Luminata Forte Western HRP substrate (EMD Millipore, Billerica, MA,
USA), followed by exposure to film. The relative intensity of the
bands was quantified using ImageJ software version 1.41 (NIH,
Bethesda, MD, USA).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from MKN28 and BGC823 cells
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. Reverse transcription was
performed to produce complementary (c) DNA with 1 µg RNA incubated
with the specific primers and reaction buffer of the Superscript
system (Invitrogen; Thermo Fisher Scientific, Inc.) at 16°C for 25
min, 42°C for 30 min and 85°C for 5 min. PCR primers for miRNA-34a
and U6 RNA were purchased from GeneCopoeia, Inc. (Guangzhou,
China). The following PCR primers were used in qPCR: E2F5 forward,
5′-CCTGTTCCCCCACCTGATG-3′ and reverse,
5′-TTTCTGTGGAGTCACTGGAGTCA-3′; and β-actin forward,
5′-CTGGAACGGTGAAGGTGACA-3′ and reverse,
5′-AAGGGACTTCCTGTAACAATGCA-3′. Primers were synthetized by Shanghai
ShengGong Biology Engineering Technology Service, Ltd. (Shanghai,
China). miRNA-34a expression was determined using Hairpin-it TM
miRNAs qPCR kit (Shanghai GenePharma Co., Ltd., Shanghai, China).
U6 RNA was used as an endogenous control. The mRNA levels of E2F5
and β-actin were detected using the SYBR green PCR kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.). qPCR was performed on
an ABI-7500 PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Quantitative PCR thermocycling conditions were:
95°C for 10 min initially, followed by 35 cycles of 95°C for 15
sec, and 60°C for 45 sec. Data were analyzed using the
2−ΔΔCq method, as previously described (15).
MTT assay
MKN45 and BGC823 cells were plated in 96-well plates
at a density of 5×103 cells/well and cultured at 37°C
for 18 h. Following transfection with miR-SCR or miR-34a inhibitor
for 24 h, followed by treatment with 100 nM paclitaxel for another
16 h, cellular viability was determined using the MTT assay.
Following this treatment for 16 h, 5 µg/ml of MTT (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) was added into each well, and
incubated at 37°C for 2 h. The supernatant was then removed, and
100 µl DMSO was added into each well to dissolve the formazan
product. The optical density at wavelength of 570 nm was determined
using the ELx808 absorbance reader (BioTek Instruments, Inc.,
Winooski, VT, USA).
5-Bromo-2-deoxyUridine (BrdU)
assay
The BrdU incorporation assay kit (Roche Applied
Science, Penzberg, Germany) was used for analyzing the
incorporation of BrdU during DNA synthesis in proliferating cells,
according to the manufacturer's protocol. A total of
2×103 MKN28 cells were cultured for 24 or 48 h, followed
by incubation at 37°C for 1 h with 10 µM BrdU (BD Pharmingen, San
Diego, CA, USA). The absorbance values were measured at a
wavelength of 450 nm with the ELx808 plate reader.
Statistical analysis
Each experiment at least was performed 3 times. Data
are presented as the mean ± standard deviation. SPSS 18.0 software
(SPSS, Inc., Chicago, IL, USA) was used to conduct statistical
analyses. Multiple comparisons were analyzed using one-way analysis
of variance followed by Tukey-Kramer post hoc analysis to test for
differences between all groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
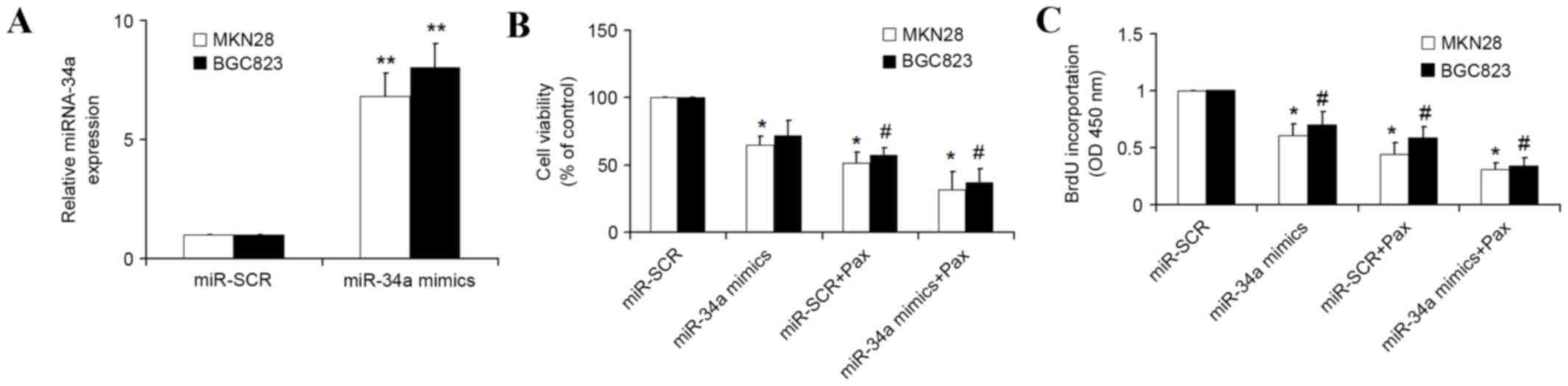
miRNA-34a mimics enhance the
chemotherapeutic efficacy of paclitaxel
As miRNA-34a is able to prevent metastasis in GC
(14), it was hypothesized that
miRNA-34a may enhance the efficacy of paclitaxel for the treatment
of GC. MTT and BrdU assays were used to detect the cellular
proliferation of GC cells, including MKN28 and BGC823 cells, which
were transfected with miRNA-34a mimics or scrambled miRNA for 24 h,
followed by treatment with paclitaxel for another 16 h. qPCR
demonstrated that transfection with miRNA-34a mimics resulted in a
significant increase (P<0.01) of miRNA-34a in MKN28 and BGC823
cells compared with the cells transfected with the scrambled miRNA
(Fig. 1A). The results of MTT assays
demonstrated that the growth rate of GC cells transfected with
miRNA-34a mimics was decreased compared with the cell transfected
with the scrambled miRNA, and the growth rate of GC co-treated with
miRNA-34a and paclitaxel was significantly decreased (P<0.05)
compared with the cell transfected with miRNA-34a mimics alone
(Fig. 1B). In addition, the BrdU
assays also demonstrated similar results, as co-treatment with
miRNA-34a and paclitaxel led to slower growth rate of GC compared
with miRNA-34a mimics (Fig. 1C).
These results demonstrated that transfection of miRNA-34a into GC
enhances the inhibition of proliferation of GC cells by
paclitaxel.
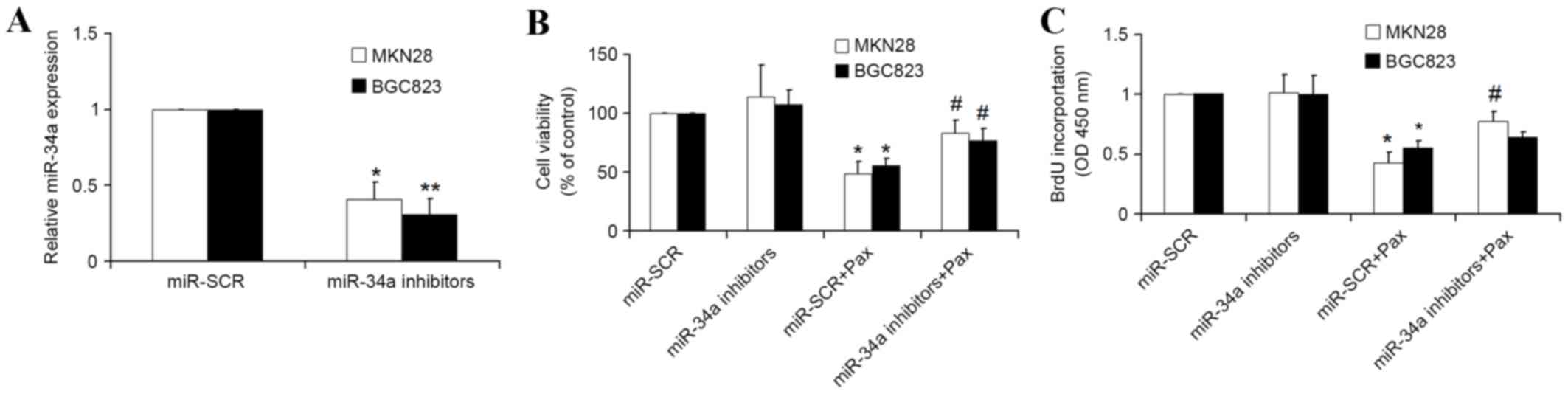
miRNA-34a inhibitors decrease the
chemotherapeutic efficacy of paclitaxel
The present study also determined whether endogenous
miRNA-34a inhibition by specific miRNA-34a inhibitors would
decrease the chemotherapeutic efficacy of paclitaxel. The specific
miRNA-34a inhibitors or scrambled miRNA-34a were transfected into
GC cells for 24 h, and followed by treatment with paclitaxel for
another 16 h. As expected, the introduction of miRNA-34a inhibitors
significantly reduced the expression of miRNA-34a in MKN28 and
BGC823 cells (Fig. 2A). The results
of MTT and BrdU assays demonstrated that treatment with paclitaxel
and miR-SCR resulted in a significant inhibition (P<0.05) of
growth rate of GC cells compared with miR-SCR alone, and
transfection miRNA-34a significantly attenuated the inhibitive
effect of paclitaxel on the growth rate of GC cells (Fig. 2B and C). Therefore, these results
indicate that miRNA-34a is involved in the chemotherapeutic
efficacy of paclitaxel.
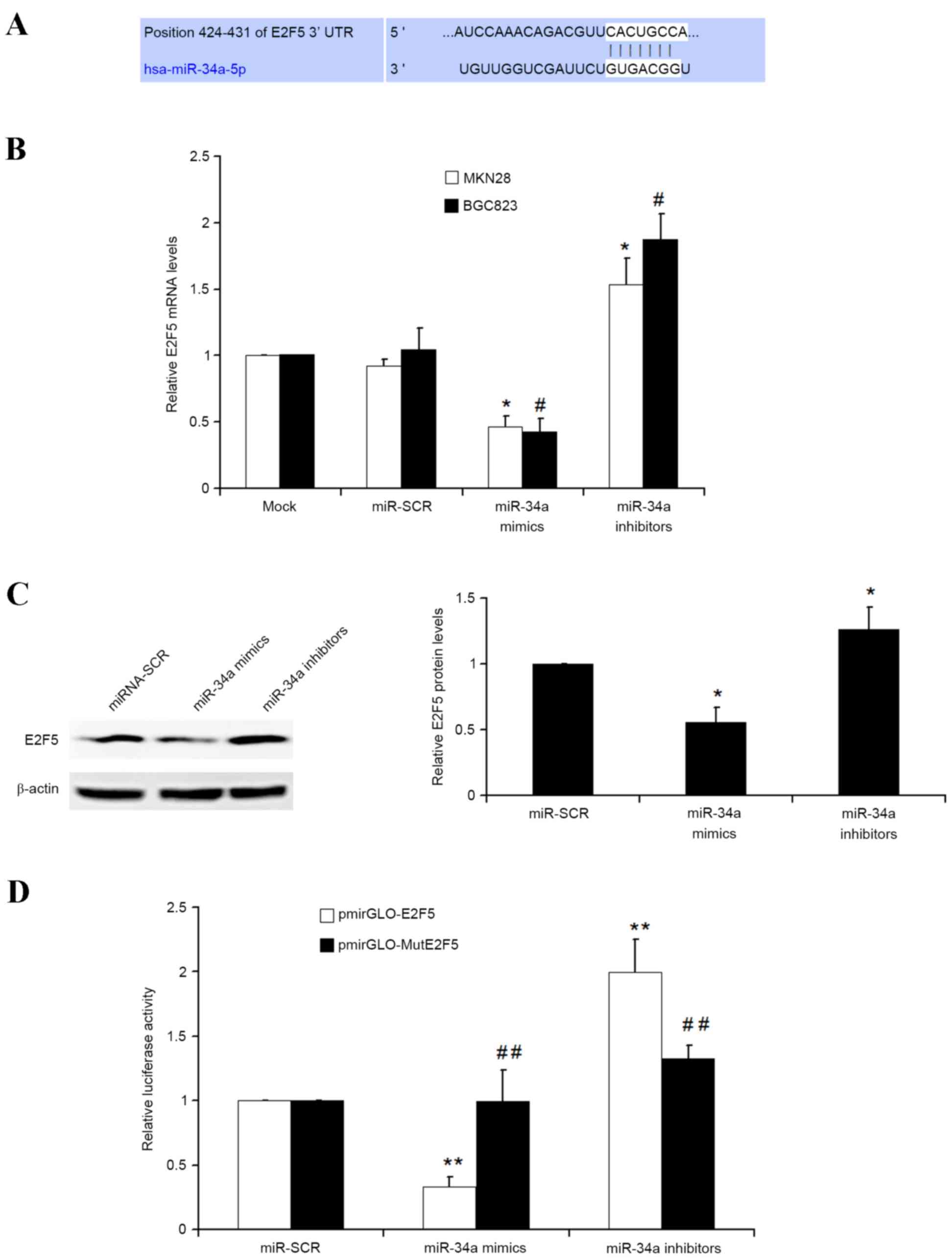
E2F5 is a direct target of
miRNA-34a
TargetScan was used to identify the potential
targets of miRNA-34a. As demonstrated in Fig. 3A, E2F transcription factor 5 protein
(E2F5) was identified as an interesting candidate, as E2F5 has
previously been observed as overexpressed in various types of
cancer, including breast (16),
epithelial ovarian (17), prostate
cancer (18) and hepatocellular
carcinoma (19), and is involved in
cellular proliferation, cell growth and cell cycle progression. To
investigate whether miRNA-34a is capable of regulating E2F5
expression in GC cells, qPCR was used. The present results
demonstrated that transfection of scramble miRNA into GC MKN28 and
BGC823 cells had no observed effect on E2F5 mRNA. However,
miRNA-34a mimics significantly inhibited E2F5 mRNA, but miRNA-34a
inhibitors significantly upregulated E2F5 mRNA (Fig. 3B). Additionally, western blot analysis
also identified similar results as transfection of miRNA-34a mimics
and miRNA-34a inhibitors into MKN28 cells could significantly
repress and increase E2F5 protein levels, respectively (Fig. 3C).
To additionally investigate whether miRNA-34a
negatively regulates E2F5 gene in GC by directly binding to the
specific miRNA-34a binding site in the 3′UTR of E2F5 gene, two
luciferase reporter recombinant vectors that contain either
wild-type (Wt) 3′UTR of E2F5 gene (pmirGLO-E2F5) or a Mut E2F5 with
mutated miRNA-34a binding sites (pmirGLO-MutE2F5) were constructed.
The luciferase reporter assays in MKN28 cells demonstrated that
miRNA-34a mimics and inhibitors significantly decreased and
increased the luciferase activity of Wt E2F5 reporter
(pmirGLO-E2F5), respectively. However, miRNA-34a and inhibitors had
no effect on the luciferase activity of E2F5 Mut reporter
(pmirGLO-MutE2F5) (Fig. 3D). Overall,
these results suggested that E2F5 is a direct target of miRNA-34a
in GC cells.
E2F5 gene knockdown enhance the
chemotherapeutic efficacy of paclitaxel
The above results demonstrated that miRNA-34a is
able to increase the chemotherapeutic efficacy of paclitaxel, and
that E2F5 is directly targeted by miRNA-34a. The present study
therefore then investigated whether E2F5 knockdown by specific E2F5
small interfering (si) RNA can also enhance the chemotherapeutic
efficacy of paclitaxel. Western blot analysis demonstrated that
E2F5 siRNA but not NC siRNA overexpression decreased E2F5 protein
level (Fig. 4A). MTT and BrdU assays
demonstrated that E2F5 knockdown by specific siRNA is capable of
significantly increasing the sensitivity of paclitaxel compared
with NC siRNA, and that E2F5 overexpression results in a
significant attenuation of the chemotherapeutic efficacy of
paclitaxel (Fig. 4B and C).
Therefore, E2F5 is also an important gene that involved in the
chemotherapeutic resistance of paclitaxel, and E2F5 gene knockdown
sensitized the GC to paclitaxel-induced proliferation
inhibition.
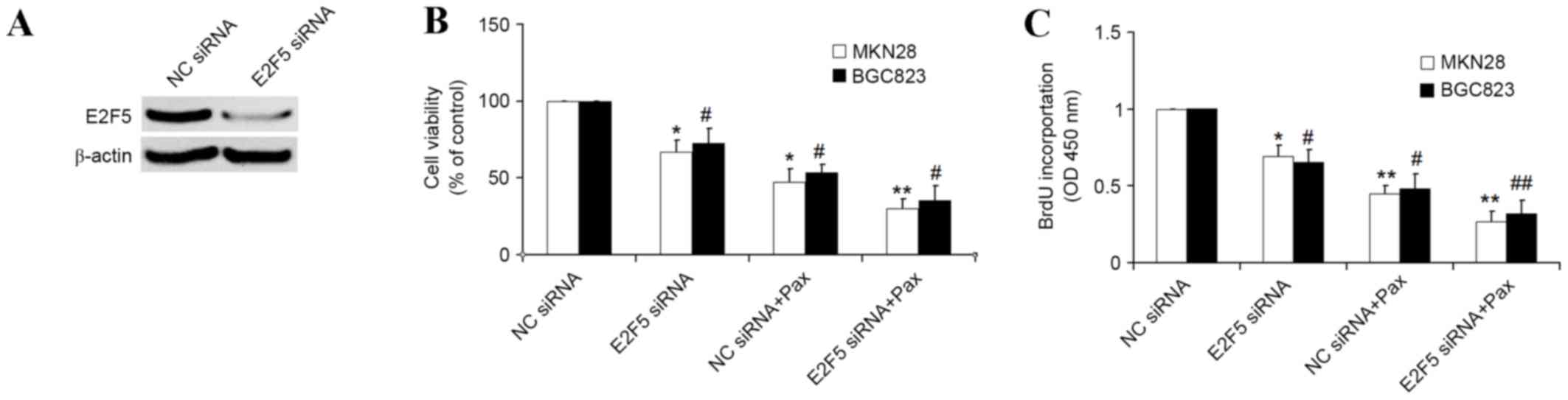 | Figure 4.E2F5 knockdown by specific siRNA in GC
enhanced the inhibitory effect of paclitaxel on cellular
proliferation. (A) Western blot analysis was used to detect E2F5
protein levels in MKN28 cells transfected with NC siRNA or E2F5
siRNA. β-actin was used as the control (B) MTT assays were used to
determine the cellular viability in MKN28 and BGC823 cells
transfected with NC siRNA or E2F5 siRNA for 24 h, followed by
treatment of paclitaxel (100 nM) for another 16 h. (C) BrdU
incorporation was used to detect the cellular proliferation in
MKN28 and BGC823 cells transfected with miR-SCR or miR-34a for 24
h, followed by treatment of paclitaxel (100 nM) for another 16 h.
*P<0.05, **P<0.01 vs. NC siRNA transfected MKN28 cells;
#P<0.05, ##P<0.01 vs. NC siRNA
transfected BGC823 cells. si, small interfering; GC, gastric
cancer; NC, negative control; BrdU, 5-Bromo-2-deoxyUridine; SCR,
scrambled; Pax, paclitaxel; OD, optical density. |
Discussion
In the present study, the effect of miRNA-34a on the
chemotherapeutic efficacy of paclitaxel was investigated. It was
observed that miRNA-34a significantly enhanced the inhibitory role
of paclitaxel in the proliferation of GC cells. In addition, the
present study identified that miRNA-34a was able to directly target
the E2F5 gene, which was demonstrated to be involved in the
chemotherapeutic resistance of paclitaxel.
An increasing number of studies have suggested that
miRNAs are a class of small highly conserved molecules that perform
a crucial role in the regulation of multiple cellular events
including cellular proliferation, differentiation, development and
apoptosis (8,20,21).
Dysregulation and dysfunction of miRNAs leads to the initiation and
development of cancers, indicating that miRNAs are important
molecules that can potentially be used for the diagnosis and
treatment of cancers (22). miRNA-34a
has previously been identified to be frequently aberrantly
expressed in human cancer, and involved in the regulation of cancer
cells as a tumor suppressor (13). A
previous study identified that miRNA-34a was significantly
downregulated in gastric cancer cells, and that miRNA-34a
suppressed EGFR-dependent MMP-7 activation (14). Additionally, another study
demonstrated that miRNA-34a was able to regulate the proliferation,
invasion and migration of GC cells by targeting survivin (23), an important anti-apoptotic gene
(24). Consistent with these studies,
the present results identified that miRNA-34a was able to inhibit
the growth rate of GC compared with scrambled miRNA. A recent study
suggested that miRNA-34a-5p can enhance chemotherapeutic
sensitivity to cisplatin in hepatocellular carcinoma MHCC-97 L
cells (25). However, whether
miRNA-34a enhances the chemotherapeutic efficacy of paclitaxel
remains unknown. The present study identified that miRNA-34a
transfection could significantly enhance the inhibitory effect of
paclitaxel on the cellular proliferation of GC, suggesting a
potential role of miRNA-34a in treatment of GC combined with
paclitaxel. However, the present study did not confirm whether
miRNA-34a is capable of enhancing the role of paclitaxel in the
inhibition of migration and invasion of GC cells. Therefore, future
investigation is warranted.
The present results demonstrated that miRNA-34a was
able to negatively regulate E2F5 by binding to the conserved
miRNA-34a binding site in the 3′UTR of E2F5. The transcription
factor E2F performs a central role in cell growth and proliferation
by controlling cell cycle (26),
which suggests that E2F5 may also be involved in cellular
proliferation and cell cycle progression as an important member of
the E2F family. Previous data has reported that E2F5 was
overexpressed in various types of human cancer including breast,
epithelial ovarian and prostate cancer and hepatocellular
carcinoma, and closely associated with cancer progression and
prognosis (16–19). However, the role of E2F5 in GC
remained unclear. The present study identified that E2F5 knockdown
by specific siRNA exerted a significant inhibitory effect on the
growth rate of GC cells, indicating that E2F5 may serve as a key
oncogene in GC. Importantly, E2F5 knockdown combined with
paclitaxel exhibited an increased inhibitory profile compared with
treatment with E2F5 siRNA or paclitaxel alone, which may provide a
more promising chemotherapeutic strategy for GC. Therefore, E2F5
may be an important target for treatment of GC. However, whether
E2F5 is also implicated in cell migration, invasion and apoptosis
in GC is unclear, and requires additional investigation.
In conclusion, the present study demonstrated the
role of miRNA-34a and its target E2F5 in the chemotherapeutic
sensitive of paclitaxel, which will not only aid future elucidation
of function of miRNA-34a/E2F5 axis in various cancers, but also
assist in the development of a promising chemotherapeutic strategy
for the treatment of GC.
References
|
1
|
Forman D and Burley VJ: Gastric cancer:
Global pattern of the disease and an overview of environmental risk
factors. Best Pract Res Clin Gastroenterol. 20:633–649. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ueda S, Kuji I, Shigekawa T, Takeuchi H,
Sano H, Hirokawa E, Shimada H, Suzuki H, Oda M, Osaki A and Saeki
T: Optical imaging for monitoring tumor oxygenation response after
initiation of single-agent bevacizumab followed by cytotoxic
chemotherapy in breast cancer patients. PLoS One. 9:e987152014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mita M, Gordon M, Rosen L, Kapoor N, Choy
G, Redkar S, Taverna P, Oganesian A, Sahai A, Azab M, et al: Phase
1B study of amuvatinib in combination with five standard cancer
therapies in adults with advanced solid tumors. Cancer Chemother
Pharmacol. 74:195–204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Le XF and Bast RC Jr: Src family kinases
and paclitaxel sensitivity. Cancer Biol Ther. 12:260–269. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu G, Qin XQ, Guo JJ, Li TY and Chen JH:
AKT/ERK activation is associated with gastric cancer cell
resistance to paclitaxel. Int J Clin Exp Pathol. 7:1449–1458.
2014.PubMed/NCBI
|
|
6
|
Zanetta G, Fei F and Mangioni C:
Chemotherapy with paclitaxel, ifosfamide, and cisplatin for the
treatment of squamous cell cervical cancer: The experience of
Monza. Semin Oncol. 27 1 Suppl 1:S23–S27. 2000.
|
|
7
|
Murray S, Briasoulis E, Linardou H,
Bafaloukos D and Papadimitriou C: Taxane resistance in breast
cancer: Mechanisms, predictive biomarkers and circumvention
strategies. Cancer Treat Rev. 38:890–903. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang C, Chen X, Alattar M, Wei J and Liu
H: MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis
of gastric cancer. Cancer Gene Ther. 22:291–301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tutar L, Tutar E, Özgür A and Tutar Y:
Therapeutic targeting of microRNAs in cancer: Future perspectives.
Drug Dev Res. 76:382–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu X, Li J, Qin F and Dai S: miR-152 as a
tumor suppressor microRNA: Target recognition and regulation in
cancer. Oncol Lett. 11:3911–3916. 2016.PubMed/NCBI
|
|
12
|
Naidu S, Magee P and Garofalo M:
MiRNA-based therapeutic intervention of cancer. J Hematol Oncol.
8:682015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Misso G, Di Martino MT, de Rosa G, Farooqi
AA, Lombardi A, Campani V, Zarone MR, Gullà A, Tagliaferri P,
Tassone P and Caraglia M: Mir-34: A new weapon against cancer? Mol
Ther Nucleic Acids. 3:e1942014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu G, Jiang C, Li D, Wang R and Wang W:
MiRNA-34a inhibits EGFR-signaling-dependent MMP7 activation in
gastric cancer. Tumour Biol. 35:9801–9806. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Polanowska J, Le Cam L, Orsetti B, Vallés
H, Fabbrizio E, Fajas L, Taviaux S, Theillet C and Sardet C: Human
E2F5 gene is oncogenic in primary rodent cells and is amplified in
human breast tumors. Genes Chromosomes Cancer. 28:126–130. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kothandaraman N, Bajic VB, Brendan PN,
Huak CY, Keow PB, Razvi K, Salto-Tellez M and Choolani M: E2F5
status significantly improves malignancy diagnosis of epithelial
ovarian cancer. BMC Cancer. 10:642010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao J, Wu XY, Ling XH, Lin ZY, Fu X, Deng
YH, He HC and Zhong W: Analysis of genetic aberrations on
chromosomal region 8q21-24 identifies E2F5 as an oncogene with copy
number gain in prostate cancer. Med Oncol. 30:4652013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang Y, Yim SH, Xu HD, Jung SH, Yang SY,
Hu HJ, Jung CK and Chung YJ: A potential oncogenic role of the
commonly observed E2F5 overexpression in hepatocellular carcinoma.
World J Gastroenterol. 17:470–477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Macfarlane LA and Murphy PR: MicroRNA:
Biogenesis, function and role in cancer. Curr Genomics. 11:537–561.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang J, Li S, Li L, Li M, Guo C, Yao J
and Mi S: Exosome and exosomal microRNA: Trafficking, sorting, and
function. Genomics Proteomics Bioinformatics. 13:17–24. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cao W, Fan R, Wang L, Cheng S, Li H, Jiang
J, Geng M, Jin Y and Wu Y: Expression and regulatory function of
miRNA-34a in targeting survivin in gastric cancer cells. Tumour
Biol. 34:963–971. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
McKenzie JA and Grossman D: Role of the
apoptotic and mitotic regulator survivin in melanoma. Anticancer
Res. 32:397–404. 2012.PubMed/NCBI
|
|
25
|
Li XY, Wen JY, Jia CC, Wang TT, Li X, Dong
M, Lin QU, Chen ZH, Ma XK, Wei LI, et al: MicroRNA-34a-5p enhances
sensitivity to chemotherapy by targeting AXL in hepatocellular
carcinoma MHCC-97L cells. Oncol Lett. 10:2691–2698. 2015.PubMed/NCBI
|
|
26
|
Chen HZ, Tsai SY and Leone G: Emerging
roles of E2Fs in cancer: An exit from cell cycle control. Nat Rev
Cancer. 9:785–797. 2009. View
Article : Google Scholar : PubMed/NCBI
|