Introduction
Glioma is one of the most common primary central
nervous system tumors with high mortality and poor 5-year survival
rate (1). Gliomas represent a
disparate group of tumors for which there are to date no cure.
Thus, there is an urgent need for novel diagnostic and therapeutic
methods based on the increased understanding of the molecular
mechanisms of glioma (2). Cancer
stem-like cells (CSCs) are poorly differentiated multipotent
tumor-propagating cells that disproportionately contribute to
therapeutic resistance and tumor recurrence (3). Research has been performed to identify
the approaches to inhibit CSCs. Suppression of stathmin, an
oncogene, inhibited invasion and enhanced chemotherapy sensitivity
of stem cells derived from glioma cell lines (4).
AKT serine-threonine kinase 1 (AKT) is a downstream
target and effector of phosphatidylinositol 3-kinase (PI3K). AKT is
considered to be a key regulator of cell growth and fate decisions
in tumors (5). It has been reported
that inhibition of the AKT pathway could suppress glioma (6). CD133 is a marker of glioma stem-like
cells. A previous study demonstrated that the CSC marker, CD133,
was associated with activated AKT and radiation resistance in colon
cancer cells (7). Zhu et al
(8) identified that overexpression of
CD133 enhanced chemoresistance to 5-fluorouracil by activating the
PI3K/AKT/ribosomal protein S6 kinase pathway in gastric cancer
cells.
MicroRNAs (miRNAs) are small non-coding RNAs which
have an important role in regulating gene expression (9). MicroRNA-200b (miR-200b) targets protein
kinase Cα and suppresses triple-negative breast cancer metastasis
(10). The restoration of miR-200b
expression may inhibit the maintenance of CSC properties and the
reverse chemoresistance of CSCs. The histone deacetylase
1/miR-200b/Suz-12-E-cadherin signaling may account for maintenance
of CSCs (11). The current study
aimed to identify the role of miR-200b in CD133+ glioma
cells.
Materials and methods
Cell culture and sorting
Cells (TC1 and TC2 cells) were isolated from primary
surgical glioma biopsy specimens in accordance with protocols as
previously described (12). The U251
glioma cell line was purchased from the Chinese Academy Medical
Science (Beijing, China). The cells were cultured in RPMI-1640
medium supplemented with 10% fetal bovine serum (FBS), penicillin
and streptomycin, all of which were obtained from Gibco (Thermo
Fisher Scientific, Inc., Waltham, MA, USA) and were cultured in a
humidified chamber with 5% CO2 at 37°C. All
transfections were performed using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). Magnetic beads (Dynabeads M-450
Epoxy; Invitrogen; Thermo Fisher Scientific, Inc.) labeled with
CD133 antibody (catalog no., PA5-38014; Thermo Fisher Scientific,
Inc.) were incubated with 2.5×106 cells in 1 ml B1
solution containing Dynabeads for 30 min at 4°C. In total, 25 µl of
labeled Dynabeads was incubated with 2.5×106 cells.
Suspension was incubated for 20 min at 4°C with gentle rotation
followed by 2 min positive isolation using an EasySep magnet (cat.
no. 18000; Stemcell Technologies, Inc., Beijing, China). Bead-bound
cells were washed 4 times using 1 ml PBS buffer.
Tissue samples
Glioma samples (n=80) and normal brain tissues
(n=20) were collected at the The First Affiliated Hospital of
Guangdong Pharmaceutical University (Guangzhou, China) with written
consent from patients or family members of patients in accordance
with institutional guidelines as approved by the First Affiliated
Hospital of Guangdong Pharmaceutical University. Pathology was
graded according to World Health Organization criteria. The RNA
were extracted from the tissues using TRIzol reagent (Thermo Fisher
Scientific, Inc.) for reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) detection.
RT-qPCR
RT of specific miRNAs (from 10 ng of total RNA) was
performed using the real-time loop primers for each type of miRNA
and the TaqMan miRNA RT kit from Applied Biosystems (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). cDNA obtained using GoScript™
Reversion Transcription Mixes (Promega Corporation, Madison, WI,
USA)was used for quantitative TaqMan PCR using the real-time
primers provided, according to the manufacturer's instructions. Cq
values were converted to fold expression changes (2−ΔΔCq
values) (13) following normalization
to U6 small nuclear RNA (primers; forward 5′-CTCGCTTCGGCAGCACA-3′,
reverse 5′-AACGCTTCACGAATTTGCGT-3′). For mRNA analysis, miR-200b
mimics, inhibitor sequences or control sequences were transfected
into glioma cells. The primer sequences were as follows:
hsa-miR-200b mimics, 5′-CAUCUUACUGGGCAGCAUUGGA-3′; miR-NC,
5′-UUCUCCGAACGUGUCACGUTT-3′; miR-200b inhibitor,
5′-UCAUCAUUACCAGGCAGUAUUA-3′; inh-NC, 5′-CAGUACUUUUGUGUAGUACAA-3′
(GenePharma, Shanghai, China). RT was performed on total RNA using
random primers (GE Healthcare Life Sciences, Chalfont, UK), and
GAPDH (forward 5′-TTGCCATCAATGACCCCTTCA-3′, reverse
5′-CGCCCCACTTGATTTTGGA-3′) were used to control for cDNA
concentration in a separate PCR reactions for each sample.
LightCycler Fast Start DNA Master SYBR Green Mix (Roche Diagnostics
GmbH, Mannheim, Germany) was added to each PCR reaction along with
cDNA and 1 pmol primer in a total volume of 10 µl.
Luciferase assays
The full-length of the 3′ untranslated regions
(UTRs) of the prominin 1 (PROM1) gene, which encodes CD133, was
amplified by PCR from U251 cells genomic DNA and inserted into the
pGL3 control vector (Promega Corporation) using the XbaI
site immediately downstream from the stop codon of luciferase.
Several inserts with deletions of 6 bp from the site of perfect
complementarity of the PROM1 gene were generated using the Qiagen
XL-Site Directed Mutagenesis kit (Qiagen, Inc., Valencia, CA, USA).
U251 cells were cotransfected using nucleoporation (Amaxa; Lonza
Group, Ltd., Basel, Switzerland) according to the manufacturer's
protocol (solution V, programme T-016) using 5 µg of the firefly
luciferase reporter vector and 0.5 µg of the control vector
containing Renilla luciferase (pRL-TK; Promega Corporation). Each
nucleoporation used 50 nM of the miR-200b or a scrambled
oligonucleotide. Firefly and Renilla luciferase activities were
measured consecutively using the dual luciferase assay and Thermo
Scientific Multiskan MK3 Microplate Reader (Thermo Fisher
Scientific, Inc.) 48 h after transfection.
Cell number counting
A total of 10,000 TC1 and TC2 cells were plated onto
a 24-well plate. Upon attachment, cells were transfected with
scramble control miRNA, miR-200b mimics, inhibitor control or
miR-200b inhibitor. Each group was evaluated in duplicate in six
wells. After being transfected for 24, 48, 72 or 96 h, cells were
digested with 100 µl trypsin, and then 0.5 ml RPMI-1640 medium
supplemented with 10% FBS, penicillin and streptomycin was added.
Upon mixing, 10 µl cell suspension was added into a slide, which
was then inserted into a cell counter (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) to count the cell number.
Bromodeoxyuridine (BrdU) cell
proliferation assay
The cell proliferation assay was performed by
measuring BrdU incorporation into the newly synthesized DNA of
replicating cells. Cells were then infected with miRNA mimics and
inhibitor. The cells were loaded with BrdU (Roche Applied Science,
Penzberg, Germany) in the last 4 h of treatment with miRNA mimics
or inhibitor. BrdU incorporation was quantified by BrdU
immunohistochemical staining kit (Abcam, Cambridge, UK) following
manufacturer's instructions. Three fields were chosen randomly from
10 sections to ensure objectivity of sampling. Digital images were
acquired using a confocal microscope. Each assay was repeated three
times. From total of 100 cells from each field the ratio of
BrdU-positive cell was calculated.
Western blot
Glioma cells were transfected with CD133 siRNA
(sense, 5′-GGCUGCUGUUUAUUAUUCUTT-3′ and antisense,
5′-AGAAUAAUAAACAGCAGCCTT-3′) and nonspecific siRNA (sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′) (Riobio, Guangzhou, China). The
proteins from glioma cell lines were extracted using RIPA lysis
buffer with a proteinase inhibitor. The protein concentration in
the lysates was measured with the Protein Bicinchoninic Acid Assay
kit (Bio-Rad Laboratories, Inc.), and 20 µg of the total protein
mixed with 2X SDS loading buffer was loaded per lane. The proteins
in the lysates were separated by 12% SDS-PAGE and transferred to
polyvinylidene difluoride (PVDF) membranes (EMD Millipore,
Billerica, MA, USA). To block nonspecific binding, the membranes
were incubated at room temperature for 1 h with 5% skim milk
powder. The PVDF membranes were then incubated for 12 h at 4°C with
an antiserum containing antibodies against CD133 rabbit monoclonal
antibody (mAb) (dilution, 1:1,000; catalog no., 5860; Cell
Signaling Technology, Inc., Danvers. MA, USA), GFAP mouse mAb
(dilution, 1:200; catalog no., sc-71143; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA), p-AKT rabbit mAb (dilution, 1:1,000;
catalog no., 4060; Cell Signaling Technology, Inc.), AKT rabbit mAb
(dilution, 1:1,000; catalog no., 4691; Cell Signaling Technology,
Inc.), Notch1 rabbit mAb (dilution, 1:1,000; catalog no., 3608;
Cell Signaling Technology, Inc.) and β-actin mouse mAb (dilution,
1:1,000; catalog no., 3700; Cell Signaling Technology, Inc.).
Secondary antibodies consisted of rabbit anti-mouse horseradish
perosidase (HRP)-conjugated (catalog no., BA1058) and goat
anti-rabbit HRP-conjugated (catalog no., BA1058) obtained from
Boster (Wuhan, China). The secondary antibodies were diluted at
1:5,000 and incubated with the membrane at room temperature for 1 h
and enhanced chemiluminescence western blot detection reagents (New
England Biolabs, Inc., Ipswich, MA, USA) were used to visualize the
target proteins, which were quantified with a Bio Image Intelligent
Quantifier 1-D (version 2.2.1; Nikon, Tokyo, Japan).
Statistical analysis
In general, significance was analyzed by unpaired
two-tailed Student t test using GraphPad InStat 5.0 software
(GraphPad Software, Inc., La Jolla, CA, USA). The significance of
CD133 expression differences in glioma samples was determined by
using Student t test (two-tailed). Results are expressed as the
mean ± standard deviation. Pearson correlation assay was used to
analyze the correlation between CD133 expression and miR-200b
expression. P<0.05 was considered to indicate a statistically
significant difference.
Results
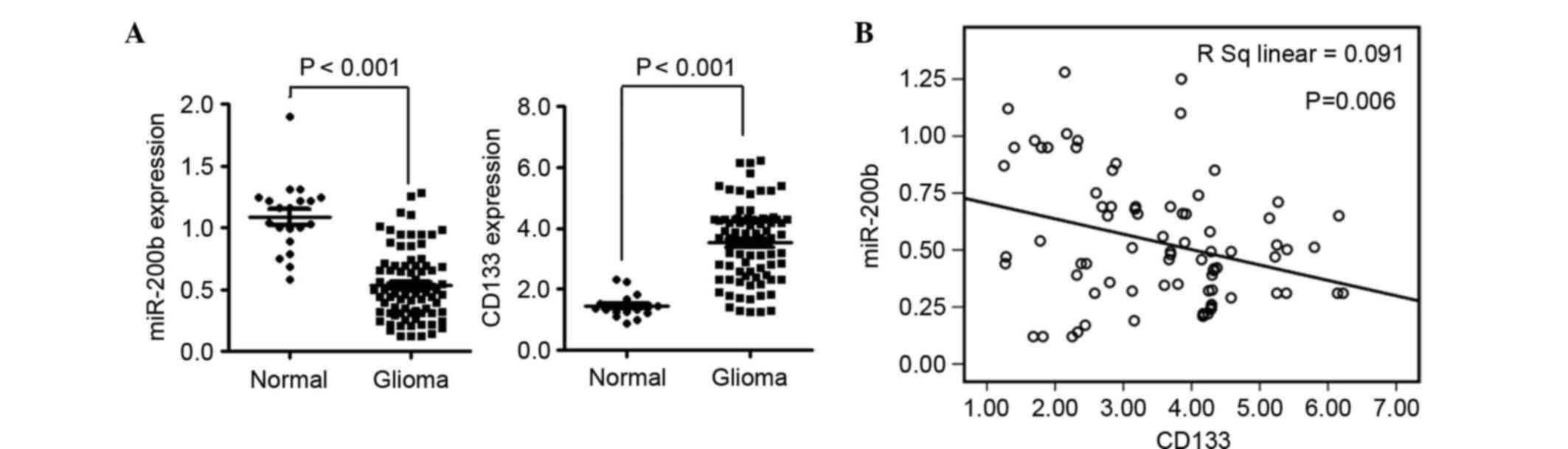
CD133 expression is elevated in
gliomas and negatively correlated with the expression of
miR-200b
Analysis of the expression of miR-200b and mRNA
levels of CD133 in glioma tissues was conducted using tissues
obtained from the First Affiliated Hospital of Guangdong
Pharmaceutical University. The expression of miR-200b was
significantly downregulated (P<0.01) and CD133 was significantly
upregulated (P<0.01) in glioma compared with normal tissues
(Fig. 1A). Statistical analysis
indicated that the expression of CD133 was negatively correlated
with miR-200b in glioma tissues (P=0.06; Fig. 1B).
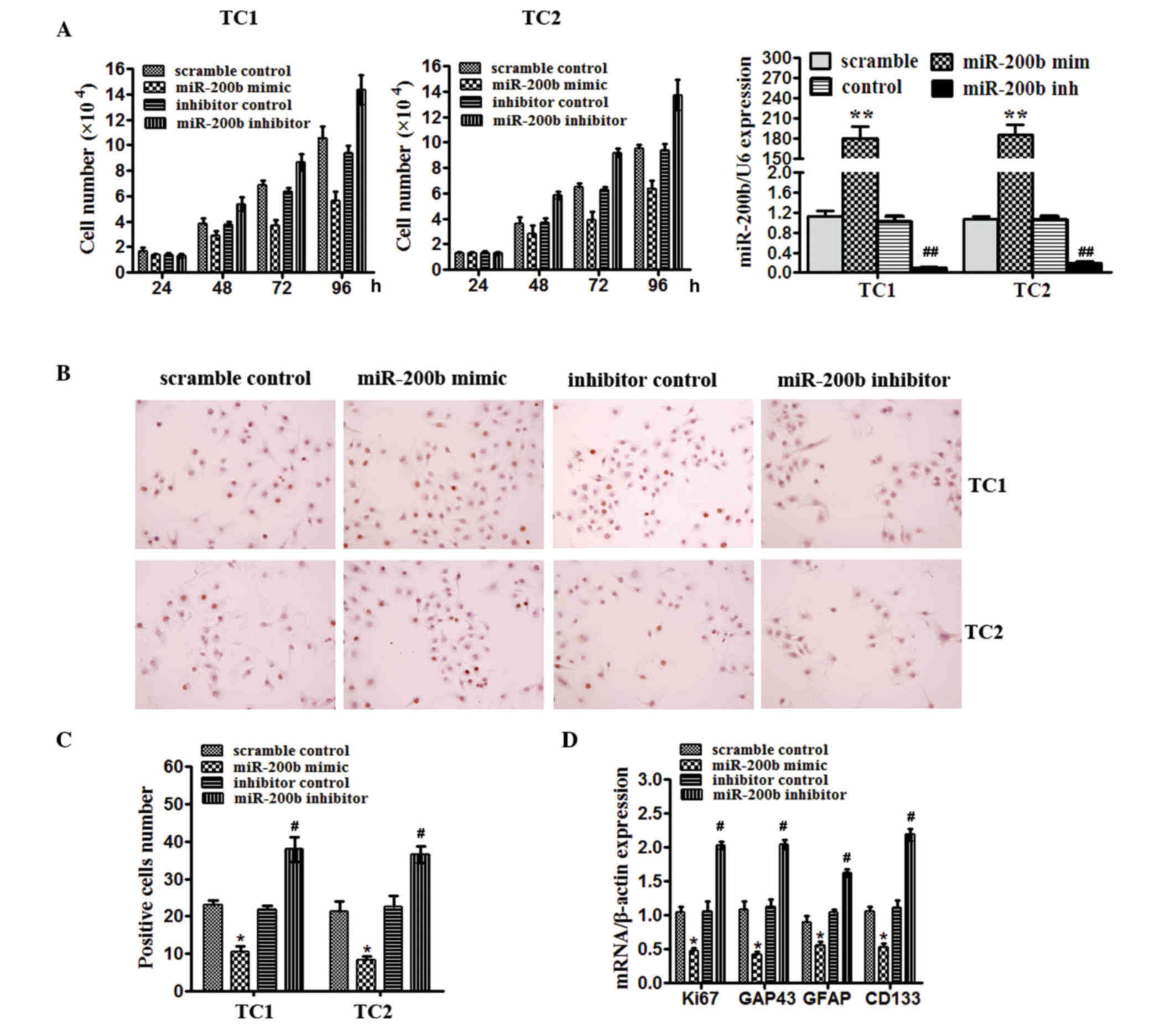
miR-200b levels are required for
proliferation and stem-like properties of human glioma cells
To address the importance of miR-200b in gliomas
in vitro, the TC1 and TC2 glioma tissues initiated cell
lines were used. Cells were transfected with miR-200b mimics, a
scrambled control, miR-200b inhibitor or a negative control and
were subsequently collected. Cell number counting assay
demonstrated that miR-200b reduced cell growth compared with the
scramble control and inhibition of miR-200b increased cell number
compared with the negative control after culture for 48 and 72 h in
TC1 and TC2 cells (Fig. 2A). In TC1
and TC2 cells, the effect of miR-200b on proliferation was
confirmed by BrdU staining (Fig. 2B and
C). The number of BrdU-positive TC1 and TC2 cells was decreased
by miR-200b mimcs compared with scramble control (P<0.05), but
increased by miR-200b inhibitor compared with the inhibitor control
(P<0.05), as detected by BrdU staining (Fig. 2C). miR-200b in TC1 cells significantly
reduced mRNA expression of the proliferation marker, Ki67
(P<0.05), and GAP43 (P<0.05), a marker of mature neurons
compared with the scramble control, and also decreased the levels
of markers known to identify normal stem-like cells and brain
tumor-initiating cells, CD133 (P<0.05), and GFAP (P<0.05).
miR-200b inhibitor had the opposite effect (Fig. 2D). These data prompted the hypothesis
that miR-200b levels may be important for proliferation and
directing the fate certain cells in glioma.
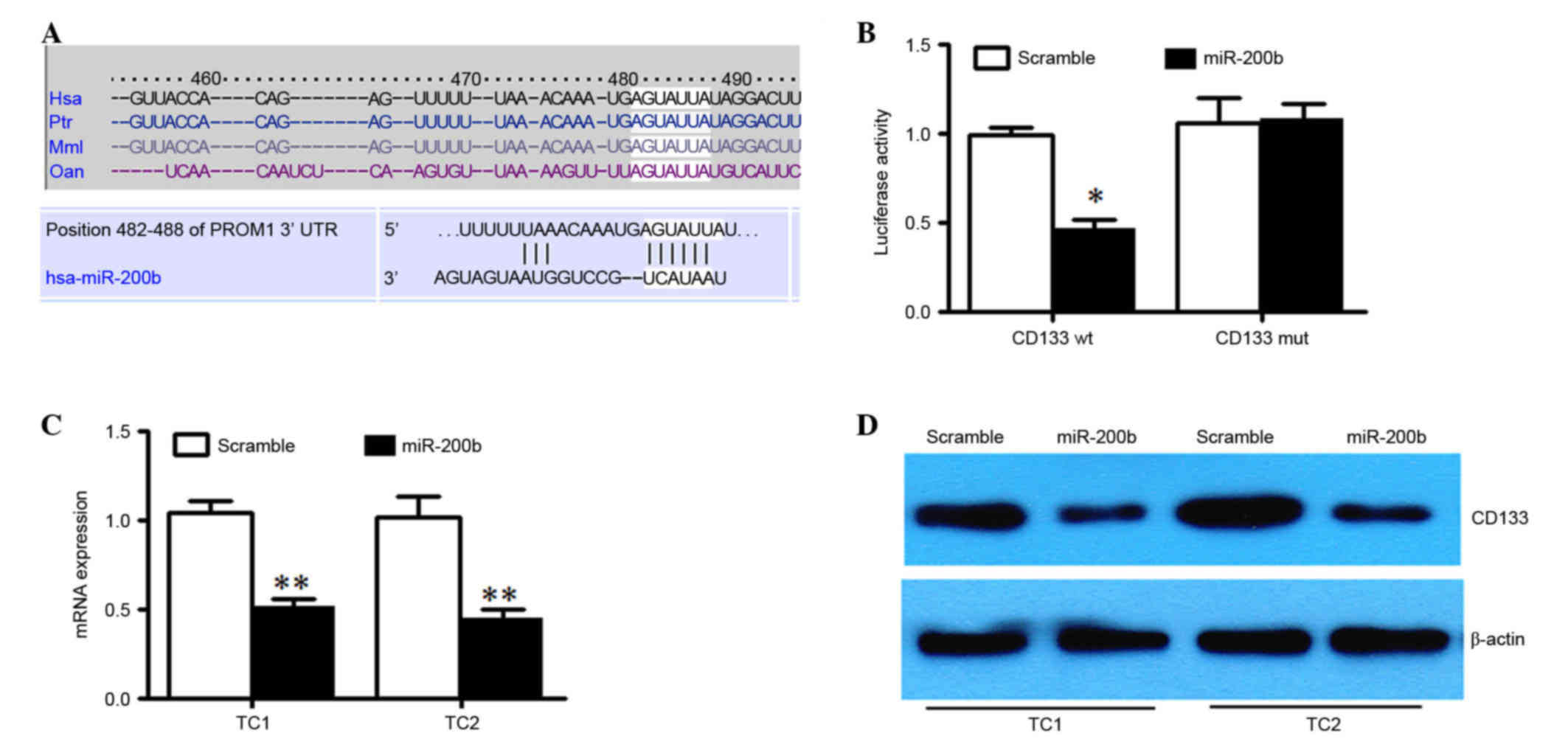
PROM1 is a direct target of
miR-200b
We used Targetscan (http://www.targetscan.org/mamm_31/) to help identify
miR-200's targets in human glioma. PROM1 was predicted (Fig. 3A), which encodes CD133 protein. The
full-length PROM1 3′-UTR was cloned downstream of the firefly
luciferase gene and cotransfected with miR-200b mimics or scrambled
oligonucleotides and LNA-modified anti-miR-200b oligonucleotide or
a control oligonucleotide. U251 cells cotransfected with wild type
PROM1 reporter constructs and miR-200b mimics exhibited a
significant reduction of luciferase activity compared with cells
transfected with scramble control miRNA (P<0.05; Fig. 3B). Additionally, mutation of the
putative miR-200b target sites in the 3′-UTR of PROM1 abrogated
luciferase the response to miR-200b (Fig.
3B). miR-200b mimic or a scrambled oligonucleotide was
transfected into TC1 and TC2 cells. The result demonstrated that
there was a marked reduction of the mRNA and protein level of CD133
in miR-200b overexpressed TC1 and TC2 cells compared with scrambled
control (Fig. 3C and D). Taken
together, these results indicated that miR-200b downregulates CD133
expression by directly targeting its 3′UTR.
miR-200b expression has a critical
role in the stemness properties and division of the CD133+ glioma
cells
TC1 cells with and without CD133 expression were
isolated. The expression of miR-200b in CD133+ TC1 cells
was higher than in CD133− TC1 cells (Fig. 4A). To determine the potential role of
miR-200b in the CD133+ glioma cells, multiple
proliferation and stem-like markers were detected. Transfection of
TC1 cells with miR-200b mimics significantly reduced the mRNA
levels of the proliferation marker Ki67, GFAP, Nestin and GAP43
(Fig. 4B and C). The miR-200b
inhibitor exerted the opposite effect (Fig. 4B and C). To determine the potential
role of miR-200b in the CD133+ populations with
neurosphere formation capacity, U251 cells were cultured and
passaged in monolayers or as spheres to promote self renewal in
vitro. Overexpression of miR-200b suppressed colony and sphere
formation of CD133+ TC1 cells. However, miR-200b
inhibitor enhanced the colony and sphere formation of
CD133+ TC1 cells (Fig. 4D and
E).
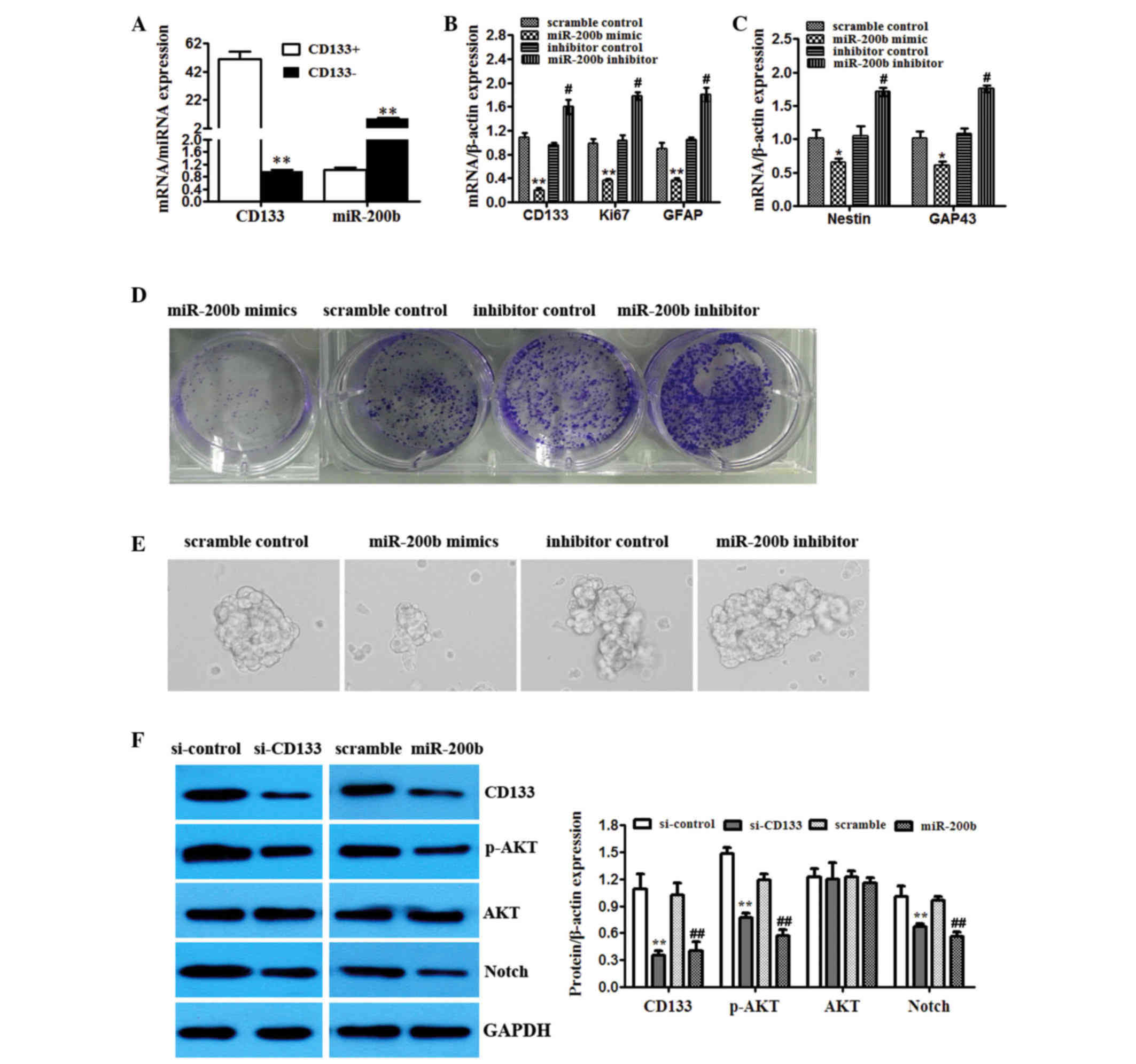 | Figure 4.miR-200b expression has a critical
role in the stemness properties and division of the
CD133+ glioma cells. (A) TC1 cells were sorted and
CD133+ and CD133− populations were analyzed
by RT-qPCR for CD133 and miR-200b levels. **P<0.01 vs.
CD133+. (B) TC1 cells were sorted for CD133. The
CD133+ population was transfected with scrambled
control, miR-200b mimics or miR-200b inhibitor. Overexpression and
knockdown confirmed by RT-qPCR. Effects on CD133, Ki67 and GFAP
mRNA levels. (C) RT-qPCR of different markers (Nestin, GAP43) from
the CD133+ population transfected with scrambled
control, miR-200b mimics or miR-200b inhibitor. *P<0.05,
**P<0.01 vs. scramble control; #P<0.05,
##P<0.01 vs. inhibitor control. (D) Clonal assay was
performed on control, miR-200b mimics and miR-200b inhibitor
transfected CD133+ TC1 cells. (E) Neurosphere formation
was detected in CD133+ TC1 cells transfected with
control, miR-200b mimics and inhibitor. (F) The activity of AKT
signaling (CD133, p-AKT, total AKT, Notch) in CD133+
cells with CD133 knockdown or miR-101 overexpression tested by
western blot. GAPDH was used as an internal control. **P<0.01
vs. si-control; ##P<0.01 vs. scramble. RT-qPCR,
reverse transcription-quantitative polymerase chain reaction; miR,
microRNA; GFAP, glial fibrillary acidic protein; GAP43,
growth-associated protein 43; si, small interfering RNA; p-,
phosphorylated; AKT, AKT serine/threonine kinase 1. |
miR-200b regulates AKT signaling by
targeting CD133 in CD133+ glioma cells
The activity of AKT signaling was detected in
CD133+ glioma cells treated with miR-200b mimics and
CD133 was knocked down. The data demonstrated that overexpression
of miR-200b and inhibition of CD133 reduced levels of
phosphorylated AKT and Notch compared with the scramble miRNA and
control si, respectively, but had no effect on total Akt (Fig. 4F).
Discussion
Many CSCs markers have been suggested to be
potential prognostic and predictive markers in glioma, including
CD133 and Nestin. CD133 and Nestin were detected in all
histological subtypes of glioma, but predominantly in WHO grade III
and IV tumors (14). The present
study identified that CD133 was overexpressed in glioma cell lines
and tissues.
miR-200b is a member of the miR-200 family that
suppress tumors. The miR-200 family members are strongly associated
with pathological epithelial to mesenchymal transitions (EMT) and
to have a metastasis suppressive role (15). miR-200b suppresses arsenic-transformed
cell migration by targeting protein kinase Cα, the Wnt5b-protein
kinase Cα positive feedback loop and inhibiting Rac1 activation
(16). miR-200b was downregulated in
the adriamycin-resistant small lung cancer H69AR cells and enforced
expression of miR-200b by miRNA mimics increased cell sensitivity
to adriamycin (17). A previous study
demonstrated that miR-200b may be a novel and valuable marker for
predicting the clinical outcome of patients with glioma (18). miR-200b was reported to be prognostic
factor and to target multiple members of the RAB GTPase family in
glioma (19). The current study
identified that miR-200b was downregulated and negatively
correlated with the expression of CD133 in glioma tissues.
CD133 is encoded by the PROM1 gene. Expression of
PROM1 in cancer is considered similar to the expression and
function observed in normal stem cells. Overexpression of PROM1 is
inversely correlated with isocitrate dehydrogenase (R132H)
mutation. These findings support thay PROM1 functions as a tumor
cell-intrinsic marker associated with glioma survival (20). Use of the online software, Targetscan,
indicated that PROM1 is a direct target of miR-200b. The the best
of our knowledge, the current study is the first to demonstrate
that PROM1 is a direct target of miR-200b.
Glioma stem cells have self-renewal capability and
are resistant to conventional chemotherapy. The role of miRNA
dysregulation in stemness and division remains to be identified. A
recent study demonstrated that targeting the miR-34a-Notch1 axis
reduces breast cancer stemness and chemoresistance (21). miR-145 is a strong repressor of EMT.
The transcriptional repressor zinc-finger E-box binding homeobox 2
(ZEB2) is a target of miR-145, and it can also regulate the
expression of miR-145. The ZEB2/miR-145 double-negative feedback
loop is important for the control of EMT and stem cell properties
during the bone metastasis of prostate cancer cells (22). miR-300 promotes self-renewal and
inhibits the differentiation of glioma stem-like cells (23). Although miR-200b was demonstrated to
be a tumor suppressor, its role in stemness properties remains
unknown. The present study initially identified that the expression
of miR-200b was essential for proliferation and stem-like
properties of human glioma cells, TC1 and TC2. Subsequently,
CD133+ cells were isolated from the glioma cell
population and revealed that miR-200b expression was important for
the stemness properties and proliferation of CD133+
glioma cells.
A previous study demonstrated that upregulation of
miR-200b in patients with biliary atresia accelerates the
proliferation and migration of hepatic stallate cells by activating
PI3K/AKT signaling (24). Zinc finger
protein, FOG family member 2 downregulation by transforming growth
factor-β1-induced miR-200b/c leads to AKT kinase activation and
glomerular mesangial hypertrophy associated with diabetic
nephropathy (25). The present study
aimed to understand the association between miR-200b and AKT
signaling in glioma stem-like cells. The data indicated that
overexpression of miR-200b and inhibition of CD133 activated AKT
signaling in CD133+ glioma cells.
Acknowledgements
This study was supported by the National Science
Foundation of China (grant no. 81173418).
References
|
1
|
Chatterjee M, Li K, Chen L, Maisano X, Guo
Q, Gan L and Li JY: Gbx2 regulates thalamocortical axon guidance by
modifying the LIM and Robo codes. Development. 139:4633–4643. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Deng Y, Wang J, Wang G, Jin Y, Luo X, Xia
X, Gong J and Hu J: p55PIK transcriptionally activated by MZF1
promotes colorectal cancer cell proliferation. Biomed Res Int.
2013:8681312013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Deplus R, Blanchon L, Rajavelu A, Boukaba
A, Defrance M, Luciani J, Rothé F, Dedeurwaerder S, Denis H,
Brinkman AB, et al: Regulation of DNA methylation patterns by
CK2-mediated phosphorylation of Dnmt3a. Cell Rep. 8:743–753. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tsai LH, Wu JY, Cheng YW, Chen CY, Sheu
GT, Wu TC and Lee H: The MZF1/c-MYC axis mediates lung
adenocarcinoma progression caused by wild-type lkb1 loss. Oncogene.
34:1641–1649. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shen Y, Pan X and Zhao H: The histone
demethylase PHF8 is an oncogenic protein in human non-small cell
lung cancer. Biochem Biophys Res Commun. 451:119–125. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang C, Lu S, Jiang J, Jia X, Dong X and
Bu P: Hsa-microRNA-101 suppresses migration and invasion by
targeting Rac1 in thyroid cancer cells. Oncol Lett. 8:1815–1821.
2014.PubMed/NCBI
|
|
7
|
Yan F, Shen N, Pang J, Xie D, Deng B,
Molina JR, Yang P and Liu S: Restoration of miR-101 suppresses lung
tumorigenesis through inhibition of DNMT3a-dependent DNA
methylation. Cell Death Dis. 5:e14132014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu Y, Yu J, Wang S, Lu R, Wu J and Jiang
B: Overexpression of CD133 enhances chemoresistance to
5-fluorouracil by activating the PI3K/Akt/p70S6K pathway in gastric
cancer cells. Oncol Rep. 32:2437–2444. 2014.PubMed/NCBI
|
|
9
|
Li J, Hart RP, Mallimo EM, Swerdel MR,
Kusnecov AW and Herrup K: EZH2-mediated H3K27 trimethylation
mediates neurodegeneration in ataxia-telangiectasia. Nat Neurosci.
16:1745–1753. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yan S, Jane DT, Dufresne MJ and Sloane BF:
Transcription of cathepsin B in glioma cells: Regulation by an
E-box adjacent to the transcription initiation site. Biol Chem.
384:1421–1427. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yin J, Wang M, Jin C and Qi Q: miR-101
sensitizes A549 NSCLC cell line to CDDP by activating caspase
3-dependent apoptosis. Oncol Lett. 7:461–465. 2014.PubMed/NCBI
|
|
12
|
Singh SK, Hawkins C, Clarke ID, Squire JA,
Bayani J, Hide T, Henkelman RM, Cusimano MD and Dirks PB:
Identification of human brain tumour initiating cells. Nature.
432:396–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yan S and Sloane BF: Molecular regulation
of human cathepsin B: Implication in pathologies. Biol Chem.
384:845–854. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sdek P, Oyama K, Angelis E, Chan SS,
Schenke-Layland K and MacLellan WR: Epigenetic regulation of
myogenic gene expression by heterochromatin protein 1 alpha. PLoS
One. 8:e583192013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Z, Humphries B, Xiao H, Jiang Y and
Yang C: MicroRNA-200b suppresses arsenic-transformed cell migration
by targeting protein kinase Cα and Wnt5b-protein kinase Cα positive
feedback loop and inhibiting Rac1 activation. J Biol Chem.
289:18373–18386. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fang S, Zeng X, Zhu W, Tang R, Chao Y and
Guo L: Zinc finger E-box-binding homeobox 2 (ZEB2) regulated by
miR-200b contributes to multi-drug resistance of small cell lung
cancer. Exp Mol Pathol. 96:438–444. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Men D, Liang Y and Chen L: Decreased
expression of microRNA-200b is an independent unfavorable
prognostic factor for glioma patients. Cancer Epidemiol.
38:152–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Q, Tang H, Liu X, Liao Y, Li H, Zhao
Z, Yuan X and Jiang W: miR-200b as a prognostic factor targets
multiple members of RAB family in glioma. Med Oncol. 31:8592014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Olausson K Holmberg, Maire CL, Haidar S,
Ling J, Learner E, Nister M and Ligon KL: Prominin-1 (CD133)
defines both stem and non-stem cell populations in CNS development
and gliomas. PLoS One. 9:e1066942014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Park EY, Chang E, Lee EJ, Lee HW, Kang HG,
Chun KH, Woo YM, Kong HK, Ko JY, Suzuki H, et al: Targeting of
miR-34a-NOTCH1 axis reduced breast cancer stemness and
chemoresistance. Cancer Res. 74:7573–7582. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ren D, Wang M, Guo W, Huang S, Wang Z,
Zhao X, Du H, Song L and Peng X: Double-negative feedback loop
between ZEB2 and miR-145 regulates epithelial-mesenchymal
transition and stem cell properties in prostate cancer cells. Cell
Tissue Res. 358:763–778. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang D, Yang G, Chen X, Li C, Wang L, Liu
Y, Han D, Liu H, Hou X, Zhang W, et al: mir-300 promotes
self-renewal and inhibits the differentiation of glioma stem-like
cells. J Mol Neurosci. 53:637–644. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xiao Y, Wang J, Chen Y, Zhou K, Wen J,
Wang Y, Zhou Y, Pan W and Cai W: Up-regulation of miR-200b in
biliary atresia patients accelerates proliferation and migration of
hepatic stallate cells by activating PI3K/Akt signaling. Cell
Signal. 26:925–932. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Park JT, Kato M, Yuan H, Castro N, Lanting
L, Wang M and Natarajan R: FOG2 protein down-regulation by
transforming growth factor-β1-induced microRNA-200b/c leads to Akt
kinase activation and glomerular mesangial hypertrophy related to
diabetic nephropathy. J Biol Chem. 288:22469–22480. 2013.
View Article : Google Scholar : PubMed/NCBI
|