Introduction
Renal cell carcinoma (RCC), which is one of the most
common types of cancer, accounts for almost 3% of all human
malignancies (1). As the most common
type of RCC, clear cell RCC (ccRCC) accounts for 70–80% of RCC
cases (2). The metastasis and
recurrence of ccRCC, as well as its poor prognosis, results in poor
survival for patients (3).
At present, with the development of microarray
technology, a large number of differentially expressed genes (DEGs)
associated with ccRCC have been identified and the genes expression
profiles have been uploaded to databases, including Gene Expression
Omnibus (GEO) and Array Express Archive for researchers to study
(4,5).
Many genes and signaling pathways involved in the metastasis of
ccRCC have been discovered. Downregulation of FOXO3a may promote
tumor metastasis in ccRCC (6). C-X-C
motif chemokine receptor 2 (CXCR2)/CXCR2 ligand biology is
important in the promotion of angiogenesis and facilitation of
tumor growth and metastasis in RCC cells (7). A previous study demonstrated that
overexpression of brain-type fatty-acid-binding protein (FABP) may
lead to the reduction of liver-type FABP in RCC, which serves a
role in cell signaling, regulation of gene expression, cell growth
and differentiation (8). Although the
above researches have identified specific genes associated with
metastasis of ccRCC, the mechanisms of ccRCC metastasis remain
unclear. Furthermore, few drugs have been developed to be effective
for treatment of metastatic ccRCC.
In the present study, in order to achieve an
improved understanding of ccRCC, early metastatic and
non-metastatic ccRCC samples were used to screen DEGs associated
with metastatic ccRCC. Ni et al (6) used P<0.05 as the criterion to screen
DEGs between metastatic and non-metastatic ccRCC samples; using
identical data, the present study screened the DEGs by stricter
cut-off criteria [false discovery rate (FDR) <0.05 and |log fold
change (FC)|>1]. Subsequently, functional and pathway enrichment
analysis was performed to predict the potential functions of DEGs.
Furthermore, a protein-protein interaction (PPI) network was
constructed to analyze the interactions between DEGs. In addition,
small drug molecules associated with ccRCC were detected. It is
anticipated that the results of the present study may lead to a
potential breakthrough in the treatment of metastatic ccRCC.
Materials and methods
Microarray data
The microarray data GSE47352 deposited by Ni et
al (6) was downloaded from the
GEO (http://www.ncbi.nlm.nih.gov/geo/) of
the National Center of Biotechnology Information. In addition,
probes annotation information was also downloaded for mapping the
probes to genes (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47352).
This dataset was generated based on the platform of GPL570
(HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array. A
total of 9 samples are enrolled in the GSE47352 dataset, including
4 early metastatic ccRCC samples (metastatic group) and 5
non-metastatic ccRCC samples (non-metastatic group). The ccRCC
tissue samples were removed from ccRCC patients who underwent
nephrectomy at the Chinese People's Liberation Army General
Hospital between January 2009 and May 2012, and were snap-frozen in
liquid nitrogen. Patients with negative abdomen and chest computed
tomography or magnetic resonance imaging and without metastatic
lesions were classed as non-metastatic ccRCC; patients with
metastatic lesions were classed as early metastatic ccRCC (6).
Data preprocessing and DEGs
screening
Based on the k-Nearest Neighbors method (9), Affymetrix (Affy) package (version
1.28.0; Affymetrix, Inc., Santa Clara, CA, USA) (10) in R language was employed to account
for the missing values in the raw data from the DNA microarray.
Subsequently, the data was normalized by the median normalization
method (11). Compared with the
non-metastatic group, the DEGs in the metastatic group were
screened using the linear model for microarray data (Limma) package
(12). The Benjamini-Hochberg method
(13) was applied to conduct multiple
testing adjustment to identify the FDR and the logFC was also
calculated. Genes with FDR<0.05 and |logFC|>1 were taken as
the DEGs between the early metastatic and non-metastatic
groups.
Comparison of gene expression between
the metastatic and non-metastatic groups
Generally, significant differences in gene
expression are observed in tissues under different disease states
(14). The gene expression values of
DEGs were extracted, and the pheatmap package (15) in R was used to perform two-way
clustering (16) based on Euclidean
distance (17).
Functional and pathway enrichment
analysis
Gene map annotator and pathway profiler (GenMAPP;
version 2.1; http://www.GenMAPP.org) was used for
visualizing, analyzing and demonstrating the microarray data in
pathways (18). The MAPPFinder was
used for coupling the annotations of the Gene Ontology (GO)
database with GenMAPP and calculated the GO-values (19). In the present study, MAPPFinder and
GenMAPP were employed separately to conduct functional and pathway
enrichment analysis for the DEGs. P<0.05 was taken as the
threshold.
PPI network construction
The Search Tool for the Retrieval of Interacting
Genes (STRING) database (http://string-db.org/) provided comprehensive
predicted PPI information (20). The
PPI pairs (combined score >0.6) were screened from the STRING
database, and the PPI network was subsequently visualized using
Cytoscape software (The Cytoscape Consortium, San Diego, CA, USA;
version 2.8; http://www.cytoscape.org) (21).
Screening of small drug molecules
The Connectivity Map (cmap; http://www.broadinstitute.org/CMAP/) database may be
used to investigate connections among small drug molecules, genes
and diseases (22,23). A higher negative score indicates a
higher correlation between the small drug molecules and the DEGs.
The DEGs were imported into cmap to screen the small drug molecules
associated with DEGs. The small drug molecules with |score|>0.8
were recorded.
Results
DEGs screening
According to the microarray data analysis between
early metastatic ccRCC and non-metastatic ccRCC samples by Limma, a
total of 359 DEGs were obtained in metastatic group, including 196
upregulated genes and 163 downregulated genes. The top ten
significantly upregulated (including vomeronasal 1 receptor 2 and
homeobox A1) and downregulated [including epiregulin and RAR
related orphan receptor A (RORA)] genes are listed in Table I.
 | Table I.Top ten up- and downregulated
genes. |
Table I.
Top ten up- and downregulated
genes.
| Downregulated
genes | Upregulated
genes |
|---|
|
|
|---|
| Gene symbol | FDR | LogFC | Gene symbol | FDR | LogFC |
|---|
| EREG | 0.0106464 | −4.63294 | VN1R2 | 0.0060399 | 4.758334 |
| CCDC158 | 0.0009937 | −4.13289 | TSPAN3 | 0.0078284 | 4.667987 |
| HMGCLL1 | 0.0024176 | −4.12305 | KCTD4 | 0.0100570 | 4.158775 |
| TRAF3IP2-AS1 | 0.0000508 | −4.10811 | CECR9 | 0.0089814 | 3.969089 |
| RORA | 0.0056259 | −4.09241 | PCDH20 | 0.0034170 | 3.889183 |
| TMEM51-AS1 | 0.0086847 | −3.87088 | FAM95A | 0.0082171 | 3.883404 |
| LOC645485 | 0.0063110 | −3.86383 | SEZ6L | 0.0078182 | 3.846624 |
| UNC93A | 0.0024362 | −3.85761 | CYLC1 | 0.0010110 | 3.833110 |
| RGPD1 | 0.0034170 | −3.78002 | SLC22A25 | 0.0069844 | 3.821152 |
| FAHD2CP | 0.0009937 | −3.75996 | HOXA1 | 0.0054553 | 3.815357 |
Comparison of gene expression between
metastatic and non-metastatic samples
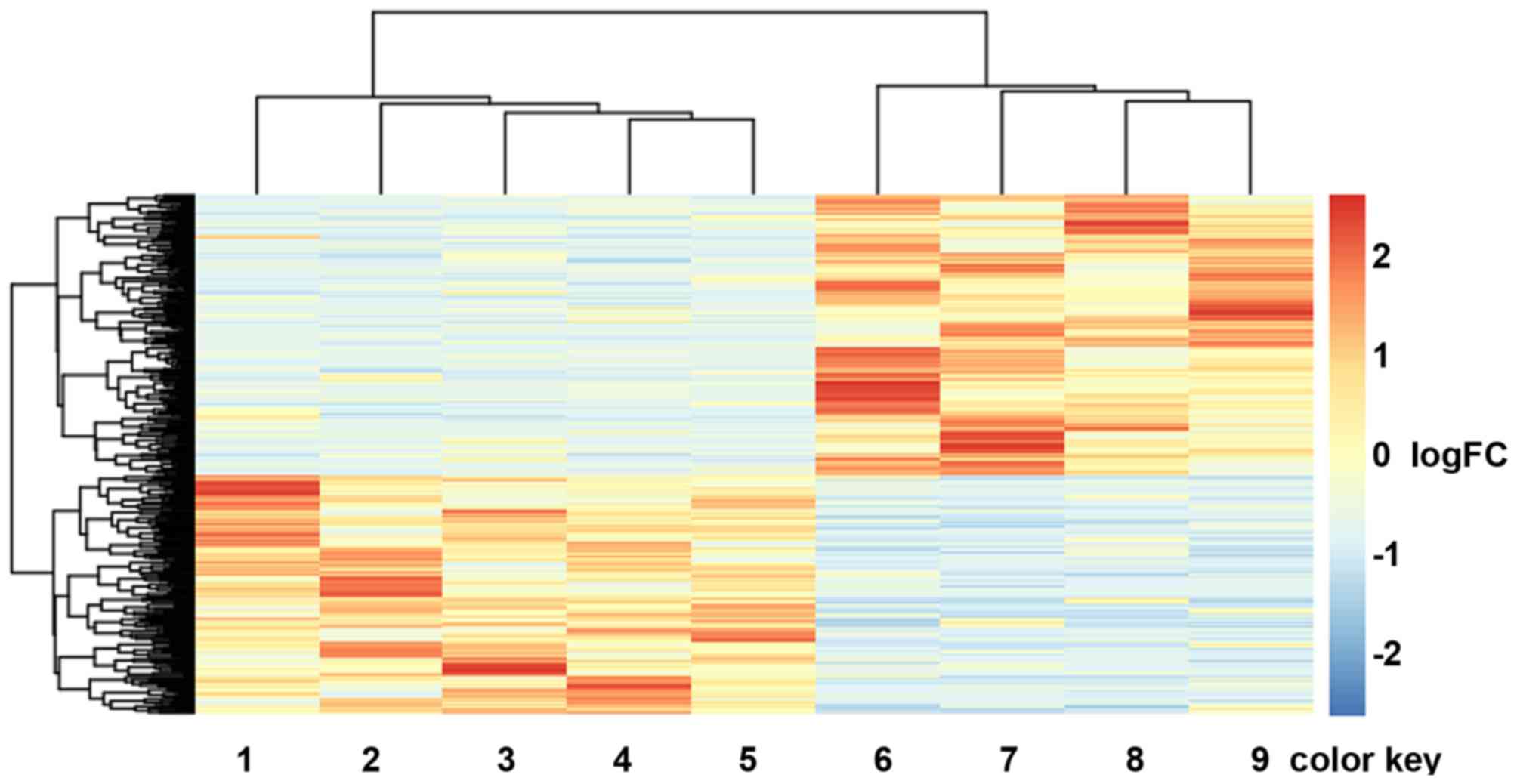
Hierarchical cluster analysis of the expression
values of DEGs revealed that the early metastatic ccRCC samples and
the non-metastatic ccRCC samples were in significantly separated
clusters (Fig. 1).
Functional and pathway enrichment
analysis
A total of five Kyoto Encyclopedia Genes and Genomes
pathways were obtained for the identified DEGs (Table II). The most significantly enriched
pathway was the renal cell carcinoma pathway (P=0.003503), which
involved 6 DEGs [endothelial PAS domain-containing protein 1
(EPAS1), ETS1, JUN, SOS2, TGFβ2, and protein tyrosine phosphatase,
non-receptor type 11 (PTPN11)]. Furthermore, these 6 DEGs were all
downregulated. In addition, 11 DEGs [including TGFβ2, nuclear
receptor subfamily 4 group A member 1 (NR4A1) and dual specificity
protein phosphatase 1 (DUSP1)] significantly participated in the
mitogen-activate protein kinase (MAPK) signaling pathway
(P=0.005407) and 5 DEGs [including laminin subunit α (LAMA) 2,
LAMA1 and LAMA4] were enriched in the extracellular matrix
(ECM)-receptor interaction pathway (P=0.034718).
 | Table II.Enriched pathways for differentially
expressed genes between early metastasis ccRCC and the
non-metastasis ccRCC samples. |
Table II.
Enriched pathways for differentially
expressed genes between early metastasis ccRCC and the
non-metastasis ccRCC samples.
|
|
|
|
| Genes |
|---|
|
|
|
|
|
|
|---|
| ID | Pathways | P-value | Count | Upregulated | Downregulated |
|---|
| hsa05211 | Renal cell
carcinoma | 0.003503 | 6 | – | EPAS1, ETS1, JUN,
TGFβ2, SOS2, PTPN11 |
| hsa04010 | MAPK signaling
pathway | 0.005407 | 11 | CACNA2D1, TNF,
PTPN5, MAPK8IP3, CACNG2 | FGF8, DUSP1, JUN,
SOS2, NR4A1, TGFβ2 |
| hsa05410 | Hypertrophic
cardiomyopathy | 0.008004 | 6 | CACNA2D1, TNF,
SGCD, CACNG2 | TGFβ2 LAMA2 |
| hsa05414 | Dilated
cardiomyopathy | 0.011083 | 6 | CACNA2D1, TNF,
SGCD, CACNG2 | TGFβ2, LAMA2, |
| hsa04512 | ECM-receptor
interaction | 0.034718 | 5 | SV2B | LAMA2, LAMA1,
LAMA4, CD36 |
The top 10 GO terms are listed in Table III, including regulation of
transcription from RNA polymerase II promoter
(P=7.26×10−6), positive regulation of the nucleic acid
metabolic process (P=3.53×10−5) and positive regulation
of the nitrogen compound metabolic process
(P=5.82×10−5). In particular, JUN, EST1, RORA and TGFβ2
were significantly enriched in the majority of the GO terms.
 | Table III.Top ten enriched functions for the
differentially expressed genes. |
Table III.
Top ten enriched functions for the
differentially expressed genes.
| ID | Gene ontology
term | Count | P-value | Genes |
|---|
| GO:0006357 | Regulation of
transcription from RNA polymerase II promoter | 29 |
7.26×10−6 | TNF, FOXK1,
ONECUT2, NR6A1, FOXK2, TP63, RORA, MED20, WT1, HOXA1, NPAS1,
SQSTM1, MED26, HSF4, RARB, DGKQ, KLF9, EPAS1, NR4A1, TEAD2, NR0B1,
IL22, SP2, ETS1, JUN, MNX1, TFAP2E, KLF4, NFIB |
| GO:0045935 | Positive regulation
of nucleic acid metabolic process | 25 |
3.53×10−5 | TNF, FOXK1,
ONECUT2, TP63, ABCA1, RORA, WT1, HOXA1, SQSTM1, H2AFX, RARB, HSF4,
EPAS1, TAF8, ESRRG, NR4A1, TEAD2, IL22, EREG, IRF6, ETS1, JUN,
TFAP2E, KLF4, NFIB |
| GO:0045893 | Positive regulation
of transcription | 21 |
5.04×10−5 | TNF, EPAS1, FOXK1,
TAF8, ONECUT2, ESRRG, TP63, NR4A1, TEAD2, RORA, IL22, WT1, HOXA1,
SQSTM1, ETS1, JUN, RARB, HSF4, TFAP2E, KLF4, NFIB |
| GO:0051254 | Positive regulation
of RNA metabolic process | 21 |
5.72×10−5 | TNF, EPAS1, FOXK1,
TAF8, ONECUT2, ESRRG, TP63, NR4A1, TEAD2, RORA, IL22, WT1, HOXA1,
SQSTM1, ETS1, JUN, RARB, HSF4, TFAP2E, KLF4, NFIB |
| GO:0051173 | Positive regulation
of nitrogen compound metabolic process | 25 |
5.82×10−5 | TNF, FOXK1,
ONECUT2, TP63, ABCA1, RORA, WT1, HOXA1, SQSTM1, H2AFX, RARB, HSF4,
EPAS1, TAF8, ESRRG, NR4A1, TEAD2, IL22, EREG, IRF6, ETS1, JUN,
TFAP2E, KLF4, NFIB |
| GO:0045944 | Positive regulation
of transcription from RNA polymerase II promoter | 18 |
6.37×10−5 | TNF, EPAS1,
ONECUT2, TP63, NR4A1, TEAD2, RORA, IL22, WT1, HOXA1, SQSTM1, ETS1,
JUN, RARB, HSF4, TFAP2E, KLF4, NFIB |
| GO:0009891 | Positive regulation
of biosynthetic process | 26 |
7.16×10−5 | TNF, FOXK1,
ONECUT2, TP63, APOC2, ABCA1, RORA, WT1, TGFβ2, HOXA1, SQSTM1, RARB,
HSF4, EPAS1, TAF8, ESRRG, NR4A1, TEAD2, IL22, EREG, IRF6, ETS1,
JUN, TFAP2E, KLF4, NFIB |
| GO:0031328 | Positive regulation
of cellular biosynthetic process | 25 |
1.51s10−4 | TNF, FOXK1,
ONECUT2, TP63, APOC2, ABCA1, RORA, WT1, HOXA1, SQSTM1, RARB, HSF4,
EPAS1, TAF8, ESRRG, NR4A1, TEAD2, IL22, EREG, IRF6, ETS1, JUN,
TFAP2E, KLF4, NFIB |
| GO:0010557 | Positive regulation
of macromolecule biosynthetic process | 24 |
1.97×10−4 | TNF, EPAS1, FOXK1,
TAF8, ONECUT2, ESRRG, TP63, NR4A1, TEAD2, RORA, IL22, WT1, TGFβ2,
HOXA1, EREG, IRF6, SQSTM1, ETS1, JUN, RARB, HSF4, TFAP2E, KLF4,
NFIB |
| GO:0010628 | Positive regulation
of gene expression | 22 |
2.62×10−4 | TNF, EPAS1, FOXK1,
TAF8, ONECUT2, ESRRG, TP63, NR4A1, TEAD2, RORA, IL22, WT1, HOXA1,
IRF6, SQSTM1, ETS1, JUN, RARB, HSF4, TFAP2E, KLF4, NFIB |
PPI network construction
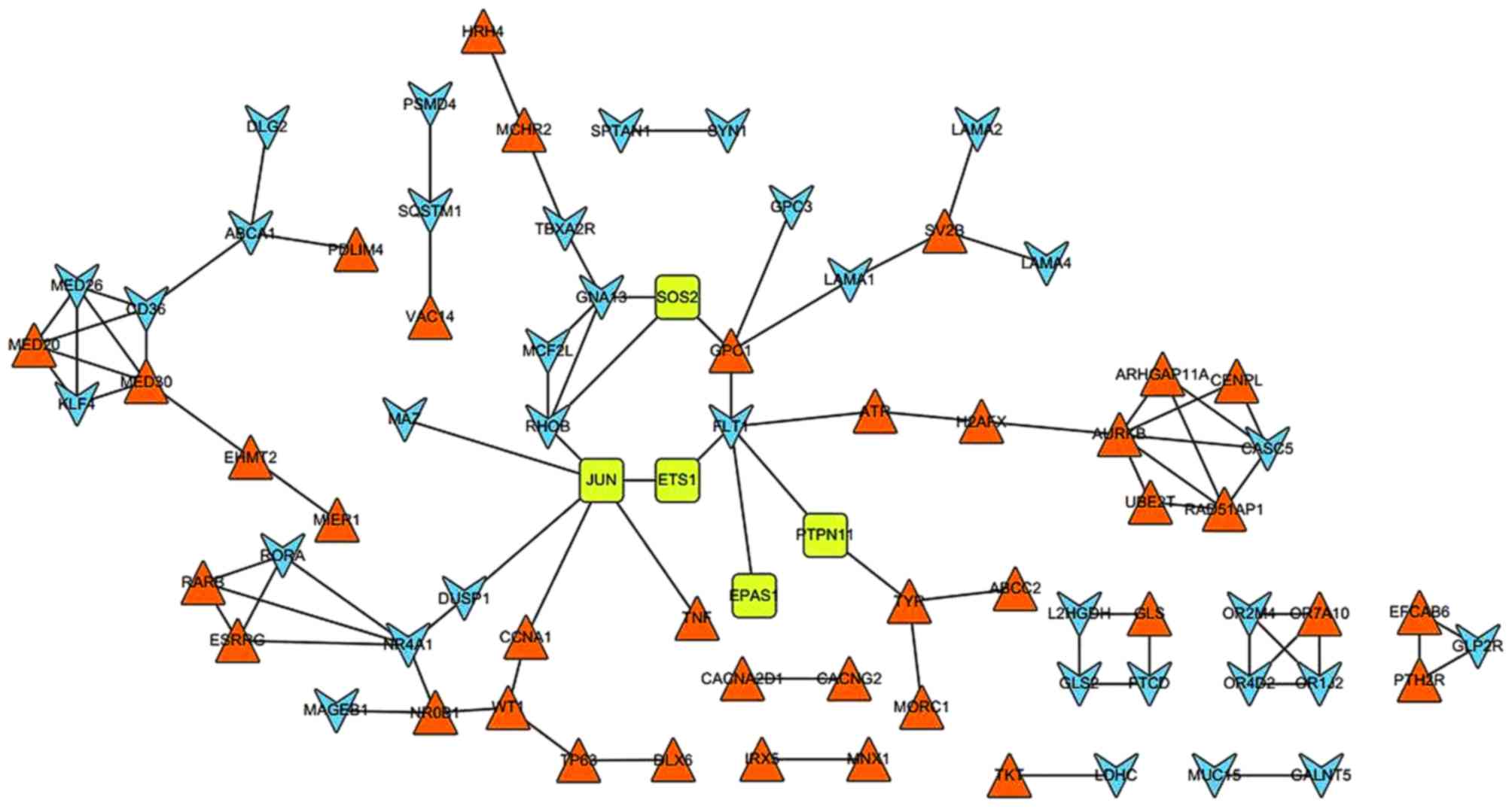
In total, 87 PPI pairs were obtained from the STRING
database. Subsequently, the six DEGs (EPAS1, ETS1, JUN, SOS2, TGFβ2
and PTPN11) that were enriched in the renal cell carcinoma pathway
were mapped to the network. The network was visualized using
Cytoscape (Fig. 2). In the PPI
network, JUN possessed the highest degree of 6; additionally,
ferritin light chain 1, NR4A1 and Ras homolog family member B
(RHOB) demonstrated degrees of 5, 5 and 4, respectively.
Furthermore, JUN could interact with ever shorter telomeres protein
1 (EST1), RHOB, DUSP1, tumor necrosis factor (TNF), MYC associated
zinc finger protein (MAZ) and cyclin A1 (CCNA1). In addition, NR4A1
demonstrated an interaction with DUSP1.
Small drug molecule screening
For the screening of small molecular drugs, 7 small
drug molecules with the |score|>0.8, including 4
negatively-correlated drugs (thapsigargin, score=−0.913; W-13,
score=−0.885; trihexyphenidyl, score=−0.839; and lovastatin,
score=−0.824) and 3 positively-correlated drugs (dioxybenzone,
score=0.825; oxybuprocaine, score=0.853; and (−)-MK-801,
score=0.887) were identified to be correlated with the DEGs
(Table IV).
 | Table IV.Small molecule drugs (|score|>0.8)
associated with the differentially expressed genes between the
primary metastatic ccRCC and the non-metastatic ccRCC samples. |
Table IV.
Small molecule drugs (|score|>0.8)
associated with the differentially expressed genes between the
primary metastatic ccRCC and the non-metastatic ccRCC samples.
| Connectivity map
name | Score | P-value |
|---|
| Thapsigargin | −0.913 | 0.00112 |
| W-13 | −0.885 | 0.02630 |
|
Trihexyphenidyl | −0.839 | 0.00839 |
| Lovastatin | −0.824 | 0.00181 |
| Dioxybenzone | 0.825 | 0.00149 |
| Oxybuprocaine | 0.853 | 0.00070 |
| (−)-MK-801 | 0.887 | 0.00016 |
Discussion
In the present study, with the investigation of the
gene expression profile between the early metastatic and
non-metastatic ccRCC using bioinformatics methods, a total of 359
DEGs were obtained, including 196 upregulated DEGs and 163
downregulated DEGs. Hierarchical cluster analysis indicated that
the metastatic ccRCC samples could be well distinguished from the
non-metastatic ccRCC samples according to the identified DEGs.
Furthermore, pathway enrichment analysis revealed that JUN was
significantly enriched in renal cell carcinoma and the MAPK
signaling pathway. Previous research has proven that MAPK serves a
key role in tumor metastasis via regulating cell migration and
apoptosis (24). Furthermore, in the
PPI network, JUN was a hub node with the highest degree of 6 and
could interact with RHOB, MAZ, DUSP1, CCNA1, TNF and EST1. JUN is
identified as oncogene, which accelerates tumor cell metastasis
(25). Zhang et al (26) demonstrated that JUN has a close
association with metastasis of cancer cells, and overexpression of
JUN may result in metastasis of breast cancer. A previous study
demonstrated that epithelial-mesenchymal transition (EMT) is a key
regulator of metastasis in cancer by conferring an invasive
phenotype via the loss of cell-cell adhesions, cell-substrates and
transition to a cell type that is capable of invading the ECM
(27). Furthermore, EMT has been
identified as a model by which the ccRCC occurs (28). In the present study, TNF and RHOB were
significantly enriched in the pathway of early metastatic ccRCC.
Previous studies have illustrated that TNF (or TNF-α) is able to
elevate the migration and invasion of ccRCC cells together with
downregulation of E-cadherin expression and promotion of EMT,
suggesting that TNF has a close association with early metastatic
ccRCC (29). RHOB is known as a tumor
suppressor and is able to affect cell adhesion and migration by
regulating surface integrin levels (30). Furthermore, it has also been observed
that RHOB serves a distinct function in EMT by regulating cell-cell
and cell- substrate contact in renal proximal tubular cells,
suggesting that RHOB has a key role in early metastatic ccRCC
(31). This appears to indicate that
JUN, along with the interaction with TNF and RHOB, may participate
in mediation of early metastatic ccRCC via regulation of cell
migration and apoptosis.
In addition, NR4A1 and TGFβ2 were significantly
enriched in the MAPK signaling pathway. Previous research has
demonstrated that the MAPK signaling pathway is involved in
inhibition of tumorigenesis, metastasis and angiogenesis in RCC via
the disruption of tumor vasculature (32). NR4A1, which belongs to the Nur nuclear
receptor family, has been implicated in cell cycle regulation,
inflammation and apoptosis (33). It
has also been reported that NR4A1 is able to promote the invasion
and metastasis of breast cancer by activating TGFβ signaling
(34). Furthermore, the loss of NR4A1
may enhance macrophage-mediated kidney injury and diseases due to a
large increase in immune cell infiltration (predominantly
macrophages, and to a lesser extent T cells and B cells) (35). Therefore, the authors hypothesized
that NR4A1 had a close association with early metastatic ccRCC.
TGFβ2, which belongs to the TGFβ family, is known to promote the
invasion of tumor cells and allow metastasis to distant organs via
induction of EMT, suppression of immune surveillance, promotion of
angiogenesis and recruitment of inflammatory cells in human cancer
cell lines and mouse tumor models (36,37).
Consequently, the present study speculated that NR4A1 and TGFβ2 may
serve a key role in the regulation of early metastatic ccRCC
through the MAPK signaling pathway.
LAMA1, LAMA2 and LAMA4, which belong to the laminins
family, were significantly enriched in the pathway of ECM-receptor
interaction. Laminins, a family of ECM glycoproteins, are the major
non-collagenous constituent of basement membranes (38). Laminins act as ECM fibers in lymph
nodes, within which tumor cell metastasis occurs (39). A previous study has also reported that
LAMA4 has a de-adhesive function and may serve a key role in
detachment, migration and invasion of renal carcinoma cells in
vivo (40). Therefore, LAMA1,
LAMA2 and LAMA4 may be potential target genes in the treatment of
early metastatic ccRCC.
Thapsigargin was discovered to have high efficiency
for the treatment of early metastasis in ccRCC. Thapsigargin is a
non-competitive inhibitor of sarco/endoplasmic reticulum
Ca2+ ATPase (41).
Thapsigargin is able to couple to a peptide carrier, producing a
soluble non-toxic pro-drug, which induces apoptosis of prostate
cancer cells (42). Research into the
use of thapsigargin as a small drug molecule for cancer treatment
has increased, including for the treatment of lung adenocarcinoma
and prostate cancer (42,43). Due to its positive effects on prostate
cancer and lung adenocarcinoma, the present authors speculated that
it may be also effective for the treatment of early metastatic
ccRCC.
In conclusion, a total of 359 DEGs were identified
in the metastatic group compared with the non-metastatic group.
Furthermore, the DEGs, including TGFβ2, JUN, NR4A1, RHOB, LAMA1,
LAMA2 and LAMA4, were involved in early metastatic ccRCC. In
addition, the present study screened a small drug molecule named
thapsigargin, which may have high efficiency for the treatment of
the early metastatic ccRCC. However, further studies to investigate
the viability of the above assumptions are required and additional
experimental research is needed to validate the results of the
present study, as well as confirm thapsigargin's safety and
efficacy for the treatment of early metastatic ccRCC.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rosner I, Bratslavsky G, Pinto PA and
Linehan WM: The clinical implications of the genetics of renal cell
carcinoma. Urol Oncol. 27:131–136. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Novara G, Ficarra V, Antonelli A, Artibani
W, Bertini R, Carini M, Cunico S Cosciani, Imbimbo C, Longo N, et
al: Validation of the 2009 TNM version in a large
multi-institutional cohort of patients treated for renal cell
carcinoma: Are further improvements needed? Eur Urol. 58:588–595.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Barrett T, Troup DB, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, et al: NCBI GEO: Archive for functional genomics data
sets-10 years on. Nucleic Acids Res. 39(Database Issue):
D1005–D1010. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Parkinson H, Kapushesky M, Shojatalab M,
Abeygunawardena N, Coulson R, Farne A, Holloway E, Kolesnykov N,
Lilja P, Lukk M, et al: ArrayExpress-a public database of
microarray experiments and gene expression profiles. Nucleic Acids
Res. 35(Database issue): D747–D750. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ni D, Ma X, Li HZ, Gao Y, Li XT, Zhang Y,
Ai Q, Zhang P, Song EL, Huang QB, et al: Downregulation of FOXO3a
promotes tumor metastasis and is associated with metastasis-free
survival of patients with clear cell renal cell carcinoma. Clin
Cancer Res. 20:1779–1790. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mestas J, Burdick MD, Reckamp K, Pantuck
A, Figlin RA and Strieter RM: The role of CXCR2/CXCR2 ligand
biological axis in renal cell carcinoma. J Immunol. 175:5351–5357.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tölle A, Jung M, Lein M, Johannsen M,
Miller K, Moch H, Jung K and Kristiansen G: Brain-type and
liver-type fatty acid-binding proteins: New tumor markers for renal
cancer? BMC Cancer. 9:2482009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang ML and Zhou ZH: A k-nearest neighbor
based algorithm for multi-label classification. Journal. 2:718–721.
2005.
|
|
10
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rao Y, Lee Y, Jarjoura D, Ruppert AS, Liu
CG, Hsu JC and Hagan JP: A comparison of normalization techniques
for microRNA microarray data. Stat Appl Genet Mol Biol. 7::
2008.PubMed/NCBI
|
|
12
|
Gentleman R, Carey VJ, Huber W, Irizarry
RA and Dudoit S: Bioinformatics and computational biology solutions
using R and Bioconductor. Springer; 2005, View Article : Google Scholar
|
|
13
|
Genovese C and Wasserman L: Operating
characteristics and extensions of the false discovery rate
procedure. J Royal Statistical Soci Series B (Statistical
Methodology). 64:499–517. 2002. View Article : Google Scholar
|
|
14
|
Ester M, Kriegel HP, Sander J and Xu X: A
density-based algorithm for discovering clusters in large spatial
databases with noise. Journal. 96:226–231. 1996.
|
|
15
|
Kolde R: Pheatmap: Pretty Heatmaps. R
Package Version 0.6.1. Journal. 2012.
|
|
16
|
Szekely GJ and Rizzo ML: Hierarchical
clustering via joint between-within distances: Extending Ward's
minimum variance method. J Classification. 22:151–183. 2005.
View Article : Google Scholar
|
|
17
|
Deza MM and Deza E: Encyclopedia of
Distances. Springer; 2009, View Article : Google Scholar
|
|
18
|
Dahlquist KD, Salomonis N, Vranizan K,
Lawlor SC and Conklin BR: GenMAPP, a new tool for viewing and
analyzing microarray data on biological pathways. Nat Genet.
31:19–20. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Doniger SW, Salomonis N, Dahlquist KD,
Vranizan K, Lawlor SC and Conklin BR: MAPPFinder: Using gene
ontology and GenMAPP to create a global gene-expression profile
from microarray data. Genome Biol. 4:R72003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39(Database issue): D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lamb J: The Connectivity Map: A new tool
for biomedical research. Nat Rev Cancer. 7:54–60. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lamb J, Crawford ED, Peck D, Modell JW,
Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et
al: The connectivity map: Using gene-expression signatures to
connect small molecules, genes and disease. Science. 313:1929–1935.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Reddy KB, Nabha SM and Atanaskova N: Role
of MAP kinase in tumor progression and invasion. Cancer Metastasis
Rev. 22:395–403. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chaturvedi MM, Sung B, Yadav VR, Kannappan
R and Aggarwal BB: NF-κB addiction and its role in cancer: ‘One
size does not fit all’. Oncogene. 30:1615–1630. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang Y, Pu X, Shi M, Chen L, Song Y, Qian
L, Yuan G, Zhang H, Yu M, Hu M, et al: Critical role of c-Jun
overexpression in liver metastasis of human breast cancer xenograft
model. BMC Cancer. 7:1452007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Davis FM, Stewart TA, Thompson EW and
Monteith GR: Targeting EMT in cancer: Opportunities for
pharmacological intervention. Trends Pharmacol Sci. 35:479–488.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tun HW, Marlow LA, Von Roemeling CA,
Cooper SJ, Kreinest P, Wu K, Luxon BA, Sinha M, Anastasiadis PZ and
Copland JA: Pathway signature and cellular differentiation in clear
cell renal cell carcinoma. PLoS One. 5:e106962010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mikami S, Mizuno R, Kosaka T, Saya H, Oya
M and Okada Y: Expression of TNF-α and CD44 is implicated in poor
prognosis, cancer cell invasion, metastasis and resistance to the
sunitinib treatment in clear cell renal cell carcinomas. Int J
Cancer. 136:1504–1514. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wheeler AP and Ridley AJ: RhoB affects
macrophage adhesion, integrin expression and migration. Exp Cell
Res. 313:3505–3516. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hutchison N, Hendry BM and Sharpe CC: Rho
isoforms have distinct and specific functions in the process of
epithelial to mesenchymal transition in renal proximal tubular
cells. Cell Signal. 21:1522–1531. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang D, Ding Y, Luo WM, Bender S, Qian
CN, Kort E, Zhang ZF, VandenBeldt K, Duesbery NS, Resau JH and Teh
BT: Inhibition of MAPK kinase signaling pathways suppressed renal
cell carcinoma growth and angiogenesis in vivo. Cancer Res.
68:81–88. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pei L, Castrillo A and Tontonoz P:
Regulation of macrophage inflammatory gene expression by the orphan
nuclear receptor Nur77. Mol Endocrinol. 20:786–794. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou F, Drabsch Y, Dekker TJ, de Vinuesa
AG, Li Y, Hawinkels LJ, Sheppard KA, Goumans MJ, Luwor RB, de Vries
CJ, et al: Nuclear receptor NR4A1 promotes breast cancer invasion
and metastasis by activating TGF-β signalling. Nat Commun.
5:33882014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Westbrook L, Johnson AC, Regner KR,
Williams JM, Mattson DL, Kyle PB, Henegar JR and Garrett MR:
Genetic susceptibility and loss of Nr4a1 enhances
macrophage-mediated renal injury in CKD. J Am Soc Nephrol.
25:2499–2510. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Janda E, Lehmann K, Killisch I, Jechlinger
M, Herzig M, Downward J, Beug H and Grünert S: Ras and TGF[beta]
cooperatively regulate epithelial cell plasticity and metastasis
Dissection of Ras signaling pathways. J Cell Biol. 156:299–313.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Heldin C-H, Vanlandewijck M and Moustakas
A: Regulation of EMT by TGFβ in cancer. FEBS Lett. 586:1959–1970.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hohenester E and Yurchenco PD: Laminins in
basement membrane assembly. Cell Adh Migr. 7:56–63. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Patarroyo M, Tryggvason K and Virtanen I:
Laminin isoforms in tumor invasion, angiogenesis and metastasis.
Semin Cancer Biol. 12:197–207. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Vainionpää N, Lehto VP, Tryggvason K and
Virtanen I: Alpha4 chain laminins are widely expressed in renal
cell carcinomas and have a de-adhesive function. Lab Invest.
87:780–791. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Rogers TB, Inesi G, Wade R and Lederer W:
Use of thapsigargin to study Ca2+ homeostasis in cardiac cells.
Biosci Rep. 15:341–349. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Denmeade SR, Jakobsen CM, Janssen S, Khan
SR, Garrett ES, Lilja H, Christensen SB and Isaacs JT:
Prostate-specific antigen-activated thapsigargin prodrug as
targeted therapy for prostate cancer. J Natl Cancer Inst.
95:990–1000. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Doan NT, Paulsen ES, Sehgal P, Møller JV,
Nissen P, Denmeade SR, Isaacs JT, Dionne CA and Christensen SB:
Targeting thapsigargin towards tumors. Steroids. 2014.PubMed/NCBI
|