Introduction
Prostate cancer (PCa) is the second most common type
of cancer affecting men worldwide, with an incidence of 1.1 million
new cases each year (1). The most
widely accepted pathological grading of PCa is the Gleason Score
(GS), which is based on architectural features of the gland and is
comprised of the sum of two Gleason Patterns (GP) (2). The GS grading system is among the most
effective predictors of clinical outcomes following radical
prostatectomy (RP), and is universally used to guide PCa treatment
(3). Within this system, GS6 PCas are
low grade, whereas GS8 or higher are classified as high grade. GS7
prostate adenocarcinomas consisting of variable proportions of GP3
and GP4 are considered to be intermediate grade, and represent the
most heterogeneous group of neoplasms with diverse clinical
outcomes (4,5) Thus, GS7 PCa is the focus of the current
study, as identifying specific prognostic factors that can provide
additional information on disease progression and treatment
outcomes for patients with GS7 PCa would be of marked clinical
benefit.
GP4 is assigned to adenocarcinomas when they contain
any of the following architectural patterns: Small/large fused
glands, poorly formed glands, glomeruloid and cribriform
architecture (4–6). The cribriform pattern is commonly
associated with intraductal carcinoma (IDC), a distinct
histopathological entity characterized by malignant cells spanning
the lumen of prostate ducts and acini (7–9). Although
specific morphologies of PCa, including cribriform architecture and
IDC, have been recognized for decades, their independent clinical
significance is only now emerging (8). Studies have demonstrated that IDC in
biopsy and/or RP tissues is indicative of a possible adverse
clinical course and metastatic disease, warranting further
aggressive treatment (10–13). Similarly, cribriform architecture has
been demonstrated to be a clinically significant independent
prognosticator for PCa-specific mortality (14–17).
The unequivocal identification of these pathological
features based on the morphological criteria alone has been
reported as challenging (18,19). In this regard, molecular features such
as DNA methylation alterations may be of value. Aberrations in DNA
methylation deregulate the genome and contribute to the loss of
tissue homeostasis observed in aging and in diseases such as PCa
(20). They may constitute the driver
and the passenger events of tumorigenesis, as is evident from the
widespread hypermethylation at the promoter regions of tumor
suppressor genes, which is a hallmark of PCa (20,21).
Detection of tumor-specific DNA methylation alterations in biopsy
tissues may, in the future, serve as molecular indicators of
cribriform architecture and/or of IDC. Additionally, DNA
methylation markers are emerging as promising biomarkers of
prostate carcinogenesis, thus complementing, if not altogether
avoiding, the requirement for the histopathological examination of
biopsy tissue samples (22). The
results of our previous studies demonstrated that the DNA
hypermethylation of a panel of seven genes [adenomatous polyposis
coli (APC), cytochrome P450 family 26 subfamily A member 1
(CYP26A1), homeobox D3 (HOXD3), HOXD8, Ras
association domain family member 1 (RASSF1), T-box 15
(TBX15) and transforming growth factor-β2 (TGFβ2)] is
associated with PCa disease progression and clinical outcome
(23–25). These markers were initially identified
through a genome-wide methylation screen of low-grade and
high-grade PCas (23). Subsequently,
the prognostic potential of a subset of these markers was validated
in an independent cohort (24,25).
However, to the best of our knowledge, their association with the
tumor architectural features of aggressive PCa has not yet been
investigated.
In the present study, cribriform architecture and
IDC were characterized in GP4 tumors derived from GS7 RP specimens,
and their association with the aforementioned panel of seven DNA
methylation markers was examined.
Materials and methods
Clinical samples and information
The present study was approved by the University
Health Network Research Ethics Board. Retrospective formalin-fixed
paraffin-embedded (FFPE) RP specimens from a total of 91 patients
diagnosed with PCa between 1998 and 2001 at the University Health
Network were included in the current study. This is a subset of a
larger cohort of 246 patients (consisting of 91 GS7 tumors) that
was characterized in our previous study (17). As previously reported, GS ≥8 tumors
were limited in the present cohort and, thus, were excluded from
analysis (17). The clinical and
pathological characteristics of the cohort are listed in Table I. All samples, as well as clinical and
pathological follow-up data, were obtained according to protocols
approved by the Research Ethics Board of Mount Sinai Hospital,
Toronto, and the University Health Network (Toronto, Canada).
 | Table I.Clinicopathological characteristics
of the study cohort. |
Table I.
Clinicopathological characteristics
of the study cohort.
| Clinicopathological
characteristics | Value (range) |
|---|
| Total number of
patients | 91 |
| Gleason
scorea, no. of
patients |
|
| 7
(3+4) | 63 |
| 7
(4+3) | 28 |
| Pathological stage,
no. of patients |
|
|
pT2 | 45 |
|
pT3a | 34 |
|
pT3b | 11 |
|
pT4 | 1 |
| Surgical margin
status, no. of patients |
|
|
Negative | 65 |
|
Positive | 26 |
| Average
preoperative PSAb | 8.56
(2.51–32.70) |
| Average prostate
weight, gramsb | 50.3
(22.0–143.5) |
| Median age,
years | 63.4
(47.6–74.5) |
Histological evaluation
The complete set of hematoxylin and eosin
(H&E)-stained slides from each RP tissue sample was collected
and reviewed by an expert genitourinary pathologist in order to
assign a modified GS (International Society of Urological Pathology
2005), along with the pathological stage, prostate weight and
surgical margin status (4). For each
GS7 PCa, the most representative GP4 tumor regions were marked on
the H&E-stained slides corresponding to an area of ≥80%
neoplastic cellularity, and the cells isolated from these regions
were used for DNA methylation analysis. Only pure GP4 tissue was
analyzed. The same marked areas on the H&E-stained slides were
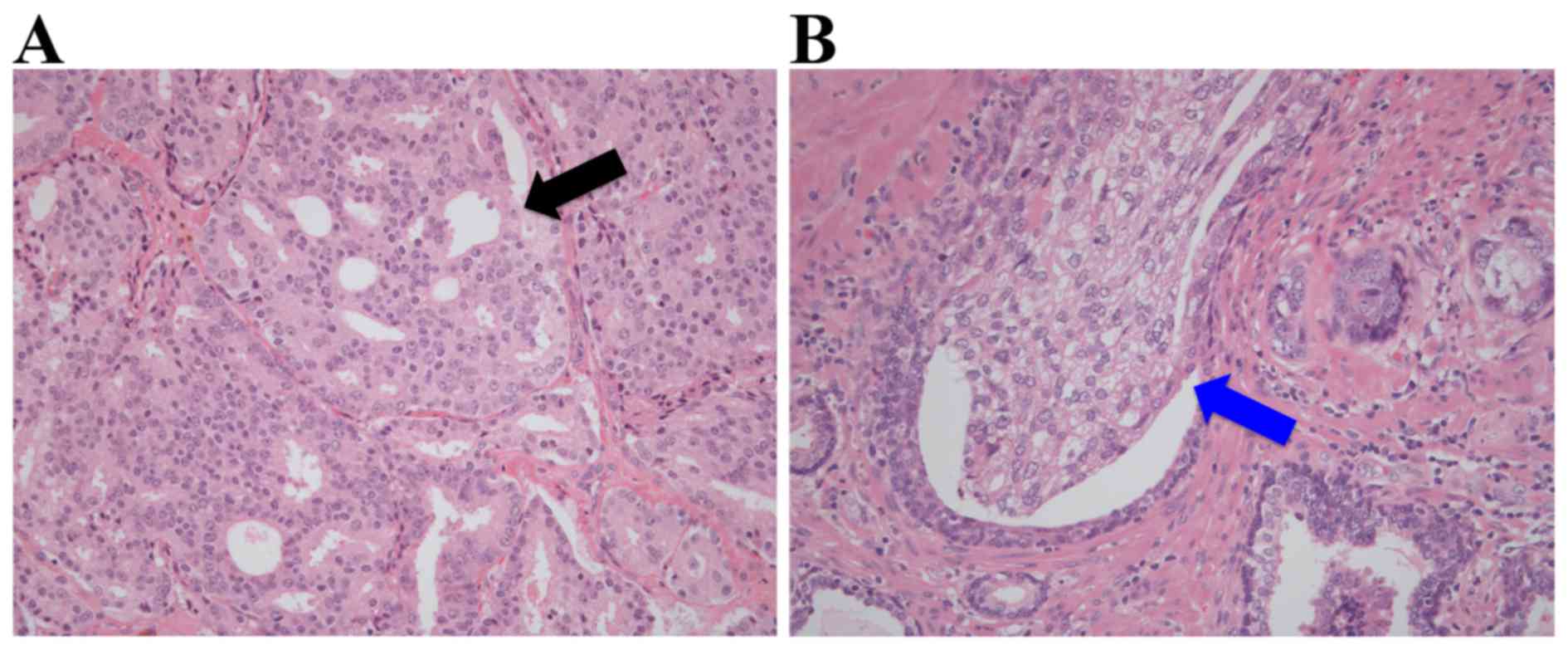
also independently reviewed by another genitourinary pathologist
for the presence or absence of cribriform architecture, which is
characterized by confluent epithelial proliferation with multiple
lumina without intervening stroma, and of IDC, which is defined as
a lumen-spanning solid or cribriform expansile neoplastic
proliferation within the prostate gland or ducts (Fig. 1). Therefore, the presence of
cribriform architecture and IDC in the same GP4 area was
investigated, an area that was manually macrodissected using a
scalpel for subsequent DNA methylation analysis, as described
previously (23–25). All GP4 lesions were determined to
contain an equal tumor cell percentage within the selected regions
of analysis.
DNA methylation analysis
DNA was extracted from 10 µm sections of previously
prepared FFPE blocks matching the selected H&E slides using a
QIAamp DNA Mini kit (Qiagen, Inc., Valencia, CA, USA), as
previously described (23–25). DNA methylation was analyzed in the
same GP4 area used for histological evaluation. Briefly, a total of
100–400 ng extracted DNA from the GP4 region for each GS7 case was
bisulfite modified using EZ DNA Methylation Gold kit (Zymo Research
Corp, Irvine, CA, USA), according to the manufacturer's
protocol.
Methylated genes were detected using a MethyLight
quantitative polymerase chain reaction (qPCR) assay that was
conducted using the TaqMan® Gold with Buffer A pack
(Thermo Fisher Scientific, Inc., Waltham, MA, USA), as described
previously (23–25). The primer and probe sequences are
presented in Table II. A percent
methylation ratio (PMR) score was calculated to assess the
methylation of each gene in GP4 separately for individual cases, as
described previously, according to the formula of Eads et al
(26): (Gene of Interest/ALU)
sample/(Gene of Interest/ALU) CpGenome ×100%. CpGenome is a
commercially available fully methylated DNA (EMD Millipore,
Billerica, MA, USA). ALU was a control reaction that
measured the level of input DNA and normalized the signal for each
methylation reaction (27).
 | Table II.Primer and probe sequences used in
the MethyLight assay for APC, CYP26A1, HOXD3, HOXD8, RASSF1,
TBX15, TGF-β genes and ALU control reaction. |
Table II.
Primer and probe sequences used in
the MethyLight assay for APC, CYP26A1, HOXD3, HOXD8, RASSF1,
TBX15, TGF-β genes and ALU control reaction.
| Genea,b | Sequence |
|---|
| ALU | Forward:
5′-GGTTAGGTATAGTGGTTTATATTTGTAATTTTAGTA-3′ |
|
| Reverse:
5′-ATTAACTAAACTAATCTTAAACTCCTAACCTCA-3′ |
|
| Probe:
5′FAM-CCTACCTTAACCTCCC-3′BHQ1 |
| APC | Forward:
5′-GAACCAAAACGCTCCCCAT-3′ |
|
ENSG000001349821 | Reverse:
5′-TTATATGTCGGTTACGTGCGTTTATAT-3′ |
| Chr5:
112737781-1127378562 | Probe:
5′FAM-CCCGTCGAAAACCCGCCGATTA-3′BHQ1 |
| CYP26A1 | Forward:
5′-TTGTAGAGATTCGACGTACGCGG-3′ |
|
ENSG00000095596 | Reverse:
5′-AAAACCTTCCGTCAAACATCCTCTACG-3′ |
| Chr10:
93069103-93069230 | Probe:
5′FAM-ACGCCCACGACGTACCCGCTTCCTTAC-3′BHQ1 |
| HOXD3 | Forward:
5′-TTAAAGGTTTATGGTTGCGC-3′ |
|
ENSG00000128652 | Reverse:
5′-TTACGAACACTAAACTACACCCG-3′ |
| Chr2:
176163186-176163274 | Probe:
5′FAM-ACAAAACGTTCCCGACGCTTCTAAAA-3′BHQ1 |
| HOXD8 | Forward:
5′-TAGTCGGTTTTGGTTCGTTGC-3′ |
|
ENSG00000175879 | Reverse:
5′-CGTTCTAAAACGAAAAAAAAAACTCGCG-3′ |
| Chr2:
176128934-176129040 | Probe:
5′FAM-TCCTCGAACAAAACGCGACTCCCGAATCTC-3′BHQ1 |
| RASSF1 | Forward:
5′-ATTGAGTTGCGGGAGTTGGT-3′ |
|
ENSG00000068028 | Reverse:
5′-ACACGCTCCAACCGAATACG-3′ |
| Chr3:
50340720-503407843 | Probe:
5′FAM-CCCTTCCCAACGCGCCCA-3′BHQ1c |
| TBX15 | Forward:
5′-GCGGTTTTGTAAGTATATTGTTGCG-3′ |
|
ENSG00000092607 | Reverse:
5′-ACTCCGAATAAAACAAAAACTAAAATCCG-3′ |
| Chr1:
118984438-118984535 | Probe: 5′FAM -
CAAATAACGCCGCCGAACGCCT-3′BHQ1 |
| TGF-β | Forward:
5′-TTTTAGGAGAAGGCGAGTCG-3′ |
|
ENSG00000092969 | Reverse:
5′-CTCCTTAACGTAATACTCTTCGTCG-3′ |
| Chr1:
218346933-218347007 | Probe:
5′FAM-TCTCGCGCTCGCAAACGACC-3′ |
Statistical analysis
The Mann-Whitney U test was performed to analyze the
differences in the median continuous PMR across different
clinicopathological categories. Pearson's Chi square test was used
to analyze proportional differences in Cribriform and/or IDC status
and clinicopathological categories. All statistical analyses were
conducted using SPSS version 21 software (SPSS, IBM Software,
Armonk, NY, USA). For all stated methods, a two-sided value of
P≤0.05 was considered to indicate a statistically significant
difference.
Results
Clinicopathological features
associated with GP4 cribriform architecture and/or IDC
The gland morphology of 91 GP4 tumor specimens
obtained from RPs with GS7 pathology composed of comparable
proportion (≥80%) of tumor cells was examined in the current study.
The presence of a cribriform growth pattern was identified in the
GP4 areas of 61/91 (67.0%) GS7 cases, whereas IDC was present in
21/91 (23.0%) GS7 cases. All 21 RP tissue specimens with IDC were
also positive for cribriform architecture. Notably, the proportion
of cases positive for cribriform architecture and/or IDC did not
significantly differ between GS7 cases composed predominantly of
GP4 (GP4+3; where the proportion of GP4 is >50% of the total
pattern seen) vs. predominantly GP3 (GP3+4); however cribriform
and/or IDC positive status was significantly associated with a more
advanced pathological stage (pT3 vs. pT2; P=0.007; Table III) and higher preoperative
prostate-specific antigen levels (PSA; P=0.022; Table III).
 | Table III.Association between cribriform and/or
IDC-positive (n=61) and negative (n=30) cases and
clinicopathological features. |
Table III.
Association between cribriform and/or
IDC-positive (n=61) and negative (n=30) cases and
clinicopathological features.
|
| Cribriform and/or
IDC |
|
|---|
|
|
|
|
|---|
| Parameter | Negative | Positive | P-value |
|---|
| Gleason score, %
cases |
|
|
|
| 7
(3+4) | 34.9 | 65.1 | 0.552a |
| 7
(4+3) | 28.6 | 71.4 |
|
| Pathological stage,
% cases |
|
|
|
|
pT2 | 46.7 | 53.3 | 0.007a |
|
pT3 | 20.0 | 80.0 |
|
| Surgical margins, %
cases |
|
|
|
|
Negative | 30.8 | 69.2 | 0.481a |
|
Positive | 38.5 | 61.5 |
|
| Age, % cases |
|
|
|
| Below
median | 32.6 | 67.4 | 0.941a |
| Above
median | 33.3 | 66.7 |
|
| Preoperative PSA, %
cases |
|
|
|
| Below
median | 45.5 | 54.5 | 0.022a |
| Above
median | 22.0 | 78.0 |
|
| Prostate weight, %
cases |
|
|
|
| Below
median | 38.1 | 61.9 | 0.542a |
| Above
median | 31.8 | 68.2 |
|
| Median PMR |
|
|
|
|
APC | 31.7 | 47.3 | 0.045b |
|
HOXD3 | 25.8 | 27.1 | 0.888b |
|
HOXD8 | 25.7 | 27.7 | 0.278b |
|
CYP26A1 | 16.3 | 18.5 | 0.426b |
|
RASSF1 | 69.5 | 99.2 | 0.007b |
|
TBX15 | 10.0 | 21.6 | 0.013b |
|
TGFβ2 |
0.0 |
0.0 | 0.287b |
DNA hypermethylation in areas with GP4
cribriform architecture and/or IDC
Gene specific DNA methylation differences were
investigated between GP4 tumor areas positive for cribriform and/or
IDC, and cases that did not harbor these features. For all genes
investigated, PMR values were increased in GP4+3 vs. GP3+4, and in
pT3 vs. pT2 cases (Table III).
However, a significant association was only observed between
CYP26A1 PMR and GS (GP4+3 vs. GP3+4; P=0.004; Table IV), as well as between RASSF1 and
TBX15, and pathological stage (pT3 vs. pT2; P=0.027 and P=0.011,
respectively; Table IV). Among the
seven genes investigated, GP4 with a cribriform/IDC component
exhibited a significant increase in the median PMR for APC, RASSF1
and TBX15 (Table IV). The median PMR
of APC in cases negative for cribriform and/or IDC features was
31.7%, whereas the median PMR was 47.3% in positive cases
(P=0.045). The median PMR of RASSF1 was 69.5% in negative cases and
99.2% in positive cases (P=0.007). TBX15 methylation also exhibited
a significantly increased PMR in cribriform/IDC positive cases, as
compared with in negative cases, with a median PMR of 10.0% in
negative cases and 21.6% in positive cases (P=0.013). An increased
median PMR for APC and TBX15, but not for RASSF1, was also detected
within GP3 areas of the same GS7 tumors that harbored
cribriform/IDC features within GP4 areas (P=0.028, P=0.025 and
P=0.280, respectively). Due to a limited number of biochemical
recurrence events in the cohort, the prognostic utility of these
biomarkers could not be comprehensively evaluated in the current
study.
 | Table IV.Median PMR values of methylation
markers and Mann-Whitney U P-values stratified according to Gleason
score and pathological stage. |
Table IV.
Median PMR values of methylation
markers and Mann-Whitney U P-values stratified according to Gleason
score and pathological stage.
|
| Median PMR
values |
|---|
|
|
|
|---|
| Parameter | APC | HOXD3 | HOXD8 | CYP26A1 | RASSF1A | TBX15 | TGFb2 |
|---|
| Gleason score |
|
|
|
|
|
|
|
| 7
(3+4) | 40.0 | 24.8 | 25.5 | 15.3 | 92.3 | 16.5 | 0.0 |
| 7
(4+3) | 52.8 | 31.6 | 28.8 | 26.0 | 93.5 | 25.7 | 0.2 |
|
Mann-Whitney U P-value | 0.120 | 0.084 | 0.492 | 0.004 | 0.523 | 0.220 | 0.779 |
| Pathological
stage |
|
|
|
|
|
|
|
|
pT2 | 32.4 | 24.9 | 25.6 | 13.8 | 82.2 | 11.4 | 0.0 |
|
pT3 | 46.6 | 27.3 | 26.9 | 18.8 | 100.1 | 27.8 | 0.0 |
|
Mann-Whitney U P-value | 0.082 | 0.272 | 0.738 | 0.227 | 0.027 | 0.011 | 0.397 |
Discussion
The effective management of patients with PCa is
hindered by a lack of optimal, highly sensitive and specific
biomarkers that are able to predict the risk of disease
aggressiveness at the time of diagnosis (28). Therefore, PCa treatment decisions are
made with limited, and at times conflicting, information (28). In the present study, prognostic DNA
methylation biomarkers were incorporated along with histological
evaluation of cribriform and IDC patterns, in order to examine
their association. To the best of our knowledge, this is the first
study to investigate DNA methylation aberrations in tumor areas
consisting of specific tumor architectural features, including
cribriform and/or IDC, which provide information on the aggressive
potential of GP4 carcinoma glands in GS7 tumors (17). Aberrations to DNA methylation in
certain GP morphologies require further investigation as it may
improve personalized patient treatment by utilizing biomarker
information. The DNA methylation of APC, CYP26A1, HOXD3, HOXD8,
RASSF1, TBX15 and TGFβ2 genes were investigated in the
current study, as they have previously been revealed as promising
prognostic biomarkers for PCa with the potential for clinical
utility in independent patient cohorts (23–25).
Of the seven genes analyzed, the median methylation
levels of APC, RASSF1 and TBX15 were significantly
associated with cribriform architecture and/or IDC within the same
GP4 regions of GS7 RP cases, harboring a similar proportion and
purity of tumor cells. Increased methylation of APC and
TBX15, but not RASSF1, was also detected within GP3
areas of the same GS7 tumors, which harbored cribriform/IDC
features within GP4 areas. Hypermethylation of APC, RASSF1
and TBX15 in the presence of cribriform architecture and/or
IDC suggests these molecular markers may serve as indicators of
these architectural features. This provides novel evidence for a
link between cribriform and/or IDC features and methylation
biomarkers, and warrants further investigation of additional hyper-
and hypo-methylation events in association with various
architectural patterns in PCa. Notably, the presence of cribriform
architecture and/or IDC, rather than the precise percentage of
cribriform cells, was associated with increased APC, RASSF1,
and TBX15 methylation within the carcinoma area (data not
presented). Given these results, it is proposed that there is an
extensive increase in DNA methylation within cribriform/IDC
architecture, as well as in the adjacent carcinoma tissue. This
concept must be evaluated in future studies, as it may have
clinically significant implications for the detection of DNA
methylation-based biomarkers as surrogate indicators of cribriform
and/or IDC patterns in biopsy tissue samples. This requires further
investigation into the sensitivity and specificity of APC,
RASSF1 and TBX15 PMR, in order to discriminate between
cribriform and/or IDC positive and negative GP4 cases. The
detection of increased DNA methylation of APC, RASSF1 and
TBX15, indicating high-grade disease, may facilitate
personalized treatment strategies, including intense follow-up or a
lower threshold for initiating adjuvant radiotherapy for patients.
Future studies are also necessary to explore the potential for the
identification of a cribriform/IDC-associated DNA methylation
signature in easily accessible biofluids, such as urine samples, as
it may serve as a surrogate to examine epigenetic events heralding
cribriform/IDC architecture in the prostate. In particular, due to
the likely potential of IDC cells to travel into the prostatic
urethra via the antecedent prostatic ductal system, it may be
valuable to investigate APC, RASSF1 and TBX15
hypermethylation in association with IDC features in post-digital
rectal exam urine samples obtained from patients with PCa.
Notably, not all of the seven methylation biomarkers
were associated with cribriform and/or IDC architecture, suggesting
unique epigenetically regulated pathways may be involved in PCa
progression and its associated morphology. However, the potential
biological roles of APC, RASSF1 and TBX15 methylation
in the formation of these clinicopathological entities are not yet
well characterized, and must be investigated in further studies. It
has been demonstrated that DNA methylation at the promoters of
APC and RASSF1 tumor suppressor genes can decrease
gene expression levels, thus inducing signaling pathways important
in cancer biology, including MYC, ERK1/2 and p21; however, the role
of TBX15 in cancer has not yet been characterized (29–34).
Conversely, it is possible that DNA methylation events are not
biologically associated with cribriform and/or IDC architecture,
but are separate hallmarks of (epi)genetic derangement in PCa.
One limitation to the current study is that it was
carried out using a restricted number of RPs. Another potential
limitation of the study is that it is an association study between
biomarkers and clinical indices, not PCa-associated mortality that
may predict clinical benefit. This was an exploratory study aimed
at acquiring novel insight into DNA methylation aberrations in
specific PCa tumor architectures. Therefore, only RP tissue samples
with an abundant quantity of carcinoma were used. The results of
the current study warrant future analysis of larger multi-site
retrospective cohorts in order to elucidate the clinical utility
and predictive clinical benefit of DNA methylation and
cribriform/IDC architecture in GP4 PCa, especially in a biopsy
setting.
In summary, the association of seven DNA methylation
markers with cribriform and IDC architecture was assessed in a
series of 91 GP4 carcinoma glands in RP tissues with GS7 PCa.
Presence and prevalence of cribriform and/or IDC architecture was
confirmed in prostate tumors, similar to previous studies on
architectural patterns in PCa RPs (10–17).
Furthermore, a significant increase in APC, RASSF1 and
TBX15 methylation in association with cribriform/IDC
features was demonstrated. Future studies are required to further
validate the clinical utility and functional relevance of DNA
methylation events in cribriform and IDC architecture, and whether
it should be routinely included in the RP pathology reports. Due to
the challenges in the unequivocal identification of these
clinicopathological entities by certain pathologists, the
evaluation of these morphological features must be incorporated in
the rapidly developing field of digitized histopathology (18,19,34). This
may provide clinically translatable imaging markers for PCa
prognosis. External validation of these findings within a larger
population with a longer follow-up time is, therefore,
essential.
Acknowledgements
This study was supported by the National Cancer
Institute of Canada/the Canadian Prostate Cancer Research
Initiative (grant no. 18568); Movember Translation Acceleration
Grant from Prostate Cancer Canada (grant no. 2014-01); the Ontario
Graduate Scholarships; and the Campbell Family Fellowship award.
The authors thank Mr Michael Nesbitt for organizing and maintaining
the patient clinical information that was used in this study.
Glossary
Abbreviations
Abbreviations:
|
APC
|
adenomatous polyposis coli
|
|
CYP26A1
|
cytochrome P450 family 26 subfamily A
member 1
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
GP
|
Gleason pattern
|
|
GS
|
Gleason score
|
|
H&E
|
hematoxylin and eosin
|
|
HM
|
high methylation
|
|
HOX
|
homeobox
|
|
IDC
|
intraductal carcinoma
|
|
LM
|
low methylation
|
|
PCa
|
prostate cancer
|
|
PMR
|
percent methylation ratio
|
|
PSA
|
prostate-specific antigen
|
|
RASSF1
|
Ras association domain family member
1
|
|
RP
|
radical prostatectomy
|
|
TBX15
|
T-box 15
|
|
TGF
|
intraductal carcinoma
|
|
LM
|
loβ2
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gleason DF: Classification of prostatic
carcinomas. Cancer Chemother Rep. 50:125–128. 1966.PubMed/NCBI
|
|
3
|
Albertsen PC, Hanley JA, Gleason DF and
Barry MJ: Competing risk analysis of men aged 55 to 74 years at
diagnosis managed conservatively for clinically localized prostate
cancer. JAMA. 280:975–980. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Epstein JI, Allsbrook WC Jr, Amin MB and
Egevad LL; ISUP Grading Committee, : The 2005 international society
of urological pathology (ISUP) consensus conference on gleason
grading of prostatic carcinoma. Am J Surg Pathol. 29:1228–1242.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Epstein JI, Partin AW, Sauvageot J and
Walsh PC: Prediction of progression following radical
prostatectomy. A multivariate analysis of 721 men with long-term
follow-up. Am J Surg Pathol. 20:286–292. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Siadat F, Sykes J, Zlotta AR, Aldaoud N,
Egawa S, Pushkar D, Kuk C, Bristow RG, Montironi R and van der
Kwast T: Not all Gleason pattern 4 prostate cancers are created
equal: A study of latent prostatic carcinomas in a
cystoprostatectomy and autopsy series. Prostate. 75:1277–1284.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kovi J, Jackson MA and Heshmat MY: Ductal
spread in prostatic carcinoma. Cancer. 56:1566–1573. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
McNeal JE and Yemoto CE: Spread of
adenocarcinoma within prostatic ducts and acini. Morphologic and
clinical correlations. Am J Surg Pathol. 20:802–814. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo CC and Epstein JI: Intraductal
carcinoma of the prostate on needle biopsy: Histologic features and
clinical significance. Mod Pathol. 19:1528–1535. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Robinson BD and Epstein JI: Intraductal
carcinoma of the prostate without invasive carcinoma on needle
biopsy: Emphasis on radical prostatectomy findings. J Urol.
184:1328–1333. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cohen RJ, Chan WC, Edgar SG, Robinson E,
Dodd N, Hoscek S and Mundy IP: Prediction of pathological stage and
clinical outcome in prostate cancer: An improved pre-operative
model incorporating biopsy-determined intraductal carcinoma. Br J
Urol. 81:413–418. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wilcox G, Soh S, Chakraborty S, Scardino
PT and Wheeler TM: Patterns of high-grade prostatic intraepithelial
neoplasia associated with clinically aggressive prostate cancer.
Hum Pathol. 29:1119–1123. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Van der Kwast T, Al Daoud N, Collette L,
Sykes J, Thoms J, Milosevic M, Bristow RG, van Tienhoven G, Warde
P, Mirimanoff RO and Bolla M: Biopsy diagnosis of intraductal
carcinoma is prognostic in intermediate and high risk prostate
cancer patients treated by radiotherapy. Eur J Cancer.
48:1318–1325. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dong F, Yang P, Wang C, Wu S, Xiao Y,
McDougal WS, Young RH and Wu CL: Architectural heterogeneity and
cribriform pattern predict adverse clinical outcome for Gleason
grade 4 prostatic adenocarcinoma. Am J Surg Pathol. 37:1855–1861.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kryvenko ON, Gupta NS, Virani N, Schultz
D, Gomez J, Amin A, Lane Z and Epstein JI: Gleason score 7
adenocarcinoma of the prostate with lymph node metastases: Analysis
of 184 radical prostatectomy specimens. Arch Pathol Lab Med.
137:610–617. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kweldam CF, Wildhagen MF, Steyerberg EW,
Bangma CH, van der Kwast TH and van Leenders GJ: Cribriform growth
is highly predictive for postoperative metastasis and
disease-specific death in Gleason score 7 prostate cancer. Mod
Pathol. 28:457–464. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Trudel D, Downes MR, Sykes J, Kron KJ,
Trachtenberg J and van der Kwast TH: Prognostic impact of
intraductal carcinoma and large cribriform carcinoma architecture
after prostatectomy in a contemporary cohort. Eur J Cancer.
50:1610–1616. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shah RB, Magi-Galluzzi C, Han B and Zhou
M: Atypical cribriform lesions of the prostate: Relationship to
prostatic carcinoma and implication for diagnosis in prostate
biopsies. Am J Surg Pathol. 34:470–477. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Iczkowski KA, Egevad L, Ma J,
Harding-Jackson N, Algaba F, Billis A, Camparo P, Cheng L, Clouston
D, Comperat EM, et al: Intraductal carcinoma of the prostate:
Interobserver reproducibility survey of 39 urologic pathologists.
Ann Diagn Pathol. 18:333–342. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
You JS and Jones PA: Cancer genetics and
epigenetics: Two sides of the same coin? Cancer Cell. 22:9–20.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sharma S, Kelly TK and Jones PA:
Epigenetics in cancer. Carcinogenesis. 31:27–36. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Strand SH, Orntoft TF and Sorensen KD:
Prognostic DNA methylation markers for prostate cancer. Int J Mol
Sci. 15:16544–16576. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kron KJ, Liu L, Pethe VV, Demetrashvili N,
Nesbitt ME, Trachtenberg J, Ozcelik H, Fleshner NE, Briollais L,
van der Kwast TH and Bapat B: DNA methylation of HOXD3 as a marker
of prostate cancer progression. Lab Invest. 90:1060–1067. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu L, Kron KJ, Pethe VV, Demetrashvili N,
Nesbitt ME, Trachtenberg J, Ozcelik H, Fleshner NE, Briollais L,
van der Kwast TH and Bapat B: Association of tissue promoter
methylation levels of APC, TGFβ2, HOXD3 and RASSF1 with prostate
cancer progression. Int J Cancer. 129:2454–2462. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kron K, Liu L, Trudel D, Pethe V,
Trachtenberg J, Fleshner N, Bapat B and van der Kwast T:
Correlation of ERG expression and DNA methylation biomarkers with
adverse clinicopathologic features of prostate cancer. Clin Cancer
Res. 18:2896–2904. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Eads CA, Danenberg KD, Kawakami K, Saltz
LB, Blake C, Shibata D, Danenberg PV and Laird PW: MethyLight: A
high-throughput assay to measure DNA methylation. Nucleic Acids
Res. 28:E322000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Campan M, Weisenberger DJ, Trinh B and
Laird PW: MethyLight. Methods Mol Biol. 507:325–337. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Van der Kwast TH: Prognostic prostate
tissue biomarkers of potential clinical use. Virchows Arch.
464:293–300. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
He TC, Sparks AB, Rago C, Hermeking H,
Zawel L, da Costa LT, Morin PJ, Vogelstein B and Kinzler KW:
Identification of c-MYC as a target of the APC pathway. Science.
281:1509–1512. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Thaler S, Hähnel PS, Schad A, Dammann R
and Schuler M: RASSF1 mediates p21Cip1/Waf1-dependent cell cycle
arrest and senescence through modulation of the Raf-MEK-ERK pathway
and inhibition of Akt. Cancer Res. 69:1748–1757. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Farin HF, Mansouri A, Petry M and Kispert
A: T-box protein Tbx18 interacts with the paired box protein Pax3
in the development of the paraxial mesoderm. J Biol Chem.
283:25372–25380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mayanil CS, George D, Freilich L, Miljan
EJ, Mania-Farnell B, McLone DG and Bremer EG: Microarray analysis
detects novel Pax3 downstream target genes. J Biol Chem.
276:49299–49309. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Esteller M, Sparks A, Toyota M,
Sanchez-Cespedes M, Capella G, Peinado MA, Gonzalez S, Tarafa G,
Sidransky D, Meltzer SJ, et al: Analysis of adenomatous polyposis
coli promoter hypermethylation in human cancer. Cancer Res.
60:4366–4371. 2000.PubMed/NCBI
|
|
34
|
Pfeifer GP and Dammann R: Methylation of
the tumor suppressor gene RASSF1 in human tumors. Biochemistry
(Mosc). 70:576–583. 2005. View Article : Google Scholar : PubMed/NCBI
|