Introduction
Lung cancer is ranked as the highest malignant tumor
that threatens human health in the world. The morbidity and
mortality associated with lung cancer increases year by year with
an increase in environmental pollution as well as the age of the
population (1). The early detection
of lung cancer is hindered due to a lack of biochemical markers
with typical clinical features, and high sensitivity and
specificity. In addition, the imaging examination often lags behind
the occurrence and development of tumor, all of which are important
factors that can lead to a poor survival prognosis (2). The transforming growth factor-β1
(TGF-β1), as a type of multifunctional cytokine which plays
important role in inducing local angiogenesis, extracellular matrix
secretion, immune evasion, cell heterogeneity adhesion, cell
proliferation, invasion, metastasis and other aspects (3). The TGF-β1 gene, which is located on
chromosome 19 of the long arm, contains seven exons and six
introns. Among them, the first exon has a higher gene mutation rate
and single-nucleotide polymorphism (SNP), which was confirmed to be
closely related to the occurrence of a variety of malignant tumors,
such as esophageal, breast, prostate, liver cancer and others
(4,5).
Based on this, our study analyzed the relationship between
polymorphisms of −800G/A and +915G/C in the TGF-β1 gene and lung
cancer susceptibility in order to provide a reference for the early
diagnosis of lung cancer.
Subjects and methods
Subject information
A total of 156 patients that were admitted to
Cangzhou Central Hospital and diagnosed with non-small cell lung
cancer (NSCLC) from January 2013 to January 2016, were selected as
part of the observation group, and 156 patients with pneumonia and
tuberculosis during the same period were selected as the control
group. Surgery, radiotherapy, chemotherapy and other treatments
were not carried out. Tissue samples were obtained, and the tissue
sections were created using the conventional method and stored at
−80°C. The ratio of age and gender of the patients in both groups
was 1:1 according to the proximal matching principle. In the
observation group, there were 82 males and 74 females, aged 42–77
years, with an average age of 62.3±14.5 years. Based on the tumor
pathologic type, there were 95 cases with squamous carcinoma and 61
cases with adenocarcinoma. Based on the clinical TNM stage, there
were 15 cases in stage I, 46 cases in stage II, 62 cases in stage
III and 33 cases in stage IV, with a maximum diameter of 0.5–3.6
cm, and an average of 2.4±1.2 cm. In the control group, there were
80 males and 76 females, aged 40–80 years, with an average age of
63.5±15.7 years. The study was approved by the Ethics Committee at
Cangzhou Central Hospital and written informed consent rights were
obtained from the patients or their families.
Research methods
The sequence-specific primer polymerase chain
reaction (PCR-SSP) technique was used to test the polymorphisms of
the first exon −800G/A and +915G/C in the TGF-β1 gene. The
expression levels of TGF-β1 in peripheral blood were detected using
ELISA.
PCR-SSP technique
Tissue DNA was extracted using the kit purchased
from Sigma (St. Louis, MO, USA). The main steps were conducted as
follows: 20 mg tissue were taken, and after being ground, 500 µl of
tissue lysate were added. The solution was soaked in the water at
50°C for 1 h. Then, proteinase K was added to reach a final
concentration of 100 µg/ml with soaking in 50°C water for 3 h.
Then, the extraction was respectively performed using an equal
volume of saturated phenol, phenol-chloroform (volume ratio as 1:1)
and chloroform-isoamyl alcohol each time. Sodium ethylate (1/10
volume) and ice ethanol (2-fold volume) were added to precipitate
the DNA. The appropriate amount of TE solution was added to
dissolve the precipitation. Subsequently, DNA concentration and
purity were detected using an ultraviolet spectrophotometer
(Applied Biosystems Life Technologies, Foster City, CA, USA). The
primers were designed by referring to the sequence on PubMed, and
the primer design was synthesized by Shanghai Bioengineering Co.,
Ltd. (Shanghai, China), which is shown in Table I. A total of 25 µl of the PCR
amplification system included 10X buffer 2.5 µl + dNTPs (2.5
mmol/l) 2.0 µl + upstream and downstream primers (20 pmol for each)
+ cDNA 200 ng + Taq polymerase 1.25U and distilled water. The
reaction conditions were as follows: 94°C for 5 min, 94°C for 40
sec, annealing for 1 min (temperature is shown in Table I), 72°C for 1 min, 35 cycles in total,
and 72°C for 5 min. The enzyme digestion system included −800G/A
and +915G/C gene expansion products (10 µl for each). The
restriction enzyme (as shown in Table
I; was used, after which the enzymatic digestion was performed
in water at 37°C for 3 h. The product, which was detected by 2%
agarose gel electrophoresis and ethidium bromide staining method,
was identified under ultraviolet light.
 | Table I.Primer sequences of polymorphisms of
−800G/A and +915G/C in TGF-β1 gene. |
Table I.
Primer sequences of polymorphisms of
−800G/A and +915G/C in TGF-β1 gene.
|
| Sequence | Reaction temperature
(°C) | Enzyme digestion | Length (bp) |
|---|
| −800G/A | F (G):
5′-ACAGTTGGCACGGGCTTTCG-3′ | 57 | HpyCh4 IV | G: 182 and 206 |
|
| F (A):
5′-CAGACTCTAGAGACTGTCAG-3′ |
|
| A: 388 |
|
|
R:
5′-GTCACCAGAGAAAGAGGAC-3′ |
|
|
|
| +915G/C | F (G):
5′-GTGCTGACGCCTGGCCG-3′ | 58 | Bsu36 I | G: 233 |
|
| F (C):
5′-GTGCTGACGCCTGGCCC-3′ |
|
| C: 233 |
|
|
R:
5′-GGCTCCGGTTCTGCACTC-3′ |
|
|
|
ELISA method
Peripheral blood (5 ml) was collected and performed
by centrifugation at 3,000 × g for 20 min. The upper layer of the
serum was taken and stored at −20°C. The kits were purchased from
Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA), and the
microplate reader was purchased from the Bio-Rad Laboratories, Inc.
(Hercules, CA, USA). The steps were carried out strictly in
accordance with the specifications.
Statistical analysis
The normal data were expressed as mean ± standard
deviation, t-test was used for comparison, while the measurement
data were expressed by rate. The Chi-square test was used for
comparison. The exposure risk was tested using a single factor
logistic model and expressed by the odds ratio (OR) value.
P<0.05 indicated that the difference was statistically
significant. The SPSS 20.0 software (IBM, Armonk, NY, USA) was
applied for statistical analysis.
Results
Analysis of the polymorphisms of
−800G/A and +915G/C gene
The proportion of −800G/A gene AA subtype and A
allelic gene in the observation group was significantly higher than
that in the control group, while the proportion of +915G/C gene CC
subtype and C allelic gene was also significantly higher than that
in the control group; differences were statistically significant
(P<0.05). The cancer risk (OR) of patients with A allelic gene
in −800G/A gene was 4.8 (117×96/39×60) (95% CI=2.563–6.537,
P<0.05) while the cancer risk (OR) of the patients with C
allelic gene in +915G/C gene was 4.7 (102×111/54×45), (95%
CI=2.317–5.864, P<0.05) (Table
II).
 | Table II.Analysis on the polymorphisms of
−800G/A and +915G/C gene [n (%)]. |
Table II.
Analysis on the polymorphisms of
−800G/A and +915G/C gene [n (%)].
|
|
| −800G/A | +915G/C |
|---|
|
|
|
|
|
|---|
| Groups | Cases | GG | GA | AA | G | A | GG | GC | CC | G | C |
|---|
| Observation
group | 156 | 28 | 11 | 106 | 39 (25.0) | 117 (75.0) | 31 | 23 | 79 | 54
(34.6) | 102 (65.4) |
| Control group | 156 | 70 | 26 | 34 | 96 (61.5) | 60
(38.5) | 96 | 15 | 30 | 111 (71.2) | 45
(28.8) |
| χ2 |
|
| 60.471 |
| 42.423 |
| 56.794 |
| 41.793 |
| P-value |
|
| <0.001 |
| <0.001 |
| <0.001 |
| <0.001 |
Comparison of serum TGF-β1 expression
levels
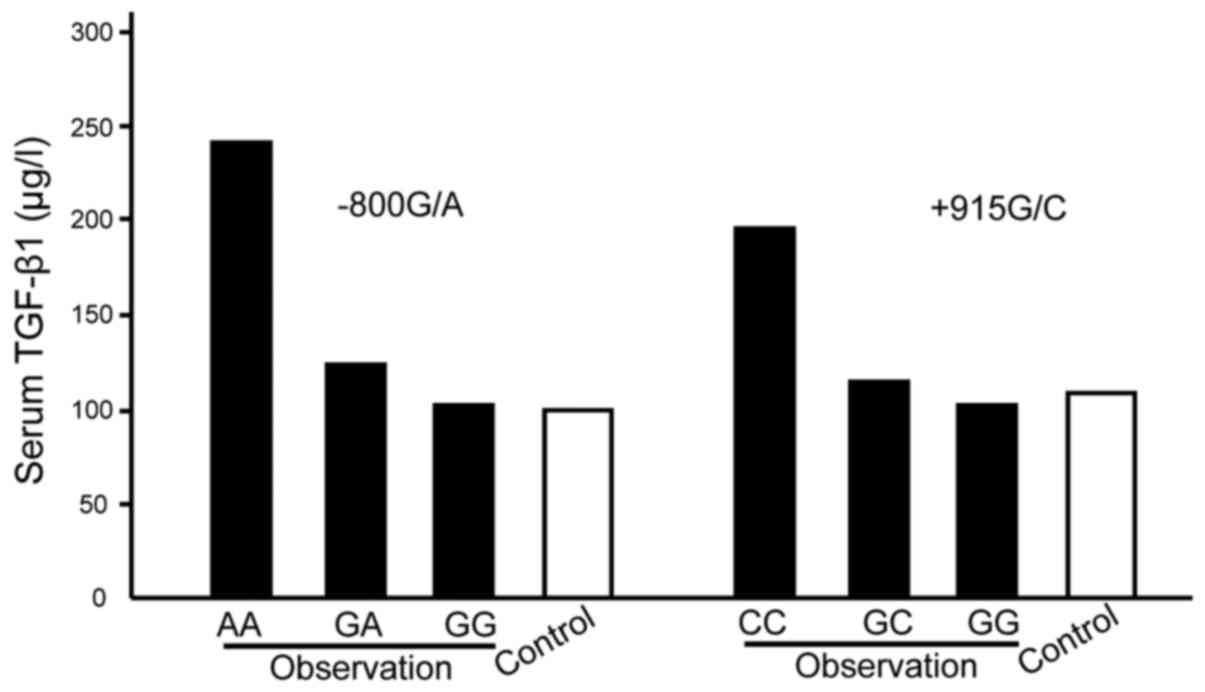
The TGF-β1 expression levels of −800G/A gene AA
subtype serum in the observation group were significantly higher
than GG type, GA type and the control group, while the TGF-β1
levels of +915G/C gene CC subtype were significantly higher than GG
type, GC type and the control group, and the differences were
statistically significant (P<0.05) (Fig. 1).
Discussion
SNP is characterized in the third generation of
genetic markers found currently. A previous study has shown that
there are 10 TGF-β1 gene SNPs at least, including −988 (C>A),
+72 (insC), −509 (C>T), −800 (G>A), codon 327 (Thr>Pro),
codon 10 (Leu>Pro), codon 47 (Gly>Glu), codon 263
(Thr>Ile), codon 25 (Arg>Pro) and 713-8delC (6). The detection methods of SNPs include
restriction fragment length polymorphism (RFLP), single-stranded
conformation polymorphism (SSCP) and allele-specific
oligonucleotide (ASO) probe.
A study has demonstrated that the number of TGF-β1
SNP sites and the pattern of manifestation are specific to both the
population and disease. The TGF-β1 promoter −800G/A and −509C/T
polymorphisms were related to the occurrence of esophageal cancer
(7), the codon 10 site was correlated
with the occurrence of breast cancer (8), and −509 (C>T) and codon 10
(Leu>Pro) polymorphisms were associated with the occurrence of
bladder cancer (9). This study
indicates that the proportion of −800G/A gene AA subtype and A
allelic gene in the observation group were significantly higher
than that in the control group, while the proportion of +915G/C
gene CC subtype and C allelic gene were significantly higher than
that in the control group; differences were statistically
significant. The cancer risk (OR) of patients with A allelic gene
in −800G/A gene was 4.8, while the cancer risk (OR) of patients
with C allelic gene in +915G/C gene was 4.7, which suggests that
−800G/A (A>G) and +915G/C (C>G) are closely related to the
lung cancer susceptibility. The serum TGF-β1 expression levels of
−800G/A gene AA subtype in the observation group were significantly
higher than GG type, GA type and the control group, while TGF-β1
levels of +915G/C gene CC subtype were significantly higher than GG
type, GC type and the control group; differences were statistically
significant. High levels of TGF-β1 play an important role in
promoting the occurrence and development of lung cancer (10). Through the reconstruction of the
eukaryotic expression recombinant of TGF-β1 promoter codon 10 in
vitro and the transfection of HeLa cell line, it was found that
the mutation of single SNP site could lead to abnormally high
expression of circulating TGF-β1 levels, the pathogenesis of which
is closely related to the processing, modification, transport and
release of proteins after translation rather than the
transcriptional activity of the gene. In addition, the different
expression of amino acids caused by an adjustment of polymorphism
of relative sites would form different primary peptide chain
structures, which would eventually decide the biological behavior
of the translated protein (11).
The study on the mechanism of polymorphic sites and
tumorigenesis has considered that activator protein 1 (AP1) and
hypoxia-inducible factor-1 (HIF-1) are important factors in the
regulation of the expression of TGF-β1 (12). When −800G/A (A>G) exist, the
binding activity of AP1 with this site is increased, which would
promote the transcription and translation of the TGF-β1 gene
(12). For HIF-1, the presence of
either A or G is not relevant and can be combined, however, for G,
the competitive binding with AP1 would be formed (13). TGF-β1 gene polymorphism is also
associated with self-autoimmune diseases, repair in trauma,
inflammatory reaction, organ fibrosis and other diseases (14,15). After
HBV infection, secondary cirrhosis may be related to the SNP of
codon 10, and the possibility of codon 10 (Leu>Pro) developed to
liver cirrhosis would be increased, while its severity may be
associated with the SNP of −509 site, and the disease condition at
−509 (C>T) may become more serious (16).
References
|
1
|
Yung KW, Yung TT, Chung CY, Tong GT, Liu
Y, Henderson J, Welbeck D and Oseni S: Principles of cancer
staging. Asian Pac J Surg Oncol. 1:1–16. 2015.
|
|
2
|
Ghoneum M, Felo N, Nwaogu OM, Fayanju IY,
Jeffe JA and Margenthaler DB: Clinical trials in surgical oncology.
Asian Pac J Surg Oncol. 1:73–82. 2015.
|
|
3
|
Eberlein C, Rooney C, Ross SJ, Farren M,
Weir HM and Barry ST: E-cadherin and EpCAM expression by NSCLC
tumour cells associate with normal fibroblast activation through a
pathway initiated by integrin αvβ6 and maintained through TGFβ
signalling. Oncogene. 34:704–716. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wan PQ, Wu JZ, Huang LY, Wu JL, Wei YH and
Ning QY: TGF-β1 polymorphisms and familial aggregation of liver
cancer in Guangxi, China. Genet Mol Res. 14:8147–8160. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Amani D, Khalilnezhad A, Ghaderi A,
Niikawa N and Yoshiura K: Transforming growth factor beta1 (TGFβ1)
polymorphisms and breast cancer risk. Tumour Biol. 35:4757–4764.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou TB, Zhao HL, Fang SL and Drummen GP:
Association of transforming growth factor-β1 T869C, G915C, and
C509T gene polymorphisms with rheumatoid arthritis risk. J Recept
Signal Transduct Res. 34:469–475. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xiao Y, Yuan X, Qiu H and Li Q:
Single-nucleotide polymorphisms of TGFβ1 and ATM associated with
radiation-induced pneumonitis: A prospective cohort study of
thoracic cancer patients in China. Int J Clin Exp Med.
8:16403–16413. 2015.PubMed/NCBI
|
|
8
|
Parvizi S, Mohammadzadeh G, Karimi M,
Noorbehbahani M and Jafary A: Effects of two common promoter
polymorphisms of transforming growth factor-β1 on breast cancer
risks in Ahvaz, West South of Iran. Iran J Cancer Prev.
9:e52662016.PubMed/NCBI
|
|
9
|
Gautam KA, Pooja S, Sankhwar SN, Sankhwar
PL, Goel A and Rajender S: c.29C>T polymorphism in the
transforming growth factor-β1 (TGFB1) gene correlates with
increased risk of urinary bladder cancer. Cytokine. 75:344–348.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shen ZT, Shen JS, Ji XQ, Li B and Zhu XX:
TGF-β1 rs1982073 polymorphism contributes to radiation pneumonitis
in lung cancer patients: A meta-analysis. J Cell Mol Med.
20:2405–2409. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou J, You W, Sun G, Li Y, Chen B, Ai J
and Jiang H: The marine-derived oligosaccharide sulfate MS80, a
novel transforming growth factor β1 inhibitor, reverses epithelial
mesenchymal transition induced by transforming growth factor-β1 and
suppresses tumor metastasis. J Pharmacol Exp Ther. 359:54–61. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shah R, Hurley CK and Posch PE: A
molecular mechanism for the differential regulation of TGF-beta1
expression due to the common SNP −509C-T (c. −1347C>T). Hum
Genet. 120:461–469. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang HB, Song WG, Liu HQ, Fang F and Xiao
Y: Role of TGFB1 polymorphism in the development of metastatic
brain tumors in non-small cell lung cancer patients. Genet Mol Res.
14:3545–3550. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Saad MN, Mabrouk MS, Eldeib AM and Shaker
OG: Effect of MTHFR TGFβ1, and TNFB polymorphisms on osteoporosis
in rheumatoid arthritis patients. Gene. 568:124–128. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Doğru-Abbasoğlu S, Vural P, Baki M,
Özderya A, Karadağ B and Uysal M: Arg25Pro (c.915G>C)
polymorphism of transforming growth factor β1 gene increases the
risk of developing Graves' disease. Int Immunopharmacol.
20:366–369. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu XD, Zeng K, Gong CS, Chen J and Chen
YQ: Transforming growth factor-β genetic polymorphisms on
development of liver cirrhosis in a meta-analysis. Mol Biol Rep.
40:535–543. 2013. View Article : Google Scholar : PubMed/NCBI
|