Introduction
Malignant tumors belong to priority areas of concern
of any responsible society and its scientific community. For
decades, the most common malignant disease in female patients has
been breast cancer (1). The
pathological evaluation of benign and malignant breast lesions
underwent remarkable changes with the introduction of molecular
diagnostic methods, and thus, increased knowledge about the
biological nature of individual lesions (2).
Mammary carcinoma is a cancer that most commonly
affects women, and dissemination of tumor cells by the
lymph-vascular pathway at an early stage of development is
considered a decisive factor of mortality (3). An early diagnosis of breast carcinoma
favors a better prognosis. Breast cancer, at a very early stage of
its development, has already cell clones with such a severe genetic
defect that can result in metastatic potential and formation of
metastases in secondary sites (4,5). In ≤30%
of breast cancer patients diagnosed with distant metastasis,
conventional treatment methods fail to stop the disease
progressing, which suggests an early event of lymph node invasion
(5,6).
Thus, the detection of occult invasion and lymph node metastasis
prediction requires novel, and preferentially more sensitive,
methods such as molecular genetics.
By using these methods, structural changes in
different genes associated with alterations in the function of
proteins can be observed. Such changes may alter or reduce the
levels of certain gene protein products, including tumor-suppressor
or metastatic-suppressor genes, thus leading to neoplasia or
metastasis formation in secondary sites (7). Currently, part of the standard
diagnostic tools monitoring breast carcinoma are biological markers
with good informative value, including estrogen receptor (ER),
progesterone receptor (PR) and lymph nodes status, which are
important prognostic and predictive biomarkers (8). However, although the presence or absence
of metastases in axillary lymph nodes is the strongest prognostic
factor for patients with primary breast cancer, it only indirectly
reflects the tendency of the cancer to spread (9).
Histological examination of axillary lymph nodes,
including lymphatic mapping of metastatic spread of the disease, is
essential for the detection or exclusion of any tumor cells in the
node (10). False-negative
examinations of lymph nodes may have severe clinical consequences
for the patient. Based on false-negative results, regional
lymphadenectomy or systematic chemotherapy is not indicated, and
thus, lymph nodes affected by metastatic turnover change may result
in untreated disease progression (11). For this reason, nodal status must be
investigated more thoroughly than only by using conventional
staining techniques of lymph nodes with hematoxylin and eosin,
which could be inadequate (12).
Currently, there are four standard methodological approaches for
surgical identification of sentinel lymph nodes (SLNs): i)
99mTc-nanocolloid lymphoscintigraphy; ii) blue color
tracking methods such as Patent Blue V; iii) a combined method
involving the use of both the above substances at the same time;
and iv) a paramagnetic method using iron oxide nanoparticles
(13).
However, there is still uncertainty and no optimal
method for precise metastatic event detection, particularly at the
level of micrometastases and isolated tumor cells. This assessment
has been recently made by using immunohistochemistry and multilevel
serial incision of lymph nodes (14).
Axillary lymph node dissection (ALND) has traditionally been a
routine component of the management of early breast cancer. The
benefits of ALND include its impact on disease control (axillary
recurrence and survival), its prognostic value and its role in
treatment selection. However, the anatomic disruption caused by
ALND may result in lymphedema, nerve injury and shoulder
dysfunction, which compromise functionality and quality of life
(15). ALND is the typical approach
for women who have clinically palpable axillary nodes or positive
nodes confirmed by methods such as ultrasound-guided fine needle
aspiration. For patients who have clinically negative axillary
lymph nodes, SLN biopsy is a method of staging the axilla with less
morbidity than that of ALND (16).
Logically, sentinel lymph node biopsy (SLNB) without ALND has been
recommended as the standard procedure for the management of
SLN-negative patients with early breast cancer; however, the
efficiency of SLNB for SLN-positive patients remains unclear
(16). Pathologists have limited time
and ability to perform a precise node examination under surgery.
However, the performance of nodal assessment several days prior to
the main surgery is now available, and allows pathologists to have
a detailed assessment of the lymphatic tissue, either performed as
a fine needle aspiration biopsy or as a complete SNB (17–20).
Isolated tumor cells are clusters usually diagnosed
by immunohistochemistry and molecular biology methods. It is
considered that these cells have no metastatic activity, and the
histopathological staging is designated as pN0mi in the regional
lymph nodes (21). Currently,
micrometastases and isolated tumor cells are undergoing renewed
scientific focus in order to identify their prognostic value and
clinical outcomes. In general, an urgent requirement to define
their prognostic value by promoter methylation status assessment of
the Ras association domain family 1 isoform A (RASSF1A) gene
in the affected nodes both at the level of micrometastases and
macrometastases exists (22).
Furthermore, these micrometastases can remain dormant for years
prior to re emerging as incurable secondary tumors that are
insensitive to adjuvant chemotherapies that were previously
effective against the primary tumor (21). Further experimental analyses are
required to investigate the precise function of RASSF1A
methylation in breast cancer invasion and metastasis.
As hypermethylation of tumor-suppressor genes is
considered an early event of breast carcinogenesis, the present
study detected aberrant methylation of the tumor-suppressor and
cancer-associated gene RASSF1A in order to identify its
potential correlation with an early stage of axillary nodal
affection, since hypermethylation of the RASSF1A gene
promoter has been reported to be an early event of carcinogenesis
and to participate in various gynecological neoplasia (23,24), such
as breast cancer (25,26).
As hypermethylation is a transient and markedly
sensitive event, the present study used rapid assays for the
detection of small levels of heterogeneous methylated alleles in
breast cancer patients based on a methylation-sensitive
high-resolution melting (MS-HRM) technology and a
methylation-specific polymerase chain reaction (PCR) (MSP)
approach. These methods were applied to assess the possible role of
RASSF1A gene hypermethylation in early axillary nodal
affection in women with breast cancer.
Materials and methods
Clinical specimens
Formalin-fixed, paraffin-embedded tissue sections
from 116 breast cancer patients operated on between June 2013 and
June 2016 at the Department of Obstetrics and Gynaecology,
Jessenius Faculty of Comenius University and University Hospital
Martin (Martin, Slovakia) were evaluated. The histopathological
data reflecting cancer biology, including lymph node involvement,
were obtained from the medical database at the Department of
Pathology, Jessenius Faculty of Comenius University and University
Hospital Martin. All participants were of Caucasian origin and
residents in the geographic area of Slovakia (Table I). The Regional Ethics Committee of
Jessenius Faculty of Medicine (registered under IRB00005636 at the
Office for Human Research Protection, USA Department of Health and
Human Services) approved the present study protocol (code no. EK
1269/2013). Written informed consent was obtained from all
patients.
 | Table I.Clinicopathological characteristics
of the cohort (n=116 patients). |
Table I.
Clinicopathological characteristics
of the cohort (n=116 patients).
| Characteristic | Patients, n
(%) |
|---|
| Age, years |
|
|
<50 | 20 (17.3) |
|
≥50 | 96 (82.7) |
| Stage |
|
| 1 | 65 (56.0) |
| 2 | 34 (29.3) |
| 3 | 7 (6.0) |
| 4 | 6 (5.2) |
| X | 4 (3.5) |
| Histological
grade |
|
| 1 | 21 (18.1) |
| 2 | 42 (36.2) |
| 3 | 48 (41.4) |
| X | 5 (4.3) |
| Lymph node
metastases |
|
|
Positive | 52 (44.8) |
|
Negative | 64 (55.2) |
| ER status |
|
|
Positive | 95 (81.9) |
|
Negative | 21 (18.1) |
| PR status |
|
|
Positive | 89 (76.7) |
|
Negative | 27 (23.3) |
| HER-2
amplification |
|
|
Positive | 23 (19.8) |
|
Negative | 93 (80.2) |
| Molecular
subtype |
|
| Luminal
A (ER+ and/or PR+, HER-2−) | 86 (74.1) |
| Luminal
B (ER+ and/or PR+, HER-2+) | 10 (8.6) |
|
Basal-like (ER−,
PR−, HER-2−) | 8 (6.9) |
| HER-2
type (ER−, PR−, HER-2+) | 12 (10.4) |
| Tumor type |
|
|
Ductal | 104 (89.7) |
|
Lobular | 7 (6.0) |
|
Other | 5 (4.3) |
Histopathological analysis
Tumor and lymph node specimens were fixed in
formalin and embedded in paraffin; basic histological examination
was performed on 4–5-µm-thick slides stained with hematoxylin and
eosin. In selected cases, lymph nodes were stained
immunohistochemically (cytokeratin 19) to detect potential isolated
tumor cells or micrometastases. Classical morphological indicators
such as tumor type and histological grade, were evaluated according
to the World Health Organization criteria and Nottingham grading
modification (27,28). The pathologic (p) stage of breast
cancer takes into consideration the characteristics of the tumor
(T) and the presence of any lymph nodes metastases (N) or distant
organ metastases (M). These major tumor characteristics were
assessed according to the criteria of the latest
tumor-node-metastasis classification (29). Biological parameters, including
estrogen receptor (ER), progesterone receptor (PR) and human
epidermal growth factor receptor (HER)-2, were detected
immunohistochemically, and their interpretation was based on the
American Society of Clinical Oncology/College of American
Pathologists criteria from 2010 and 2013 (30–32).
Briefly, immunohistochemistry. For ER, PR and HER-2 was performed
concurrently on serial sections with ready-to-use (RTU) reagents
using an automated immunostainer Autosteiner Link 48 (Dako; Agilent
Technologies, Inc., Santa Clara, CA, USA).
Primary ER (Flex Monoclonal Rabbit ERα; clone EP1,
RTU; cat. no. IR08461) and PR antibody (Flex Monoclonal Mouse;
clone PgR636, RTU; cat. no. IS0683) were supplied by Dako; Agilent
Technologies, Inc. Antigen retrieval was performed using
EnVision™ Flex Target Retrieval High pH solution (pH
9.0) for 20 min at 97–98°C in a PT Link instrument (both Dako;
Agilent Technologies, Inc.). Endogenous peroxidase activity was
blocked by incubating sections with 3% hydrogen peroxide for 10
min, followed by primary antibody incubation for 20 min at room
temperature. The EnVision Flex/HRP High pH kit (cat. no. K8000;
Dako; Agilent Technologies, Inc.) was used to visualize staining
according to the manufacturer's protocol.
The immunohistochemistry for HER-2 was performed
using a HercepTest™ Breast+Gastric kit (cat. no. SK001;
Dako; Agilent Technologies, Inc.). Antigens were retrieved in
HercepTest Epitope Retrieval Solution (pH 6.0), using PT Link for
40 min at 97–98°C. Sections were blocked for endogenous peroxidase
in 3% hydrogen peroxide for 10 min, and then incubated with the
primary antibody for 30 min at room temperature. HercepTest
Visualization reagents from the kit were used 30 min at room
temperature according to the manufacturer's protocol.
Tumors were considered as ER and PR positive if ≥1%
of neoplastic cells stained positively. HER-2-expressing tumors had
to exhibit a 3+ reaction in ≥10% of neoplastic cells to be
considered positive. Cases with 2+ reaction of HER-2 staining were
considered as equivocal and were analyzed by fluorescent in
situ hybridization to confirm or exclude HER-2 gene
amplification. Briefly, slides were hybridized with probes to
locus-specific identifier, HER2/neu and centromere 17 using the
PathVysion HER-2 DNA Probe kit (Abbott Pharmaceutical Co. Ltd.,
Lake Bluff, IL, USA) according to manufacturer's protocol.
Definite positivity of HER-2 status in tumors was
defined as a HER-2/chromosome enumeration probe 17 ratio of ≥2.0 or
an average HER-2 copy number of ≥6.0 signals per cell (31,32).
DNA extraction and bisulfite
modification
Paraffin sections of tissue were subjected to
deparaffinization by incubation with an organic solvent (xylene)
and a decreasing series of 96, 80 and 70% ethanol solutions.
Subsequently, the ethanol was removed from the sections by drying
the samples at room temperature until the ethanol had evaporated
completely. Tissues were suspended in 200 µl of lysis buffer
(Buffer AL) and digested using proteinase K (both Qiagen GmbH,
Hilden, Germany) for 3 days, or longer if necessary, at 56°C.
Subsequent genomic DNA extraction was performed using DNeasy Blood
& Tissue kit (Qiagen GmbH) according to the manufacturer's
recommendations. Bisulfite modification of 116 target DNA samples
(≤3–5 µg) was performed with the EpiTect Bisulfite kit (Qiagen
GmbH) according to the manufacturer's protocol with minor changes.
Instead of incubating the columns for 5 min at 56°C in a heating
block, the columns were incubated for 15 min at 56°C in a
thermostat in the present study.
As positive (methylated) and negative (unmethylated)
controls, commercially available EpiTect methylated and
unmethylated controls (Qiagen GmbH) were used, which contained 0.1
µg/µl methylated and fully unmethylated DNA, respectively.
MSP
The first step of MSP was performed with 2.0 µl of
bisulfite-modified DNA template in 25 µl of reaction mixture
containing 2.5 mmol/l MgCl2, 10 pmol/l of each forward and reverse
external primers, 0.5 mmol/l of each of the four deoxynucleotides
and 2.5 mmol/l of 10X ReddyMix PCR buffer (ABgene; Thermo Fisher
Scientific, Inc.). Negative-control samples without DNA target were
included. The external primers used in first MSP step were forward,
5′-TTGAGTTGYGGGAGTTGGTAT-3′ and reverse,
5′-CCCAAATAAAATCRCCACAAAAAT-3′. The amplification reaction was
performed with a hot start at 95°C for 8 min, followed by 45 cycles
of denaturation at 95°C for 30 sec, annealing temperature for
external primers of 60°C for 30 sec, extension at 72°C for 30 sec
and a final step of 8 min at 72°C. In total, 9 µl of each reaction
were loaded onto a 1.5% agarose gel stained with Gel
Red™ (Biotium, Inc., Hayward, CA, USA) and visualized
under ultraviolet light. The PCR product for the external primer
had a length of 198 bp. The second step of MSP was performed using
1 µl of the PCR product (10–50 ng cDNA) obtained in the first step
of the reaction with internal primers, diluted in a 25 µl reaction
volume. For methylated DNA targets, the following primers were
used: Forward, 5′-GTGTTAACGCGTTGCGTATC-3′ and reverse,
5′-AACCCCGCGAACTAAAAACGA-3′. For unmethylated DNA targets, the
following primers were used: Forward, 5′-TTTGGTTGGAGTGTGTTAATGTG-3′
and reverse, 5′-CAAACCCCACAAACTAAAAACAA-3′. The reamplification
products were analyzed on a Gel Red™-stained agarose gel
and subsequently validated by bisulfite sequencing.
Bisulfite sequencing
To validate the results from MSP, DNA sequencing was
performed on PCR-reamplified MSP products. The PCR products were
purified with the NucleoSpin Gel and PCR Clean-up kit
(Macherery-Nagel, GmbH, Düren, Germany) according to the
manufacturer's recommendations. The purified PCR products were
amplified in a sequencing reaction with BigDye Terminator v1.1
Cycle Sequencing kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) and subsequently purified on a Sepharose™
SigmaSpin Post-Reaction Clean-Up Column (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). The products were denatured and then analyzed
by capillary electrophoresis in a 3500 Genetic Analyzer (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The resulting
sequences were analyzed by using Chromas software 2.0 (Technelysium
Pty Ltd., South Brisbane, Australia) and compared with the sequence
of the gene RASSF1A (AF132675.1) in the GenBank database
(https://www.ncbi.nlm.nih.gov/gene?Cmd=DetailsSearch&Term=11186)
(Fig. 1).
MS-HRM
MS-HRM detected ≤1% methylated DNA in a background
of unmethylated DNA. The technology is sensitive, inexpensive and
thus likely to become an appropriate technique for a diagnosis of
methylation of the RASSF1A gene in breast cancer patients as
a predictor of bilateral nodal spread (33). To compare the sensitivity of detection
of methylation by various methods, MS-HRM was used, since this
method is able to define more precisely the extent of methylation
in the sample than MSP (34). MS-HRM
was conducted on a LightCycler® 480 System (Roche
Diagnostics, Basel, Switzerland). PCR products (5–10 ng cDNA) of
the first step of MSP were diluted to the desired concentration in
a 10 µl reaction volume, and the standards were prepared by mixing
methylated DNA (0.1 µg/µl) with unmethylated DNA (0.1 µg/µl) (both
Qiagen GmbH) to obtain 100, 50, 25 and 0% methylated/unmethylated
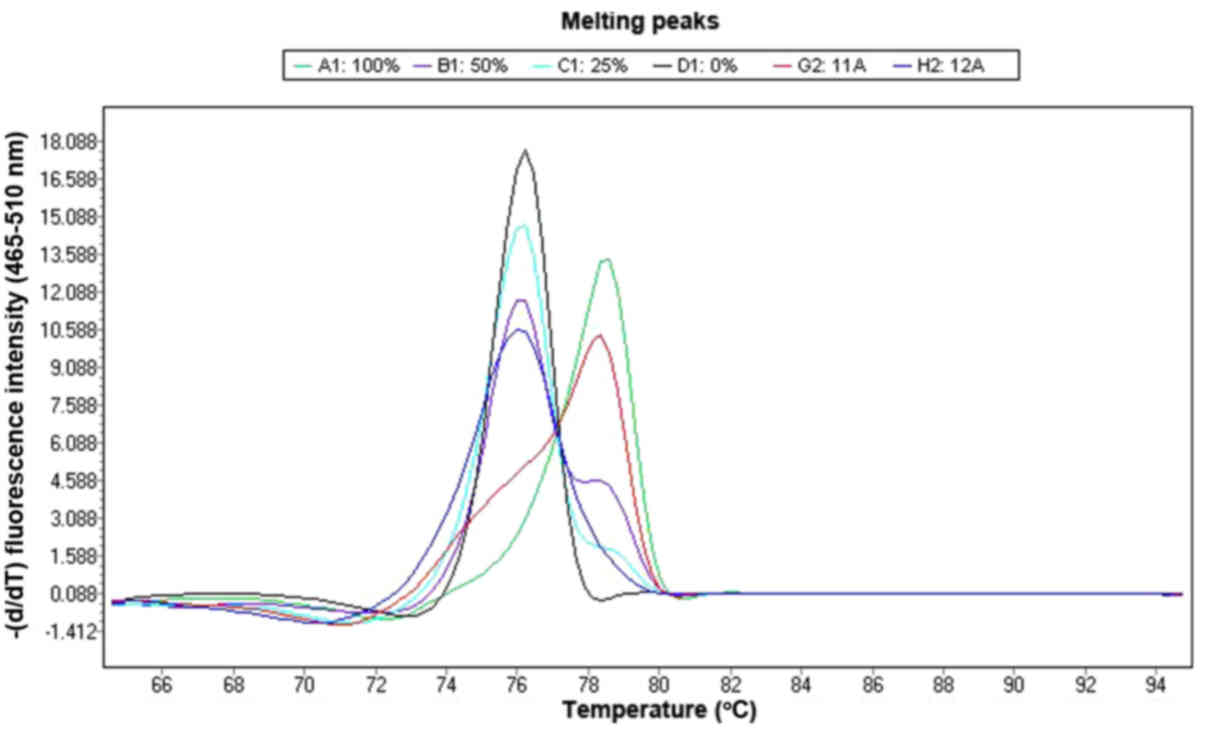
DNA dilutions (Fig. 2). MS-HRM was
performed with the same internal primers as described earlier.
MS-HRM was performed in a total volume of 10 µl of reaction mixture
containing 2X EpiTect HRM Master Mix (Qiagen GmbH), 10 µM of each
primer, 1 µl of the diluted PCR product from the first step of MSP
(5–10 ng per reaction) and RNase-free water to a final volume of 10
µl. Amplification consisted of an initial denaturation step at 95°C
for 5 min, followed by 45 cycles of the following steps:
Denaturation for 10 sec at 95°C, annealing for 30 sec at 61°C and
extension for 10 sec at 72°C. To perform high-resolution melting
analysis, the temperature was increased from 65 to 95°C. The
fluorescence of the binding fluorescent dye was measured
continuously as the temperature was increased at a speed of
0.02°C/sec and was plotted against the temperature.
Statistical analysis
All statistical tests were performed using R
software (version 3.2.3) (35).
Pearson's χ2 test with Yates continuity correction was
used to test the compliance of the two methods used (MSP vs.
MS-HRM) (Fig. 3). A multinomial
logistic regression model was used to model the dependence of the
methylation status levels derived from MS-HRM on the patients'
clinicopathological characteristics. P<0.05 was considered to
indicate a statistically significant difference.
Results
Overall methylation results
The present study used MSP and MS-HRM to examine the
methylation status of the promoter region of the RASSF1A
gene in paraffin sections of 116 patients with breast cancer
(Table I). Promoter region CpG
hypermethylation was identified by MSP in 97 of 116 (83.6%) primary
tumors, while hypermethylation of RASSF1A was confirmed by
MS-HRM in 107 (92.2%) of cases.
MS-HRM for RASSF1A vs. lymph node
status
The examined methylation status of RASSF1A
included 64 breast tumor samples with stage N0 (negative lymph
nodes) and 52 samples with stages N1, N2 and N3 (positive lymph
nodes). The percentage of RASSF1A promoter methylation had a
distinct range in tissues from patients with different lymph node
metastatic stage. Based on the result of the multinomial logistic
regression model, there was no significant difference between the
frequency of RASSF1A promoter methylation among lymph
node-positive and node-negative patients in general. However,
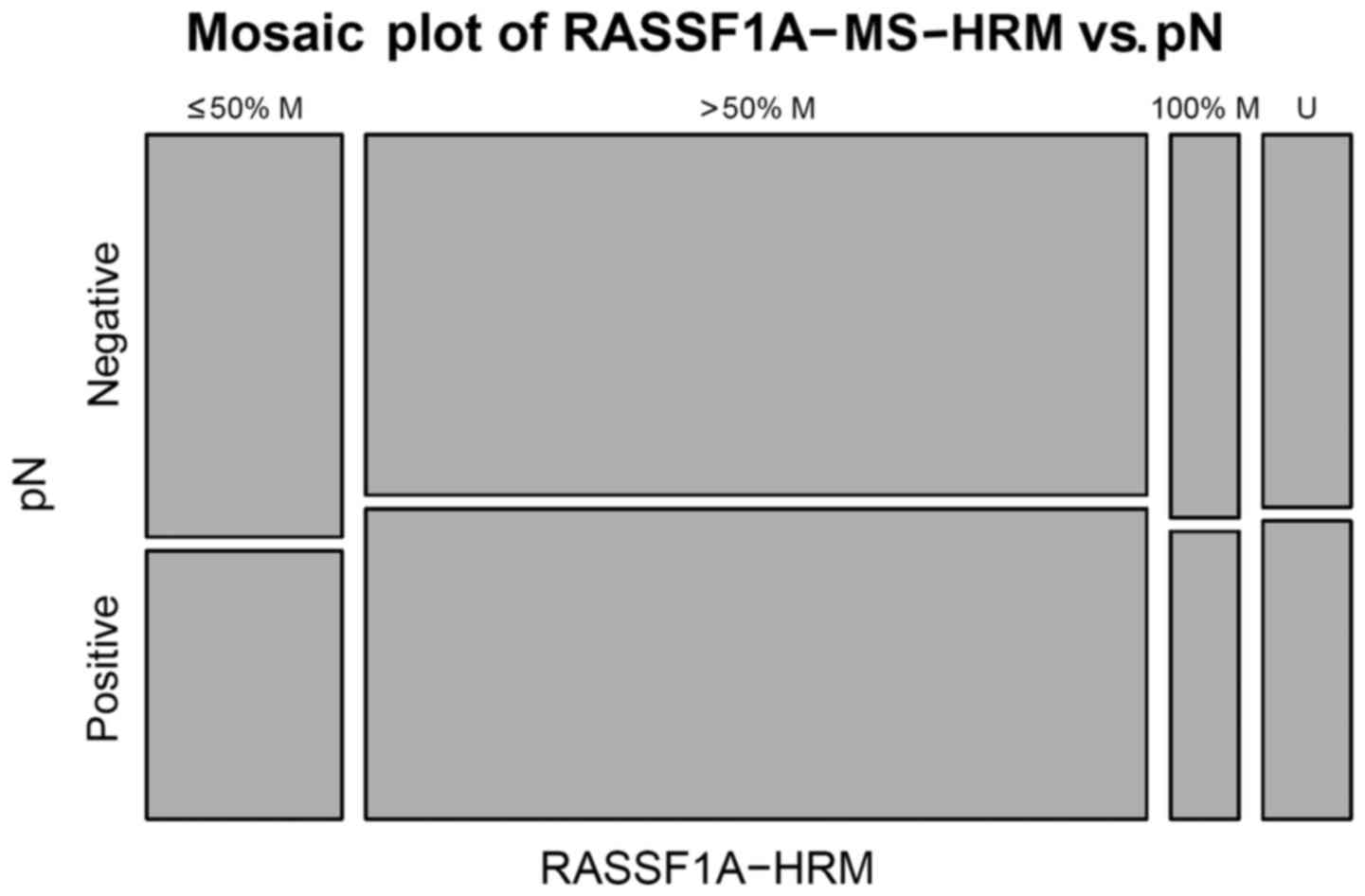
unexpectedly, an association between pN0 lymph node-negative status
(without axillary metastases) and percentage of methylation was
detected by MS-HRM in two groups of methylated alleles for
RASSF1A: ≤50% methylated group (P<0.05) and >50%
heterogeneous methylated group, where a stronger significant
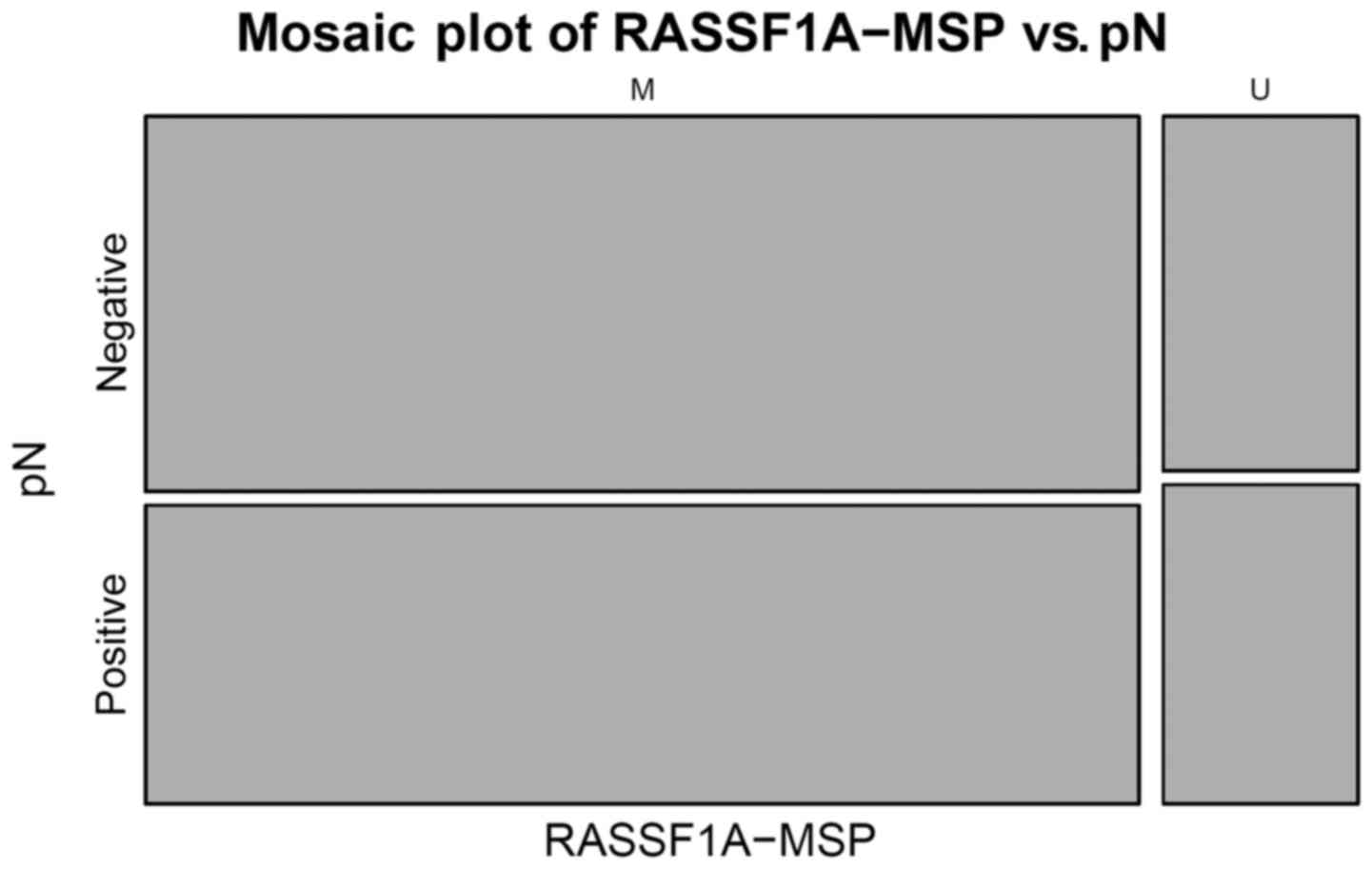
association was observed (P<0.0001) (Fig. 4). The MSP method did not identify any
significant association (Fig. 5).
MS-HRM vs. MSP results
In the breast cancer samples, comparable results
were obtained with the two assays used. More specifically, 48
samples were observed to be methylated and 4 unmethylated in lymph
node-positive cases by MS-HRM, while 59 samples were observed to be
methylated and 5 unmethylated in lymph node-negative cases. There
were only small differences in the second method used. Only 43
samples were identified as methylated and 9 as unmethylated in
lymph node-positive samples by MSP, while 54 samples were
methylated and 10 samples were unmethylated in the lymph
node-negative cases of breast cancer by MSP.
Statistical outputs
In the pN-negative group, the risk of having >50%
methylated alleles was identified as 8.6. In the pN-positive group
the risk was 9.25. The comparison of the risks, as estimated by the
odds ratio, indicates that if a patient's nodal status changes from
pN- to pN+ then the risk of possessing >50% methylated alleles
increases by 7%. This is in contrast with the results from the MSP,
where moving from pN-negative to pN-positive decreases the risk of
possessing >50% methylated alleles by 6%.
Discussion
The tumor-suppressor RASSF1A gene is the
first identified RASSF family member that is frequently
epigenetically inactivated in a wide range of cancer types
(36). RASSF1A has been
reported to be epigenetically inactivated in lung, ovary, bladder,
kidney, endometrium and breast tumor tissue (37), and is methylated in ~60–70% of breast
cancers (38,39). As a tumor-suppressor gene,
RASSF1A regulates the activation of cell death (40), cell cycle (41) and microtubule formation (42). The methylation signature of
RASSF1A is considered to be one among the earliest cellular
changes in tumorigenesis (38,39).
At present, DNA methylation is a widely studied
epigenetic event (43). A previous
study indicated that formalin-fixed, paraffin-embedded tissue is a
valuable source for breast cancer biomarkers, for its biologic
profiling or validation of certain signaling pathways (44,45). Blocs
can be also used for the detection of promoter hypermethylation as
a diagnostic and prognostic biomarker in various cancers (46,47). The
methylation status of particular tumor-suppressor genes identified
in paraffin-tissue samples displayed higher sensitivity for breast
cancer origin than conventional biomarkers (43). Similarly, methylation occurs at the
early stages of breast cancer development (48), and it may potentially reflect its
metastatic potential into lymph nodes (49). Therefore, the present study used MSP
and MS-HRM assays to identify a potential association between
methylation of RASSF1A in breast cancer tissue and bilateral
axillary nodal involvement. Bisulfite sequencing was used to
validate the results.
Previous studies have identified RASSF1A
promoter methylation as a potentially useful breast cancer
biomarker for the presence of invasiveness of disease (44,50). In
addition, other studies have reported that methylation of the
RASSF1A promoter provides an important prognostic
information in operative breast tumors, and that methylation serves
an important role in the clinical behavior of breast cancer
(26). Although there is a
significant effect of RASSF1A methylation on the biological
characteristics of breast tumors, the association between
methylation of CpG islands of this gene in breast tumor tissue
obtained from paraffin sections and prognosis prediction by
assessment of nodal affection has not been fully established yet.
The prognostic value of aberrant methylation of RASSF1A in
breast tumors has been demonstrated in cell-free DNA circulating in
serum prior to therapy, and RASSF1A has been reported to be
one among the 39 genes with prognostic significance in association
with unfavorable development of the disease (45). According to that study, the results on
RASSF1A methylation from paraffin sections also provide
important prognostic information, since patients with
RASSF1A methylation in the promoter region had a shorter
disease-free interval than those with absence of methylation in
this gene (45). It is likely that
RASSF1A gene silencing due to promoter methylation causes
deactivation of its tumor-suppressor role, and is therefore a
possible contributor to short survival in patients with breast
cancer.
Further studies demonstrated that RASSF1A
methylation confers poor prognosis (44,45,51) and
significantly higher methylation with increasing tumor stage (from
in situ to stage III) was observed, with a trend towards
HER-2+ tumors, in women who were lymph node-positive at
the time of diagnosis (52). Another
study supported these results, since it observed that
RASSF1A was frequently methylated in metastatic lymph nodes
(53). Involvement of axillary lymph
node metastasis is one of the single most important prognostic
factors in the management of patients with primary breast cancer,
and is considered to be a predictor of disease-free survival and
overall survival in breast cancer (54,55). Only
20–30% of node-negative patients will develop recurrence within 10
years, compared with ~70% of patients with axillary nodal
involvement (55,56). In general, patients with ≥4 involved
nodes at initial diagnosis have a worse outcome upon relapse than
patients with negative lymph nodes (56). Furthermore, nodal metastasis is not
only a marker of diagnosis at a later point in the natural history
of breast cancer, but also a marker of an aggressive phenotype
(57). Similarly, micrometastases
have been associated with decreased survival in the early stage of
breast cancer (9). Despite negative
SLN-findings, metastases were detected in 7% of patients (58). In another study, 6 patients were
identified with lymph node-negative,
ER+/HER-2− breast cancer, and low 21-gene
expression assay results (recurrence score of 0–17) were able to
determine the risk of distant recurrence within 5 years of their
breast cancer diagnosis (59).
The present study is in agreement with the above
studies, since hypermethylation of RASSF1A was mostly
observed in lymph node-positive cases. In addition, methylation
occurred even in lymph node-negative cases, which suggests the
onset of an epigenetic process in early breast carcinogenesis.
Using the MS-HRM method, the results of the current study revealed
that RASSF1A methylation correlated with SLN metastasis,
while no significant association with SNL metastasis was observed
using the MSP method. These findings suggest that the silencing of
the RASSF1A gene is consistent with its role as a tumor
suppressor in breast carcinogenesis. The present study provides
methylation data in correlation with lymph node status in breast
cancer, suggesting that promoter hypermethylation of the
RASSF1A gene is a molecular predictor of early disease
progression.
A great advantage of using MS-HRM is its ability to
detect a methylated template in an unmethylated background, with a
sensitivity similar to that of MSP. Furthermore, MS-HRM-based
methylation screening is cost-, labor- and time-efficient, in
contrast to direct bisulfite sequencing, which therefore, is
unsuitable as a screening method. However, it still requires to
determine the methylation status of individual CpG sites (60,61).
Compared with MSP, the MS-HRM method provides
comparable but not consistent results. The differences between
MS-HRM and MSP can be explained by the different principles on
which these methods are based (62).
In MSP, a positive signal is obtained only in cases where the
specific designed methylated primers bind a specific CpG island
site in the sequence. However, it is known that different specimens
may have different methylation sites in a specific sequence of the
promoter region. For instance, if a sample is methylated in
positions 2, 5 and 8, and the MSP primers are designed to discern
methylation of CpG sites in positions 3, 4 and 7, MSP will provide
a negative result, while MS-HRM will provide a positive result,
since it is affected by the presence of any methylated CpG island
that is located between the primers. On the other side, if the
methylation sites that are recognized by the MSP primers are not
included in the region amplified by the MS-HRM primers, a sample
detected as positive by MSP will be detected as negative by MS-HRM
(62). Furthermore, these differences
were demonstrated in the results of the methylation analyses in the
present study. In addition, the methylation status in the promoter
region of the RASSF1A gene could be detected by MS-HRM with
higher precision than by MSP.
In conclusion, RASSF1A is one of the most
frequently hypermethylated tumor-suppressor genes detected in
breast cancer, and the present results are consistent with those
from previous studies (25,26,36). These
findings suggest the importance of RASSF1A methylation in
breast cancer. Furthermore, the association of RASSF1A
hypermethylation with known clinicopathological features, including
lymph node metastasis, provides a better understanding of breast
aggressiveness, and it could serve as an important prognostic
marker during the treatment of breast cancer patients. Based on the
current results, it can be assumed that heterogeneous methylation
of the RASSF1A gene in breast carcinoma may indicate a
potential connection with early-stage metastasis and invasion in
ipsilateral axillary lymph nodes, even at a low level. However,
this should be demonstrated by using detailed analytical methods,
thus increasing the accuracy of this assumption. Such studies must
focus mainly on geno-proteomic comparisons between node-positive
and node-negative cases in order to examine the same events in
metastatic tissue from the affected lymph nodes and the primary
tumor. Particularly, based on the results from the IBCSG 23–01 and
Z0011 studies, this biological activity and extension assessment is
relevant for patient management, and thus axillary dissection could
be avoided for patients with limited SN involvement (63,64).
Additionally, the results derived from such molecularly focused
studies may lead to an improvement in the early detection of
axillary metastatic spread of breast cancer in women compared with
that of current diagnostic procedures.
Acknowledgements
The present study was supported by the Slovak
Research and Development Agency (contract no. APVV-14-0815), the
Scientific Grant Agency of the Ministry of Education, Science,
Research and Sport of the Slovak Republic and the Slovak Academy of
Sciences (grant no. 1/0243/12) and the Comenius University in
Bratislava (grant no. UK/156/2010).
References
|
1
|
Tao Z, Shi A, Lu C, Song T, Zhang Z and
Zhao J: Breast cancer: epidemiology and etiology. Cell Biochem
Biophys. 72:333–338. 2014. View Article : Google Scholar
|
|
2
|
Lester SC and Hicks DG: Diagnostic
Pathology: Breast. 2nd. Elsevier Health Sciences; 2016
|
|
3
|
Lyman GH, Temin S, Edge BS, Newman LA,
Turner RR, Weaver DL, Benson AB III, Bosserman LD, Burstein HJ,
Cody H III, et al: Sentinel lymph node biopsy for patients with
early-stage breast cancer: American Society of Clinical Oncology
clinical practice guideline update. J Clin Oncol. 32:1365–1383.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weigelt B, Hu Z, He X, Livasy C, Carey LA,
Ewend MG, Glas AM, Perou CM and Van't Veer LJ: Molecular portraits
and 70-gene prognosis signature are preserved throughout the
metastatic process of breast cancer. Cancer Res. 65:9155–9158.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Weigelt B, Peterse JL and van 't Veer LJ:
Breast cancer metastasis: Markers and models. Nat Rev Cancer.
5:591–602. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jones TI, Neboori H, Wu H, Yang Q, Haffty
BG, Evans S, Higgins S and Moran MS: Are breast cancer subtypes
prognostic for nodal involvement and associated with
clinicopathologic features at presentation in early-stage breast
cancer? Ann Surg Oncol. 20:2866–2872. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Byler S, Goldgar S, Heerboth S, Leary M,
Housman G, Moulton K and Sarkar S: Genetic and epigenetic aspects
of breast cancer progression and therapy. Anticancer Res.
34:1071–1077. 2014.PubMed/NCBI
|
|
8
|
Weigel MT and Dowsett M: Current and
emerging biomarkers in breast cancer: Prognosis and prediction.
Endocr Relat Cancer. 17:R245–R262. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
de Boer M, van Dijck JA, Bult P, Borm GF
and Tjan-Heijnen VC: Breast cancer prognosis and occult lymph node
metastases, isolated tumor cells, and micrometastases. J Natl
Cancer Inst. 102:410–425. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu M, Bardia A, Wittner BS, Stott SL, Smas
ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, et al:
Circulating breast tumor cells exhibit dynamic changes in
epithelial and mesenchymal composition. Science. 339:580–584. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Harlow SP and Weaver DL: Diagnosis,
staging and the role of sentinel lymph node biopsy in the nodal
evaluation of breast cancer. http://www.uptodate.com/contents/overview-of-sentinel-lymph-node-biopsy-in-breast-cancerDecember
12–2016
|
|
12
|
Veta M, Pluim JP, Van Diest PJ and
Viergever MA: Breast cancer histopathology image analysis: A
review. IEEE Trans Biomed Eng. 61:1400–1411. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Karakatsanis A, Christiansen PM, Fischer
L, Hedin C, Pistioli L, Sund M, Rasmussen NR, Jørnsgård H,
Tegnelius D, Eriksson S, et al: The Nordic SentiMag trial: A
comparison of super paramagnetic iron oxide (SPIO) nanoparticles
versus Tc(99) and patent blue in the detection of sentinel node
(SN) in patients with breast cancer and a meta-analysis of earlier
studies. Breast Cancer Res Treat. 157:281–294. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Upender S, Mohan H, Hand U and Attri AK:
Intraoperative evaluation of sentinel lymph nodes in breast
carcinoma by imprint cytology, frozen section and rapid
immunohistochemistry. Diagn Cytopathol. 37:871–875. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Soares EW, Nagai HM, Bredt LC, da Cunha AD
Jr, Andrade RJ and Soares GV: Morbidity after conventional
dissection of axillary lymph nodes in breast cancer patients. World
J Surg Oncol. 12:672014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Z, Wu LC and Chen JQ: Sentinel lymph
node biopsy compared with axillary lymph node dissection in early
breast cancer: A meta-analysis. Breast Cancer Res Treat.
129:675–689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gipponi M, Fregatti P, Garlaschi A,
Murelli F, Margarino C, Depaoli F, Baccini P, Gallo M and Friedman
D: Axillary ultrasound and Fine-Needle Aspiration Cytology in the
preoperative staging of axillary node metastasis in breast cancer
patients. Breast. 30:146–150. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Co M and Kwong A: Preoperative Sentinel
Node Mapping in Sentinel Node Biopsy in Early Breast Cancers-Is It
Cost-Effective? Clin Breast Cancer. 2016.(Epub ahead of print).
PubMed/NCBI
|
|
19
|
Chirappapha P, Lohsiriwat V, Kongdan Y,
Lertsithichai P, Sukarayothin T, Supsamutchai C, Talakhadze N and
Zurrida S: Sentinel lymph node biopsy under local anesthesia in
patients with breast cancer. Gland Surg. 1:151–155. 2012.PubMed/NCBI
|
|
20
|
Luini A, Gatti G, Frasson A, Naninato P,
Magalotti C, Arnone P, Viale G, Pruneri G, Galimberti V, De Cicco C
and Veronesi U: Sentinel lymph node biopsy performed with local
anesthesia in patients with early-stage breast carcinoma. Arch
Surg. 137:1157–1160. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Leidenius MH, Vironen JH, Heikkilä PS and
Joensuu H: Influence of isolated tumor cells in sentinel nodes on
outcome in small, node-negative (pT1N0M0) breast cancer. Ann Surg
Oncol. 17:254–262. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gobardhan PD, Elias SG, Madsen EV, Bongers
V, Ruitenberg HJ, Perre CI and Van Dalen T: Prognostic value of
micrometastases in sentinel lymph nodes of patients with breast
carcinoma: A cohort study. Ann Oncol. 20:41–48. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cul'bová M, Lasabová Z, Stanclová A,
Tilandyová P, Zúbor P, Fiolka R, Danko J and Visnovský J:
Methylation of selected tumor-supressor genes in benign and
malignant ovarian tumors. Ceska Gynekol. 76:274–279.
2011.PubMed/NCBI
|
|
24
|
Fiolka R, Zubor P, Janusicova V, Visnovsky
J, Mendelova A, Kajo K, Lasabova Z, Plank L and Danko J: Promoter
hypermethylation of the tumor-suppressor genes RASSF1A, GSTP1 and
CDH1 in endometrial cancer. Oncol Rep. 30:2878–2886.
2013.PubMed/NCBI
|
|
25
|
Alvarez C, Tapia T, Cornejo V, Fernandez
W, Muñoz A, Camus M, Alvarez M, Devoto L and Carvallo P: Silencing
of tumor suppressor genes RASSF1A, SLIT2 and WIF1 by promoter
hypermethylation in hereditary breast cancer. Mol Carcinog.
52:475–487. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rauscher GH, Kresovich JK, Poulin M, Yan
L, Macias V, Mahmoud AM, Al-Alem U, Kajdacsy-Balla A, Wiley EL,
Tonetti D and Ehrlich M: Exploring DNA methylation changes in
promoter, intragenic, and intergenic regions as early and late
events in breast cancer formation. BMC Cancer. 15:8162015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lakhani SR, Ellis IO, Schnitt SJ, Tan PH
and van de Vijver MJ: WHO Classification of Tumours of the Breast.
4th. Lyon: IARC; 2012
|
|
28
|
Elston CW and Ellis IO: Pathological
prognostic factors in breast cancer. I. The value of histological
grade in breast cancer: experience from a large study with
long-term follow-up. Elston C.W..Ellis I. O.: Histopathology.
1991.19:403–410. Histopatology. 41:151–153. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: the 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hammond ME, Haves DF, Wolff AC, Mangu PB
and Temin S: American Society of Clinical Oncology/College of
American Pathologists guideline recommendations for
immunohistochemical testing of estrogen and progesterone receptors
in breast cancer. J Oncol Pract. 6:195–197. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wolff AC, Hammond ME, Hicks DG, Dowsett M,
McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M,
Fitzgibbons P, et al: Recommendations for human epidermal growth
factor receptor 2 testing in breast cancer: American Society of
Clinical Oncology/College of American Pathologists clinical
practice guideline update. Arch Pathol Lab Med. 138:241–256. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wolff AC, Hammond ME, Hicks DG, Dowsett M,
McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M,
Fitzgibbons P, et al: Recommendations for human epidermal growth
factor receptor 2 testing in breast cancer: American Society of
Clinical Oncology/College of American Pathologists clinical
practice guideline update. J Clin Oncol. 31:3997–4013. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang KT, Mikeska T, Li J, Takano EA,
Millar EK, Graham PH, Boyle SE, Campbell IG, Speed TP, Dobrovic A
and Fox SB: Assessment of DNA methylation profiling and copy number
variation as indications of clonal relationship in ipsilateral and
contralateral breast cancers to distinguish recurrent breast cancer
from a second primary tumour. BMC Cancer. 15:6692015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wojdacz TK and Dobrovic A:
Methylation-sensitive high resolution melting (MS-HRM): A new
approach for sensitive and high-throughput assessment of
methylation. Nucleic Acids Res. 35:e412007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
R Core Team. R, . A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna, Austria: https://www.R-project.org/2015
|
|
36
|
Dammann R, Schagdarsurengin U, Seidel C,
Strunnikova M, Rastetter M, Baier K and Pfeifer GP: The tumor
suppressor RASSF1A in human carcinogenesis: An update. Histol
Histopathol. 20:645–663. 2005.PubMed/NCBI
|
|
37
|
Agathanggelou A, Honorio S, Macartney DP,
Martinez A, Dallol A, Rader J, Fullwood P, Chauhan A, Walker R,
Shaw JA, et al: Methylation associated inactivation of RASSF1A from
region 3p21. 3 in lung, breast and ovarian tumours. Oncogene.
20:1509–1518. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Agathanggelou A, Cooper WN and Latif F:
Role of the Ras-association domain family 1 tumor suppressor gene
in human cancers. Cancer Res. 65:3497–3508. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dammann R, Yang G and Pfeifer GP:
Hypermethylation of the cpG island of Ras association domain family
1A (RASSF1A), a putative tumor suppressor gene from the 3p21. 3
locus, occurs in a large percentage of human breast cancers. Cancer
Res. 61:3105–3109. 2001.PubMed/NCBI
|
|
40
|
Ghazaleh HA, Chow RS, Choo SL, Pham D,
Olesen JD, Wong RX, Onyskiw C and Baksh S: 14-3-3 mediated
regulation of the tumor suppressor protein, RASSF1A. Apoptosis.
15:117–127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shivakumar L, Minna J, Sakamaki T, Pestell
R and White MA: The RASSF1A tumor suppressor blocks cell cycle
progression and inhibits cyclin D1 accumulation. Mol Cell Biol.
22:4309–4318. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dallol A, Agathanggelou A, Fenton SL,
Ahmed-Choudhury J, Hesson L, Vos MD, Clark GJ, Downward J, Maher ER
and Latif F: RASSF1A interacts with microtubule-associated proteins
and modulates microtubule dynamics. Cancer Res. 64:4112–4116. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Duffy MJ, Sturgeon CM, Sölétormos G, Barak
V, Molina R, Hayes DF, Diamandis EP and Bossuyt PM: Validation of
new cancer biomarkers: A position statement from the European group
on tumor markers. Clin Chem. 61:809–820. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lewis CM, Cler LR, Bu DW, Zöchbauer-Müller
S, Milchgrub S, Naftalis EZ, Leitch AM, Minna JD and Euhus DM:
Promoter hypermethylation in benign breast epithelium in relation
to predicted breast cancer risk. Clin Cancer Res. 11:166–172.
2005.PubMed/NCBI
|
|
45
|
Müller HM, Widschwendter A, Fiegl H,
Ivarsson L, Goebel G, Perkmann E, Marth C and Widschwendter M: DNA
methylation in serum of breast cancer patients: An independent
prognostic marker. Cancer Res. 63:7641–7645. 2003.PubMed/NCBI
|
|
46
|
Marino AL, Evangelista AF, Vieira RA,
Macedo T, Kerr LM, Abrahão-Machado LF, Longatto-Filho A, Silveira
HC and Marques MM: MicroRNA expression as risk biomarker of breast
cancer metastasis: A pilot retrospective case-cohort study. BMC
Cancer. 14:7392014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sequeiros T, García M, Montes M, Oliván M,
Rigau M, Colás E, De Torres I, Morote J, Reventós J and Doll A:
Molecular markers for prostate cancer in formalin-fixed
paraffin-embedded tissues. Biomed Res Int. 2013:2836352013.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
van Hoesel AQ, Sato Y, Elashoff DA, Turner
RR, Giuliano AE, Shamonki JM, Kuppen PJ, van de Velde CJ and Hoon
DS: Assessment of DNA methylation status in early stages of breast
cancer development. Br J Cancer. 108:2033–2038. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Schrijver WA, Jiwa LS, van Diest PJ and
Moelans CB: Promoter hypermethylation profiling of distant breast
cancer metastases. Breast Cancer Res Treat. 151:41–55. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Fackler MJ, McVeigh M, Evron E, Garrett E,
Mehrotra J, Polyak K, Sukumar S and Argani P: DNA methylation of
RASSF1A, HIN-1, RAR-β, Cyclin D2 and Twist in in situ and invasive
lobular breast carcinoma. Int J Cancer. 107:970–975. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fackler MJ, McVeigh M, Mehrotra J, Blum
MA, Lange J, Lapides A, Garrett E, Argani P and Sukumar S:
Quantitative multiplex methylation-specific PCR assay for the
detection of promoter hypermethylation in multiple genes in breast
cancer. Cancer Res. 64:4442–4452. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Swift-Scanlan T, Vang R, Blackford A,
Fackler MJ and Sukumar S: Methylated genes in breast cancer:
Associations with clinical and histopathological features in a
familial breast cancer cohort. Cancer Biol Ther. 11:853–865. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Feng W, Orlandi R, Zhao N, Carcangiu ML,
Tagliabue E, Xu J, Bast RC Jr and Yu Y: Tumor suppressor genes are
frequently methylated in lymph node metastases of breast cancers.
BMC Cancer. 10:3782010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Rouzier R, Extra JM, Klijanienko J, Falcou
MC, Asselain B, Vincent-Salomon A, Vielh P and Bourstyn E:
Incidence and prognostic significance of complete axillary
downstaging after primary chemotherapy in breast cancer patients
with T1 to T3 tumors and cytologically proven axillary metastatic
lymph nodes. J Clin Oncol. 20:1304–1310. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fisher ER, Anderson S, Redmond C and
Fisher B: Pathologic findings from the national surgical adjuvant
breast project protocol B-06: 10-year pathologic and clinical
prognostic discriminants. Cancer. 71:2507–2514. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Fitzgibbons PL, Page DL, Weaver D, Thor
AD, Allred DC, Clark GM, Ruby SG, O'Malley F, Simpson JF, Connolly
JL, et al: Prognostic factors in breast cancer: College of American
Pathologists consensus statement 1999. Arch Pathol Lab Med.
124:966–978. 2000.PubMed/NCBI
|
|
57
|
Jatoi I, Hilsenbeck SG, Clark GM and
Osborne CK: Significance of axillary lymph node metastasis in
primary breast cancer. J Clin Oncol. 17:2334–2340. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Weiss M, Meyer M, Siegert S, Bartenstein P
and Pfluger T: Metastases in patients with breast cancer despite of
negative sentinel lymph node. Has the concept to be changed?
Nuklearmedizin. 52:14–20. 2013.PubMed/NCBI
|
|
59
|
Wen HY, Krystel-Whittemore M, Patil S,
Pareja F, Bowser ZL, Dickler MN, Norton L, Morrow M, Hudis CA and
Brogi E: Breast carcinoma with an Oncotype Dx recurrence score<
18: Rate of distant metastases in a large series with clinical
follow-up. Cancer. 123:131–137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wojdacz TK, Møller TH, Thestrup BB,
Kristensen LS and Hansen LL: Limitations and advantages of MS-HRM
and bisulfite sequencing for single locus methylation studies.
Expert Rev Mol Diagn. 10:575–580. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wojdacz TK, Windeløv JA, Thestrup BB,
Damsgaard TE, Overgaard J and Hansen L: Identification and
characterization of locus-specific methylation patterns within
novel loci undergoing hypermethylation during breast cancer
pathogenesis. Breast Cancer Res. 16:R172014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Dobrovic A: Analysis of DNA methylation in
clinical samples: Methods and applicationsmolecular pathology in
cancer research. pp. 261–277. Springer; New York: 2016, View Article : Google Scholar
|
|
63
|
Galimberti V, Cole BF, Zurrida S, Viale G,
Luini A, Veronesi P, Baratella P, Chifu C, Sargenti M, Intra M, et
al: Axillary dissection versus no axillary dissection in patients
with sentinel-node micrometastases (IBCSG 23-01): A phase 3
randomised controlled trial. Lancet Oncol. 14:297–305. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Giuliano AE, Ballman K, McCall L, Beitsch
P, Whitworth PW, Blumencranz P, Leitch AM, Saha S, Morrow M and
Hunt KK: Locoregional Recurrence After Sentinel Lymph Node
Dissection With or Without Axillary Dissection in Patients With
Sentinel Lymph Node Metastases: Long-term Follow-up From the
American College of Surgeons Oncology Group (Alliance) ACOSOG Z0011
Randomized Trial. Ann Surg. 264:413–420. 2016. View Article : Google Scholar : PubMed/NCBI
|