Introduction
Esophageal cancer is one of the most frequent
malignant tumors of the digestive tract, and China has the highest
incidence of esophageal cancer in the world (1). Currently, cisplatin-based adjuvant
chemotherapy is an important method used in the treatment of
esophageal cancer. However, this type of chemotherapy has multiple
side effects, which certain patients are unable to tolerate
(2). Therefore, the development of
low-toxicity and efficient chemotherapeutics for the treatment of
esophageal cancer is required (3,4).
Tea is a beverage made from the leaves of the
Theaceae family of plants. The annual production and consumption of
tea leaves worldwide surpasses >3 billion kg. The consumption of
green tea is popular in Asia, particularly in China. In recent
years, a number of researchers have studied various green tea
extracts (5). It has been
demonstrated that green tea is able to inhibit the occurrence and
development of the majority of human tumor types (6). Between 30 and 42% of dry green tea by
weight is formed of catechins, which include (−)-epicatechin,
(−)-epicatechin-3-gallate, (−)-epigallocatechin and
(−)-epigallocatechin-3-gallate (EGCG) (7). EGCG has been observed to exhibit
significant antitumor effects (8). It
has been demonstrated that EGCG is able to affect a number of
signaling pathways in the body (9–12);
however, the underlying molecular mechanisms remain to be
completely elucidated. In the present study, the effect of
treatment with EGCG on the human esophageal cancer cell line ECa109
was investigated and the associated mechanisms were discussed. The
aim of the present study was to provide a basis for the further
application of EGCG in the treatment of human esophageal
cancer.
Materials and methods
Cell culture
The esophageal carcinoma cell line ECa109 was
obtained from the China Center for Type Culture Collection (Wuhan,
China). Cells were cultured in Dulbecco's modified Eagle's medium
containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), 100 IU/ml penicillin and 100 µg/ml
streptomycin at 37°C in an atmosphere containing 5% CO2.
EGCG (>98% purity) was obtained from Sigma-Aldrich (Merck
Millipore, Darmstadt, Germany), solubilized in PBS and stored at
−20°C until required.
Cell viability
ECa109 cells were inoculated into 96-well plates at
a density of 5,000 cells/well and incubated at 37°C overnight. The
cells were subsequently treated with 0, 25, 50, 100 and 200 mg/l
EGCG and incubated for a further 96 h. Cell viability was measured
using the MTT (5 mg/ml) assay at 24, 48, 72 and 96 h. The assay was
incubated at 37°C for 4 h and formazan crystals were dissolved by
dimethyl sulfoxide, then the absorbance (optical density) was
measured at the wavelength of 492 nm. The percentages of viable
cells were calculated using the following formula: Viable cells
(%)=value of absorbance in experimental group/value of absorbance
in control group × 100 (13).
Flow cytometry
A total of 1×106 ECa109 cells were
inoculated into 6-well plates and treated with increasing
concentrations of EGCG for 96 h. Following treatment, the cells
were trypsinized, washed with PBS and labeled with fluorescein
isothiocyanate (FITC)-tagged Annexin V and propidium iodide
(Annexin V-FITC Apoptosis Detection kit; KeyGen Biotech Co., Ltd.,
Nanjing, China), according to the manufacturer's protocol, and the
data was analyzed and quantified by Windows Multiple Document
Interafce for Flow Cytometry (WinMDI) version 2.9.
Methylation-specific polymerase chain
reaction (MSP)
Following treatment with increasing concentrations
of EGCG for 96 h, the cells were harvested and the cell density was
5×106, subsequent to washing with PBS and DNA was
extracted using the DNeasy Tissue kit (Qiagen, Inc., Valencia, CA,
USA), according to the manufacturer's protocol. The DNA was
subsequently denatured using 0.2 M NaOH, treated with 3.3 M sodium
bisulfite and 0.66 mM hydroquinone, purified using the
Wizard® DNA Clean-Up kit (Promega Corporation, Madison,
WI, USA) and eluted with H2O. The DNA was diluted using
the NaOH to give a final concentration of 0.3 M, prior to
desulfonation and purification, and the pH value was 5.0.
The following primers were used to perform the MSP
(14): Cyclin-dependent kinase
inhibitor 2A (p16) methylated DNA forward,
5′-TTATTAGAGGGTGGGGCGGATCGC-3′ and reverse,
5′-GACCCCGAACCGCGACCGTAA-3′ (final product, 150 bp); and p16
unmethylated DNA forward, 5′-TTATTAGAGGGTGGGGTGGATTGT-3′ and
reverse, 5′-CAACCCCAAACCACAACCATAA-3′ (final product, 151 bp). The
following thermocycling conditions were used: 95°C for 30 sec; 40
cycles of 95°C for 15 min, 56°C for 50 sec and 72°C for 10 min; and
72°C for 50 sec. All assays were performed in triplicate. The PCR
products were analyzed using agarose gel (25 mg/ml) electrophoresis
with the visualization agent of bromophenol blue, and the relative
signal intensity was quantified using the G:BOX Gel Imaging and
Analysis system F3 (Syngene, Cambridge, UK).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA (2 µl) was extracted from
~5×106 ECa109 cells using TRIzol® reagent
(Promega Corporation). RT-PCR was performed by Taq DNA polymerase
(Invitrogen Platinum; Thermo Fisher Scientific, Inc.) using the
AccessQuick™ RT-PCR system (Sangon Biotech Co., Ltd., Shanghai,
China), according to the manufacturer's protocol. The following
primers were used: p16 forward, 5′-CCCAACGCACCGAATAGTTAC-3′ and
reverse, 5′-ATTCCAATTCCCCTGCAAACT-3′; and GAPDH forward,
5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-GAAGATGGTGATGGGATTTC-3′.
The conditions of reaction were as follows: 42 cycles of 95°C for
30 sec, 55°C for 45 sec and 72°C for 60 sec. Following 40
amplification, the Cq value was calculated using the
2−ΔΔCq method (15) and
the p16 mRNA expression relative to GAPDH was quantified using the
following formulae: ΔCqp16 = Cqp16
-CTGAPDH; ΔΔCq =
ΔCqp16-ΔCqGAPDH.
Western blotting
Following washing with ice-cold PBS, the cells
(1×107) were lysed for 10 min in
radioimmunoprecipitation assay buffer (Fuzhou Maixin Biotech Co.,
Ltd., Fuzhou, China). Total protein concentration was determined
using the Coomassie Brilliant Blue assay. Total protein was
separated using 10% SDS-PAGE, 30 µg per lane, and subsequently
transferred to a polyvinylidene difluoride membrane (Immobilon-P;
Merck Millipore, Shanghai, China). The membrane was blocked with 5%
skimmed milk powder in TBS at room temperature for 1 h and
subsequently incubated with rabbit anti-human p16 polyclonal
antibody (cat. no. bs-1856R; 1:500; Beijing Biosynthesis
Biotechnology Co., Ltd., Beijing, China) at 4°C overnight and
washed with TBST 3 times prior to use. GAPDH was blotted as an
internal reference using the anti-GAPDH rabbit polyclonal antibody
(cat. no. AB21612-1; 1:5,000; AbSci, LLC, Portland, OR, USA). The
membrane was washed with TBS containing Tween-20 three times and
incubated with goat anti-rabbit IgG horseradish
peroxidase-conjugated secondary antibody (cat. no. bs-0295G-HRP;
1:5,000; Beijing Biosynthesis Biotechnology Co., Ltd.) for at room
temperature for 2 h. Following washing by TBST for 3 times, protein
bands were visualized using the ECL Western Blotting Detection kit
(Zhejiang Tianhang Biological Technology Co., Ltd, Hangzhou,
China).
Statistical analysis
Values are presented as the mean ± standard
deviation. All statistical analysis was performed using SPSS
software (version 17.0; SPSS Inc., Chicago, IL, USA). Multi-group
comparisons of the means were carried out using one-way analysis of
variance and the Student-Newman-Keuls post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Treatment with EGCG inhibits ECa109
cell viability
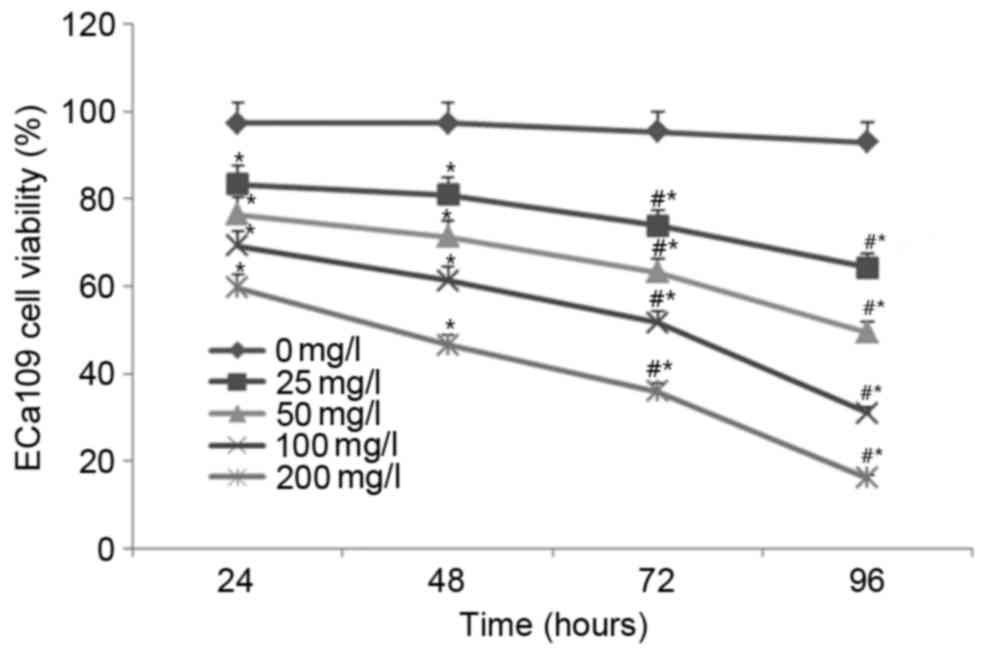
As shown in Fig. 1, no
significant difference in cell viability was observed for the
untreated (0 mg/l) cells between the time points (24, 48, 72 and 96
h). Following treatment with 25, 50, 100 and 200 mg/l EGCG, ECa109
cell viability was significantly decreased in a time- and
concentration-dependent manner compared with the untreated cells at
all time points (all P<0.05; Fig.
1). These results indicate that treatment with EGCG
significantly inhibits ECa109 cell viability.
Treatment with EGCG increases the rate
of apoptosis in ECa109 cells
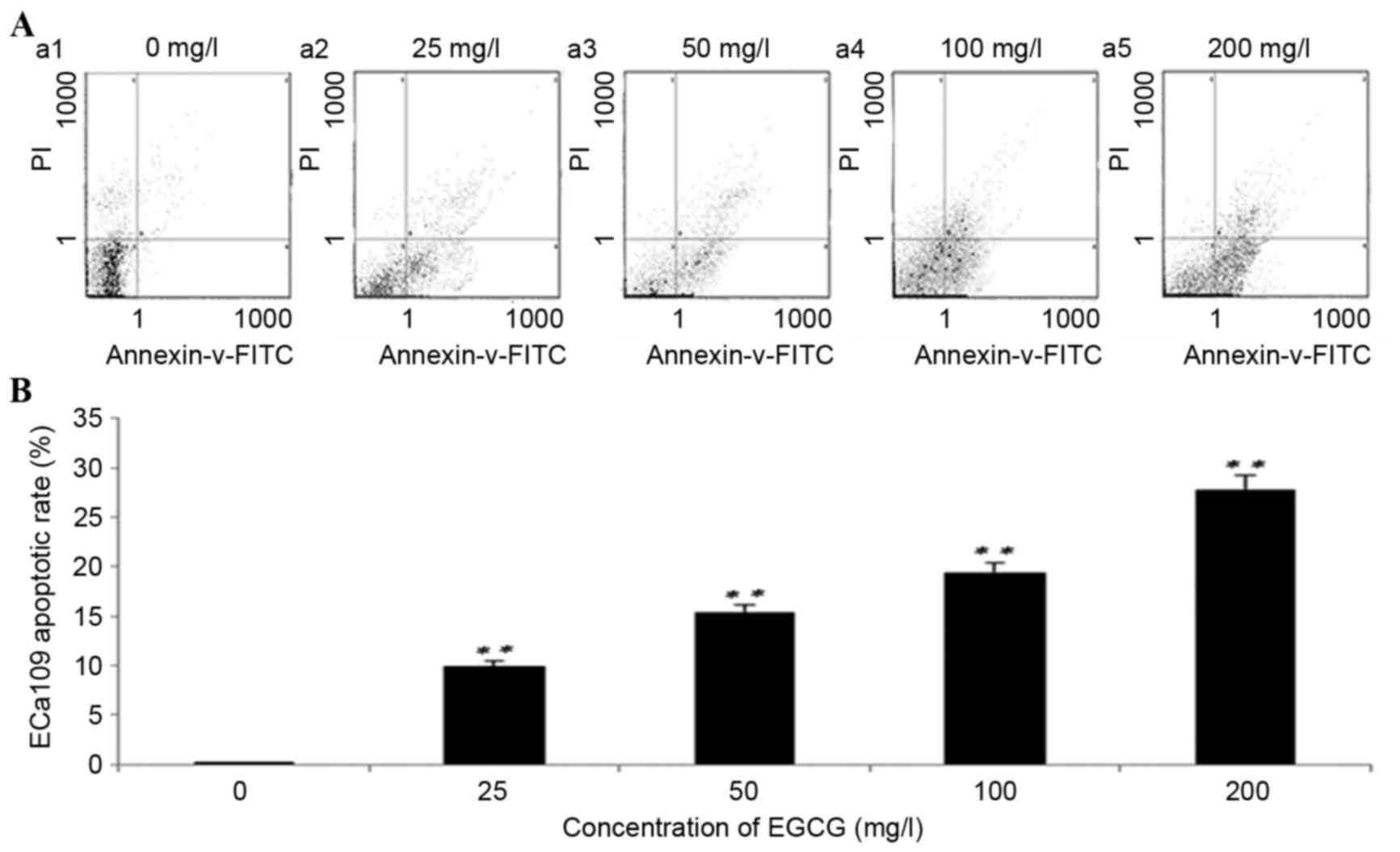
Following 96 h of treatment with EGCG, the rate of
ECa109 apoptosis increased significantly at all concentrations
compared with the untreated cells (P<0.01; Fig. 2). ECa109 apoptosis rates following
treatment with 25, 50, 100 and 200 mg/l EGCG were 9.98±0.32,
15.60±1.54, 19.40±2.89 and 27.20±2.01%, respectively. The
differences between each group were P<0.001, with the exception
of 50 mg/l vs. 100 mg/l (P=0.008). These results suggest that
treatment with EGCG significantly induces ECa109 cell
apoptosis.
Treatment with EGCG increases p16 gene
demethylation
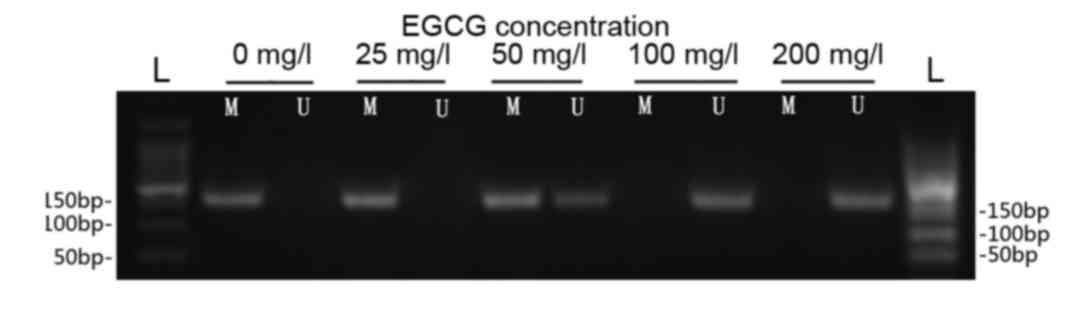
The results of the MSP analysis indicated that the
p16 gene was hypermethylated in ECa109 cells prior to treatment
with EGCG (Fig. 3). Demethylated p16
DNA levels were markedly increased following treatment with 50, 100
and 200 mg/l EGCG in a concentration-dependent manner (Fig. 3).
Treatment with EGCG increases p16 mRNA
expression
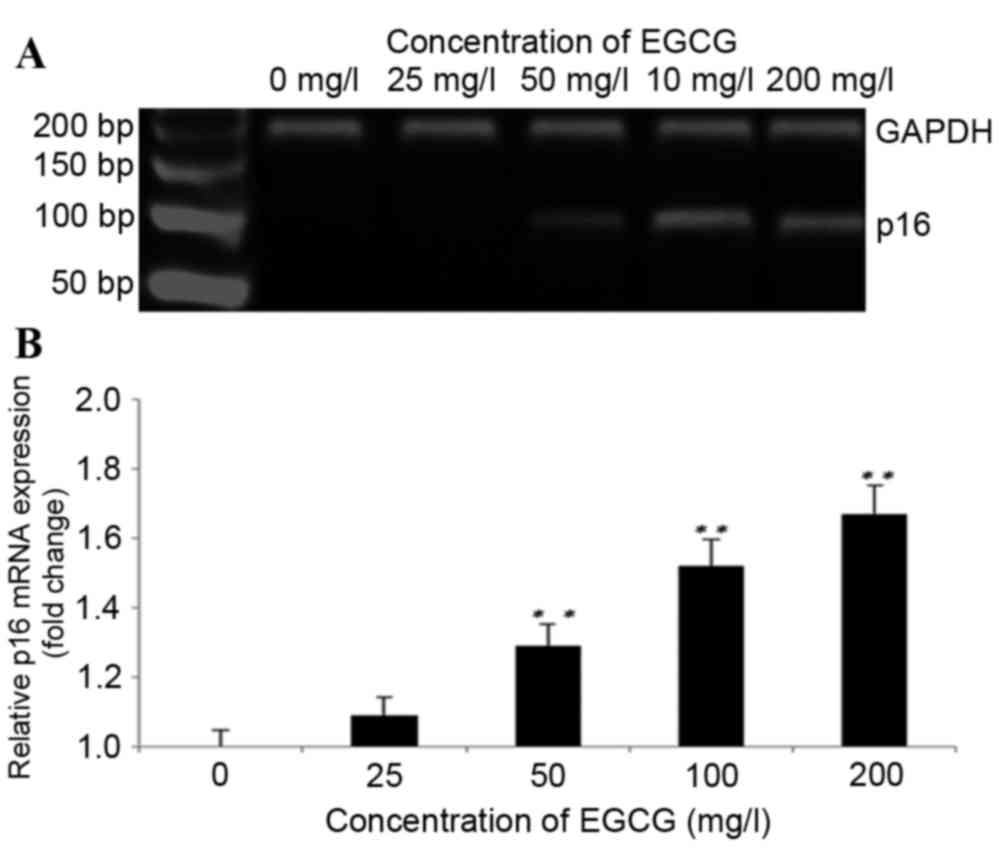
As shown in Fig. 4A and
B, treatment with EGCG induced the expression of the p16 gene
at the mRNA level. p16 mRNA levels increased significantly
following treatment with 50, 100 and 200 mg/l EGCG compared with
the untreated cells (P<0.01; Fig.
4B). The fold-changes in p16 expression levels following
treatment with 25, 50, 100 and 200 mg/l EGCG were 1.18±0.43,
1.29±0.11, 1.52±0.74 and 1.67±0.37, respectively, as compared with
the untreated cells.
Treatment with EGCG increases p16
protein expression
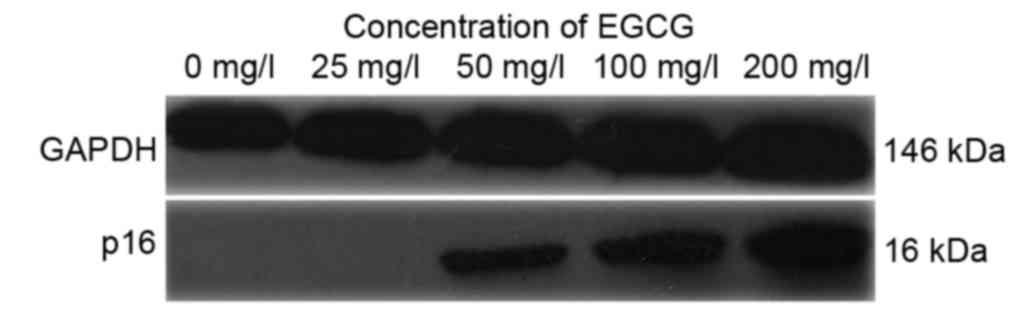
As shown in Fig. 5A and
B, p16 protein expression was increased following treatment
with EGCG in a concentration-dependent manner. P16 protein
expression was markedly increased following treatment with 50, 100
and 200 mg/ml EGCG compared with the untreated cells (Fig. 5).
Discussion
Epidemiological surveys (16,17) and
in vivo studies (18–20) have confirmed that green tea and its
extracts have antitumor effects, although the underlying molecular
mechanisms remain unclear. A previous study observed that EGCG, the
primary component of polyphenols in green tea, increased the
methylation of the estrogen receptor-α (ERα) gene, which promoted
ERα protein expression and improved the sensitivity of ERα-negative
breast cancer cells to chemotherapeutic drugs (13). Furthermore, EGCG was able to induce
the demethylation of the WNT inhibitory factor 1 and
reversion-inducing cysteine-rich protein with kazal motifs genes,
and restore their expression in pulmonary and oral squamous cell
carcinoma, thus inhibiting the growth of tumor cells (21,22).
An epigenetic study demonstrated that abnormal DNA
methylation may lead to unstable gene expression, and is involved
in the occurrence and development of tumors (23). The p16 protein, encoded for by the p16
gene, leads to G1 phase cell cycle arrest, and thus
inhibits cell proliferation and malignant transformation (24). Dysregulation of the p16 gene leads to
unregulated cell proliferation and subsequent tumorigenesis. p16
protein inactivation has a high incidence in esophageal cancer and
is caused by increased methylation of the p16 gene promoter region
(21).
The results of the present study indicate that
treatment with EGCG had a significant inhibitory effect on ECa109
cell viability, and significantly induced apoptosis in a
concentration-dependent manner. In addition, EGCG induced marked
demethylation of the p16 gene, and significantly increased p16 mRNA
and protein expression. Therefore, EGCG may inhibit ECa109 cell
viability and induce apoptosis through the reversal of p16 gene
methylation, thus increasing its protein expression.
DNA methylation is a procedure mediated by DNA
methyltransferase (DNMT). During this process, the methyl group of
an S-adenosylmethionine molecule is transferred to the fifth carbon
atom in cytosine. The methylation and demethylation of DNA are a
series of reversible enzymatic reactions. An abnormal methylation
status exists, to varying extents, in the majority of tumors, which
makes it a candidate strategy in the prevention and treatment of
tumors (25) Fang et al
(26) reported that EGCG was able to
inhibit the activity of DNMT in the esophageal cancer cell line
KYSE510, and subsequently reverse the methylation status of
O-6-methylguanine DNMT, retinoic acid receptor-β and other
tumor-associated genes. This led to increased protein expression
and induced tumor cell apoptosis. Therefore, EGCG may be a
competitive inhibitor of DNMT. However, it remains unclear whether
the reversal of p16 gene methylation following treatment with EGCG
is due to the inhibition of DNMT. In addition, it remains unclear
whether this phenomenon occurs in other tumor cell lines and
tumor-associated genes, and whether there are synergistic or
antagonistic mechanisms for p16 gene demethylation and its
induction of tumor cell apoptosis. Therefore, further
investigations are warranted.
In conclusion, the present study determined that
treatment with EGCG was able to induce apoptosis in esophageal
cancer ECa109 cells and reverse the methylation status of the tumor
suppressor gene, p16. Furthermore, the present study identified the
antitumor effects of EGCG and provided a basis for further
application of EGCG to the treatment of esophageal cancer.
References
|
1
|
Gao QY and Fang JY: Early esophageal
cancer screening in China. Best Pract Res Clin Gastroenterol.
29:885–893. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aichler M, Motschmann M, Jütting U, Luber
B, Becker K, Ott K, Lordick F, Langer R, Feith M, Siewert JR and
Walch A: Epidermal growth factor receptor (EGFR) is an independent
adverse prognostic factor in esophageal adenocarcinoma patients
treated with cisplatin-based neoadjuvant chemotherapy. Oncotarget.
5:6620–6632. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Podolski-Renić A, Andelković T, Banković
J, Tanić N, Ruždijić S and Pešić M: The role of paclitaxel in the
development and treatment of multidrug resistant cancer cell lines.
Biomed Pharmacother. 65:345–353. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ding XW, Wu JH and Jiang CP: ABCG2: A
potential marker of stem cells and novel target in stem cell and
cancer therapy. Life Sci. 86:631–637. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He X, Gao B, Zhou L and Xiong S: Green tea
polyphenol epigallocatechin-3-gallate alleviated coxsackievirus
B3-induced myocarditis through inhibiting viral replication, but
not through inhibiting inflammatory responses. J Cardiovasc
Pharmacol. 69:41–47. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang CS and Landau JM: Effects of tea
consumption on nutrition and health. J Nutr. 130:2409–2412.
2000.PubMed/NCBI
|
|
7
|
Suganuma M, Takahashi A, Watanabe T, Iida
K, Matsuzaki T, Yoshikawa HY and Fujiki H: Biophysical approach to
mechanisms of cancer prevention and treatment with green tea
catechins. Molecules. 21:pii: E15662016. View Article : Google Scholar
|
|
8
|
Balentine DA, Wiseman SA and Bouwens LC:
The chemistry of tea flavonoids. Crit Rev Food Sci Nutr.
37:693–704. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kerksick CM, Roberts MD, Dalbo VJ, Kreider
RB and Willoughby DS: Changes in skeletal muscle proteolytic gene
expression after prophylactic supplementation of EGCG and NAC and
eccentric damage. Food Chem Toxicol. 61:47–52. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shimizu M, Adachi S, Masuda M, Kozawa O
and Moriwaki H: Cancer chemoprevention with green tea catechins by
targeting receptor tyrosine kinases. Mol Nutr Food Res. 55:832–843.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Manohar M, Fatima I, Saxena R, Chandra V,
Sankhwar PL and Dwivedi A: (−)-Epigallocatechin-3-gallate induces
apoptosis in human endometrial adenocarcinoma cells via ROS
generation and p38 MAP kinase activation. J Nutr Biochem.
24:940–947. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kuhn DJ, Burns AC, Kazi A and Dou QP:
Direct inhibition of the ubiquitin-proteasome pathway by ester
bond-containing green tea polyphenols is associated with increased
expression of sterol regulatory element-binding protein 2 and LDL
receptor. Biochim Biophys Acta. 1682:1–10. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Y, Yuan YY, Meeran SM and Tollefsbol
TO: Synergistic epigenetic reactivation of estrogen receptor-α
(ERα) by combined green tea polyphenol and histone deacetylase
inhibitor in ERα-negative breast cancer cells. Mol Cancer.
9:2742010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mir MR, Shabir N, Wani KA, Shaff S,
Hussain I, Banday MA, Chikan NA, Bilal S and Aejaz S: Association
between p16, hMLH1 and E-cadherin promoter hypermethylation and
intake of local hot salted tea and sun-dried foods in Kashmiris
with gastric tumors. Asian Pac J Cancer Prev. 13:181–186. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Q, Wang Y, Ji Z, Chen X, Pan Y, Gao
G, Gu H, Yang Y, Choi BC and Yan Y: Risk factors for multiple
myeloma: A hospital-based case-control study in Northwest China.
Cancer Epidemiol. 36:439–444. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang J, Zhang W, Sun L, Yu H, Ni QX, Risch
HA and Gao YT: Green tea drinking and risk of pancreatic cancer: A
large-scale, population-based case-control study in urban Shanghai.
Cancer Epidemiol. 36:e354–e358. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu H, Xin Y, Xiao Y and Zhao J: Low-dose
docetaxel combined with (−)-epigallocatechin-3-gallate inhibits
angiogenesis and tumor growth in nude mice with gastric cancer
xenografts. Cancer Biother Radiopharm. 27:204–209. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Stearns ME, Amatangelo MD, Varma D, Varma
D, Sell C and Goodyear SM: Combination therapy with
epigallocatechin-3-gallate and doxorubicin in human prostate tumor
modeling studies: Inhibition of metastatic tumor growth in severe
combined immunodeficiency mice. Am J Pathol. 177:3169–3179. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shankar S, Marsh L and Srivastava RK: EGCG
inhibits growth of human pancreatic tumors orthotopically implanted
in Balb C nude mice through modulation of FKHRL1/FOXO3a and
neuropilin. Mol Cell Biochem. 372:83–94. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kato K, Long NK, Makita H, Toida M,
Yamashita T, Hatakeyama D, Hara A, Mori H and Shibata T: Effects of
green tea polyphenol on methylation status of RECK gene and cancer
cell invasion in oral squamous cell carcinoma cells. Br J Cancer.
99:647–654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gao Z, Xu Z, Hung MS, Lin YC, Wang T, Gong
M, Zhi X, Jablon DM and You L: Promoter demethylation of WIF-1 by
epigallocatechin-3-gallate in lung cancer cells. Anticancer Res.
29:2025–2030. 2009.PubMed/NCBI
|
|
23
|
Lima SC, Hernández-Vargas H, Simão T,
Durand G, Kruel CD, Le Calvez-Kelm F, Pinto LF Ribeiro and Herceg
Z: Identification of a DNA methylome signature of esophageal
squamous cell carcinoma and potential epigenetic biomarkers.
Epigenetics. 6:1217–1227. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rocco JW and Sidransky D: p16
(MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res. 264:42–55.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Islam M Nazmul, Yadav S, Haque M Hakimul,
Munaz A, Islam F, Al Hossain MS, Gopalan V, Lam AK, Nguyen NT and
Shiddiky MJ: Optical biosensing strategies for DNA methylation
analysis. Biosens Bioelectron. 92:668–678. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fang M, Chen D and Yang CS: Dietary
polyphenols may affect DNA methylation. J Nutr. 137 1
Suppl:223S–228S. 2007.PubMed/NCBI
|