Introduction
Colorectal cancer (CRC) is one of the most commonly
diagnosed types of cancer worldwide (1). Despite increases in early diagnosis due
to implementation of screening programmes, >5% of patients
present with distant and in the majority of cases unresectable,
metastatic lesions, representing a common cause of
cancer-associated mortality. Patients with mCRC, who are not able
to undergo radical resection of metastases, are subjected to
standard system chemotherapy with or without a targeted agent
(2). Bevacizumab is a recombinant
humanized monoclonal antibody (moAb) that blocks angiogenesis by
inhibiting vascular endothelial growth factor (VEGF) or VEGF-A.
When used in combination with standard chemotherapy
(fluorouracil/capecitabine and oxaliplatin), bevacizumab treatment
resulted in a longer progression free survival (PFS) of patients
with mCRC compared with treatment with chemotherapy alone (3). However, the benefit of the treatment is
modest and causes toxicity in patients when used unselectively.
Additionally, almost all treated patients will develop secondary
resistance to bevacizumab (3–5). Other therapeutic alternatives, including
moAb against the epidermal growth factor receptor (EGFR) cetuximab,
a fully humanised EGFR-antibody panitumumab or other targeted
agents (6,7) may be used for treatment if there is a
possibility of distinguishing between patients who may or may not
benefit from bevacizumab/5-flourouracil, leucovorin, oxaliplatin
(FOLFOX) regimen.
MicroRNAs (miRNAs) are small, non-coding RNAs that
regulate gene expression post-transcriptionally. Numerous miRNAs
function as either oncogenes or tumor suppressors (8). miRNAs serve an important role in the
regulation of the majority of cellular processes. Variability in
miRNA expression has been demonstrated to affect various cancer
types including CRC (9). miRNAs are
relatively resistant to decay and so may be detected and quantified
in tissues and body fluids (10).
This property makes miRNAs candidate biomarkers with diagnostic,
prognostic and predictive potential, and also makes them promising
therapeutic targets (11). To the
best of our knowledge, there have been two previous studies
published to date that focused on the ability of miRNAs to predict
responses to bevacizumab-based therapy in metastatic CRC (12,13).
The aim of the present study was to identify tissue
miRNAs associated with PFS in patients with mCRC treated
specifically with a bevacizumab/FOLFOX regimen in order to
establish an effective tool for distinguishing patients who would
benefit from this first-line treatment for mCRC.
Materials and methods
Patients, tissue samples and study
design
A total of 61 Caucasian patients with mCRC treated
with combined therapy bevacizumab/FOLFOX in a standard regimen as
previously described (3) were
enrolled in the study. The patients included 40 males and 21
females; the mean ± standard deviation age was 63±8 years with a
range of 34–74 years. All patients were treated for
generalized-radically inoperable metastatic colorectal cancer at
the Masaryk Memorial Cancer Institute (Brno, Czech Republic)
between January 2006 and December 2012. All patients had a
well-documented history of treatment, including the effect of
treatment and toxicity. A total of 63 tissue samples obtained from
the patients during surgical resection of the primary tumor were
used in the present study. The clinicopathological characteristics
of patients are summarized in Table
I. Response to the bevacizumab/FOLFOX treatment was assessed
according to Response Evaluation Criteria in Solid Tumors (RECIST)
criteria and duration of PFS (14).
The patients were divided into two groups: i) Complete or partial
response (responders), and ii) stable disease and progressive
disease (non-responders). The tissue samples were fixed in 10%
formalin immediately following extraction. The discovery (n=20,
including 10 responding and 10 non-responding patients) and
validation cohorts (n=41) of patients with mCRC were set up
retrospectively based on the RECIST response and PFS. The study
protocol was approved by Masaryk Memorial Cancer Institute Ethics
Committee, and written informed consent was obtained from all
patients enrolled in the study.
 | Table I.Patient characteristics. |
Table I.
Patient characteristics.
| Variable | All patients | Discovery cohort | Validation
cohort |
|---|
| Total, n | 61 | 20 | 41 |
| Gender, n |
|
|
|
| Male | 40 | 12 | 28 |
|
Female | 21 | 8 | 13 |
| Age, years |
|
|
|
| Mean ±
standard deviation | 63±8 | 60±10 | 64±6 |
|
Range | 34–74 | 34–74 | 51–74 |
| KRAS, n |
|
|
|
|
Wild-type | 33 | 9 | 24 |
|
Mutated | 19 | 6 | 13 |
| Not
available | 9 | 5 | 4 |
| RECIST response,
n |
|
|
|
|
CR/PR | 34 | 10 | 24 |
|
SD/PD | 27 | 10 | 17 |
| PFS, median
(interquartile range) |
|
|
|
|
Respondersa | 25 (20–44) | 45 (33–55) | 20 (19–25) |
|
Non-respondersb | 6 (3–8) | 3 (2–6) | 7 (4–8) |
RNA isolation
The total RNA enriched for small RNA fraction was
isolated from formalin-fixed paraffin embedded (FFPE) tissue
samples of primary CRC tumors, which were surgically resected as a
first treatment modality. The first step of the protocol was xylene
deparaffinization, followed by purification by mirVana™ miRNA
Isolation kit (Ambion; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) according to the manufacturer's protocol. The concentration
and purity of RNA was determined by UV spectrophotometry using an
ND-1000 machine (Nanodrop; Thermo Fisher Scientific, Inc.,
Wilmington, DE, USA). RNA samples with sufficient purity (260/280
ratio >2.0 and 260/230 ratio >1.8) were used for further
study.
miRNA microarray profiling
miRNA microarray profiling was conducted using
Affymetrix GeneChip miRNA 3.0 arrays (Affymetrix, Inc., Santa
Clara, CA, USA) containing 5,607 probe sets for human pre-miRNA,
miRNAs, small nuclear RNA, and small Cajal body-specific RNA. These
Affymetrix miRNA arrays provide 100% coverage of the miRNAs in the
miRBase (version 17; http://www.mirbase.org). Probe sets that were deleted
in a more recent version of miRBase (version 21) were excluded from
the analysis. All steps of the procedure were performed according
to the Affymetrix standardized protocol for miRNA 3.0 arrays.
RT-qPCR quantification
Commercial TaqMan Advanced miRNA assays for
miR-877-5p, miR-642a-3p, miR-642b-3p, miR-92b-3p, miR-3156-5p,
miR-10a-5p and miR-125a-5p, based on stem-loop reverse
transcription, and TaqMan Universal PCR Master mix (all from
Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) were used for the individual quantification of the miRNAs
selected in the discovery phase of the study. RT-qPCR was performed
with QuantStudio 12 K Flex Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using the temperature
profiles from the manufacturer's protocols. PCR was run in
triplicate. Average threshold cycles and standard deviation values
were calculated as described in the subsequent section.
Normalization and statistical
analysis
The Affymetrix raw data were normalized using the
robust multichip average algorithm from ‘oligo’ Bioconductor
package (15) in R version 3.0.1
(16). Normalized miRNA expression
data were evaluated using Bioconductor LIMMA differential
expression analysis (17). P<0.05
was selected according to the potential of identified miRNAs to
accurately discriminate responding and non-responding patients in
subsequent hierarchical clustering analysis (15). In the validation phase of the study,
mean expression levels of candidate miRNAs in RT-qPCR
quantification were normalized to the expression of miR-6098 as the
endogenous control as per a previously described method (18). miR-6098 was selected by the use of
GeneNorm and NormFinder algorithms to be uniformly expressed in all
samples in the discovery cohort (19). Normalized expression data were
evaluated by the Mann-Whitney U test, by a bidirectional stepwise
logistic regression model to establish the combined miRNA
diagnostic score (4-miRNA-based diagnostic score = 1.9086–0.0037x
miR-92a-3p −1.1517x miR-3156-5p-0.0892x miR-10a-5p-0.0986x
miR-125a-5p) and by receiver operating characteristic (ROC)
analysis to identify cut-off values and subsequently by
Kaplan-Meier analysis followed by log-rank test for survival
analysis (GraphPad Prism version 5.0; GraphPad Software, Inc., La
Jolla, CA, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
miRNAs are differentially expressed
between responders and non-responders to FOLFOX-bevacizumab
therapy
High-throughput miRNA microarray profiling was
performed on a discovery cohort of 20 patients. These individuals
were treated with a bevacizumab/FOLFOX regimen and were divided
into responding or non-responding groups. LIMMA differential
expression analysis of normalized expression data identified 67
miRNAs that were differentially expressed in responding and
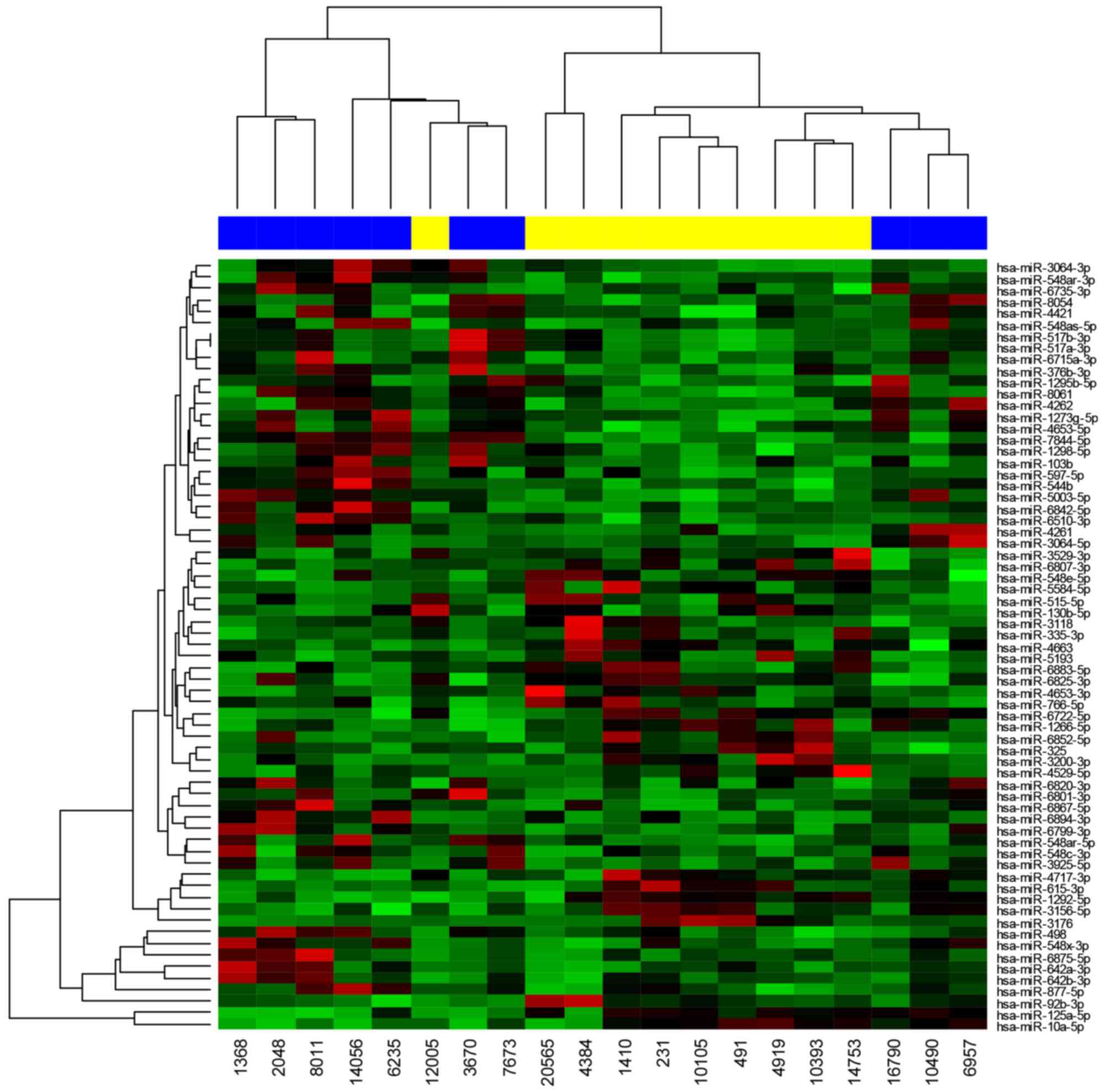
non-responding patients (Fig. 1;
Table II). A total of 7 candidate
miRNAs (miR-887-5p, miR-642a-3p, miR-642b-3p, miR-92b-3p,
miR-3156-5p, miR-10a-5p and miR-125a-5p) were chosen for further
independent validation by RT-qPCR according to the following
criteria: Fold change >2, P<0.05 and Affymetrix miRNA array
signal intensity >3.
 | Table II.miRs identified to be differentially
expressed in patients responding and non-responding to
bevacizumab/FOLFOX treatment. |
Table II.
miRs identified to be differentially
expressed in patients responding and non-responding to
bevacizumab/FOLFOX treatment.
| miR | Fold-change | P-value |
|---|
| hsa-miR-5003-5p | 1.48 | 0.001 |
| hsa-miR-7844-5p | 1.49 | 0.002 |
|
hsa-miR-548x-3p | 2.68 | 0.003 |
|
hsa-miR-3925-5p | 1.99 | 0.003 |
|
hsa-miR-6825-3p | 0.66 | 0.005 |
|
hsa-miR-1273g-5p | 1.36 | 0.006 |
| hsa-miR-4262 | 1.37 | 0.007 |
| hsa-miR-498 | 3.20 | 0.008 |
|
hsa-miR-4653-5p | 1.34 | 0.009 |
|
hsa-miR-6842-5p | 1.45 | 0.01 |
|
hsa-miR-6807-3p | 0.71 | 0.01 |
| hsa-miR-544b | 1.31 | 0.012 |
|
hsa-miR-877-5pa | 2.85 | 0.013 |
|
hsa-miR-6820-3p | 1.46 | 0.014 |
|
hsa-miR-548as-5p | 1.36 | 0.014 |
| hsa-miR-5193 | 0.62 | 0.014 |
|
hsa-miR-3529-3p | 0.72 | 0.015 |
| hsa-miR-515-5p | 0.70 | 0.017 |
|
hsa-miR-642a-3pa | 5.53 | 0.017 |
| hsa-miR-8054 | 1.32 | 0.017 |
| hsa-miR-3176 | 0.45 | 0.017 |
|
hsa-miR-3064-5p | 1.50 | 0.018 |
|
hsa-miR-548e-5p | 0.74 | 0.019 |
|
hsa-miR-4653-3p | 0.65 | 0.019 |
|
hsa-miR-6722-5p | 0.66 | 0.02 |
|
hsa-miR-6883-5p | 0.66 | 0.023 |
|
hsa-miR-517a-3p | 1.37 | 0.024 |
|
hsa-miR-517b-3p | 1.37 | 0.024 |
|
hsa-miR-548c-3p | 1.49 | 0.024 |
|
hsa-miR-548ar-5p | 1.51 | 0.024 |
| hsa-miR-8061 | 1.29 | 0.024 |
| hsa-miR-325 | 0.77 | 0.028 |
| hsa-miR-4663 | 0.72 | 0.028 |
|
hsa-miR-6894-3p | 1.70 | 0.028 |
|
hsa-miR-92b-3pa | 0.32 | 0.029 |
|
hsa-miR-3156-5pa | 0.52 | 0.029 |
| hsa-miR-615-3p | 0.57 | 0.029 |
| hsa-miR-3118 | 0.68 | 0.029 |
|
hsa-miR-6510-3p | 1.35 | 0.03 |
|
hsa-miR-4717-3p | 0.59 | 0.03 |
|
hsa-miR-6852-5p | 0.66 | 0.03 |
|
hsa-miR-548ar-3p | 1.39 | 0.031 |
| hsa-miR-597-5p | 1.32 | 0.032 |
| hsa-miR-4261 | 1.63 | 0.033 |
|
hsa-miR-6799-3p | 1.49 | 0.034 |
|
hsa-miR-1298-5p | 1.30 | 0.034 |
|
hsa-miR-6875-5p | 3.18 | 0.034 |
|
hsa-miR-130b-5p | 0.74 | 0.036 |
|
hsa-miR-6867-5p | 1.69 | 0.036 |
|
hsa-miR-5584-5p | 0.81 | 0.037 |
|
hsa-miR-642b-3pa | 3.49 | 0.037 |
|
hsa-miR-3064-3p | 1.30 | 0.038 |
|
hsa-miR-4529-5p | 0.72 | 0.038 |
| hsa-miR-103b | 1.38 | 0.038 |
|
hsa-miR-376b-3p | 1.33 | 0.038 |
| hsa-miR-766-5p | 0.67 | 0.039 |
|
hsa-miR-3200-3p | 0.75 | 0.04 |
|
hsa-miR-6735-3p | 1.27 | 0.04 |
|
hsa-miR-6801-3p | 1.38 | 0.04 |
|
hsa-miR-10a-5pa | 0.20 | 0.044 |
|
hsa-miR-1266-5p | 0.75 | 0.045 |
|
hsa-miR-1295b-5p | 1.27 | 0.047 |
| hsa-miR-335-3p | 0.68 | 0.047 |
|
hsa-miR-6715a-3p | 1.34 | 0.047 |
| hsa-miR-4421 | 1.22 | 0.049 |
|
hsa-miR-125a-5pa | 0.19 | 0.049 |
|
hsa-miR-1292-5p | 0.62 | 0.05 |
Association of miRNAs with response to
bevacizumab/FOLFOX therapy
The expression levels of 7 candidate miRNAs
identified in the discovery phase of the present study were
determined in the validation cohort of 41 patients by RT-qPCR
(P<0.01; Cq<35). A total of 4 miRNAs (miR-92b-3p,
miR-3156-5p, miR-10a-5p, and miR-125a-5p) were confirmed to be
associated with radiological response according to RECIST criteria
(P<0.005; Fig. 2A-D; Table III). All 4 miRNAs were upregulated
in responders to bevacizumab/FOLFOX therapy. The association with
RECIST response of the remaining 3 miRNAs included in the
validation phase did not reach statistical significance: miR-877-5p
(P=0.4430), miR-642a-3p (P=0.1974) and miR-642b-3p (P=0.3688).
Kaplan-Meier analysis demonstrated that higher levels of
miR-125a-5p, miR-10a-5p, miR-3156-5p and miR-92b-3p were also
significantly associated with extended PFS (Fig. 2E-H; Table
III).
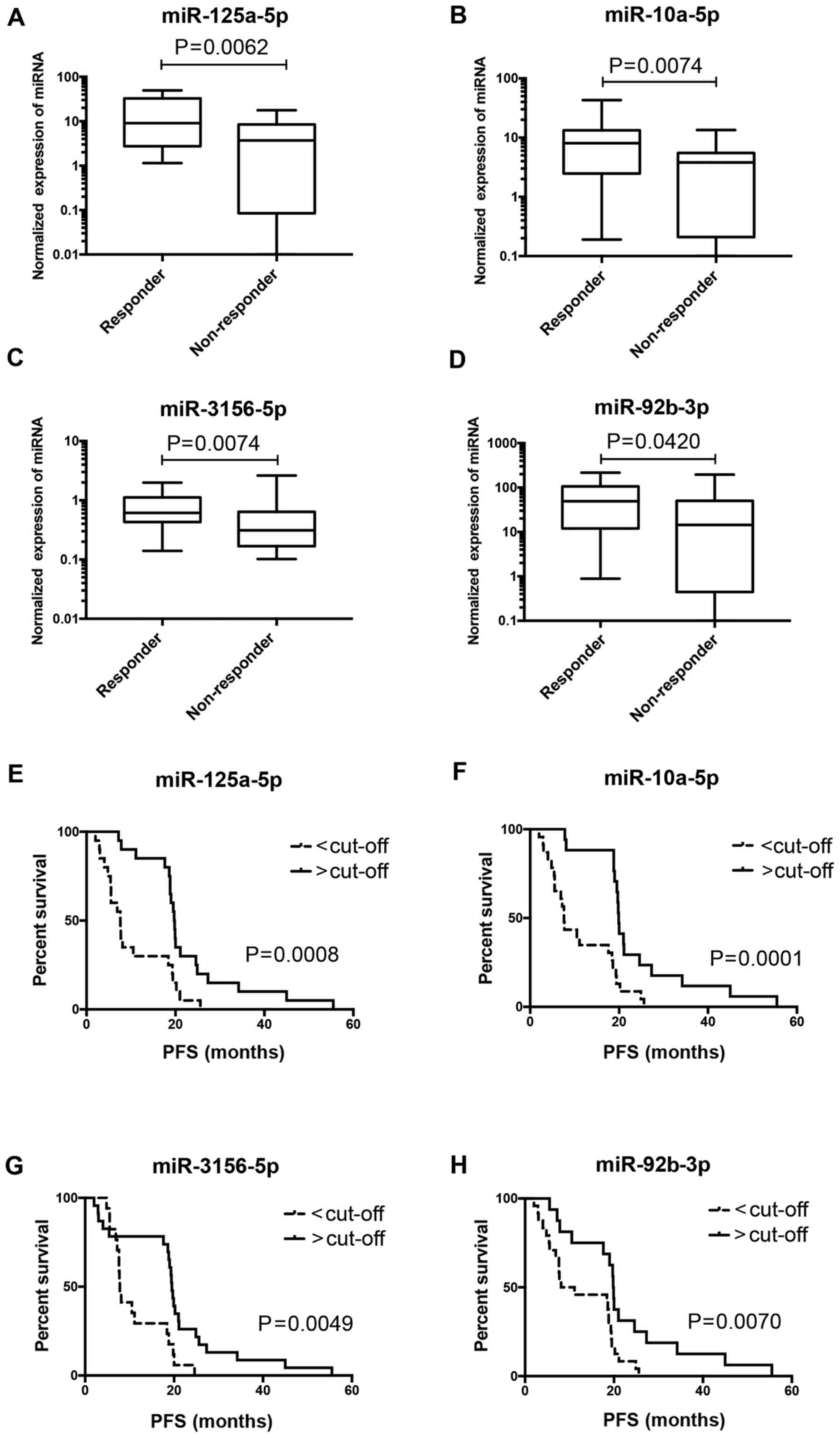 | Figure 2.miRNAs associated with therapy
response. Normalized expression of (A) miR-125a-5p, (B) miR-10a-5p,
(C) miR-3156-5p, and (D) miR-92b-3p in patients responding and
non-responding to bevacizumab/FOLFOX therapy. Quantification of
quantitative reverse transcription polymerase chain reaction was
normalized to miR-6098 expresion. Kaplan-Meier survival curves
estimating time to progression in bevacizumab/FOLFOX-treated
patients with metastatic colorectal cancer (n=41) according to (E)
miR-125a-5p, (F) miR-10a-5p, (G) miR-3156-5p, and (H) miR-92b-3p
tumor tissue expression levels. miRNA, microRNA; FOLFOX,
5-flourouracil, leucovorin, oxaliplatin; PFS, progression-free
survival. |
 | Table III.Validation of miRNAs identified in
the discovery phase (n=41) of the study. |
Table III.
Validation of miRNAs identified in
the discovery phase (n=41) of the study.
| Parameters | Number of
patients | Median PFS
(months) | Log-rank
P-value | Hazard ratio | 95% CI |
|---|
| miR-125a-5p |
|
|
|
|
|
| Low,
<8.7 | 20 |
7.6 | 0.0008 | 2.652 | 1.323–5.313 |
| High,
≥8.7 | 21 | 19.8 |
|
|
|
| miR-10a-5p |
|
|
|
|
|
| Low,
<6.2 | 17 |
7.6 | 0.0001 | 2.805 | 1.447–5.434 |
| High,
≥6.2 | 24 | 20.0 |
|
|
|
| miR-3156-5p |
|
|
|
|
|
| Low,
<0.4 | 24 |
7.8 | 0.0049 | 2.259 | 1.101–4.636 |
| High,
≥0.4 | 17 | 19.5 |
|
|
|
| miR-92b-3p |
|
|
|
|
|
| Low,
<0.4 | 17 |
9.6 | 0.0070 | 2.215 | 1.178–4.166 |
| High,
≥0.4 | 24 | 19.9 |
|
|
|
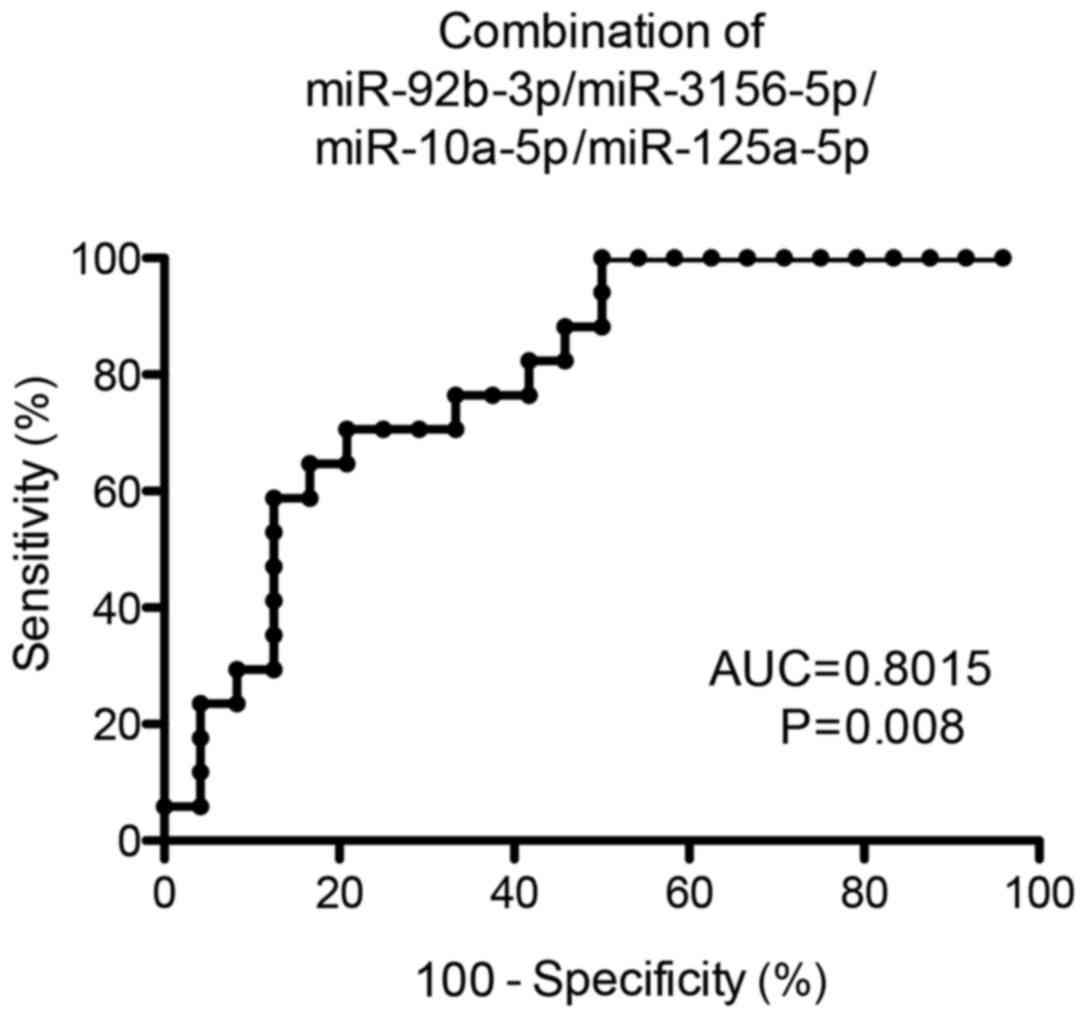
Subsequent ROC analysis revealed that the usage of
4-miRNA-combined diagnostic score, as established by a
bidirectional stepwise logistic regression, enabled the
identification of responders to bevacizumab/FOLFOX therapy with a
sensitivity of 82% and specificity of 64% [area under the curve
(AUC) = 0.8015; P=0.008, cut-off value = −0.466; Fig. 3]. Based on this diagnostic score, the
median PFS in responders was 20 months vs. 7 months for
non-responders.
Discussion
The aim of the present study was to identify
candidate miRNAs that would assist in identifying patients with
mCRC who were likely to respond to targeted antiangiogenic
treatment in combination with a FOLFOX regimen. Oxaliplatin-based
chemotherapy combined with bevacizumab is a common choice for
patients with mCRC. Unfortunately, the benefit for patients is
rather limited as at present, it has not been possible to identify
patients most likely to benefit from this regimen (3). The results of the present indicate that
specific miRNAs exhibit a predictive value in this setting.
By global miRNA microarray profiling of FFPE tumor
tissues and comparing miRNA expression between responders and
non-responders to bevacizumab/FOLFOX therapy, a total of 67 miRNAs
with significantly different expression were identified. However,
only 7 miRNAs fulfilled the criteria for selection of candidate
miRNAs for validation. These 7 miRNA were further investigated in
the validation phase of the present study. The independent
validation of these miRNAs in the cohort of 41 patients with mCRC
confirmed the ability to distinguish patients with different
responses to bevacizumab/FOLFOX treatment only in the case of 4
miRNAs (miR-125a-5p, miR-10a-5p, miR-3156-5p and miR-92b-3p). In
the group of responders, significantly higher levels of these 4
miRNAs was observed compared with non-responders. The analytical
performance of the individual miRNAs was unsatisfactory (AUC
<0.75). However when analyzed in combination, these 4 miRNAs
enabled the sensitive and specific identification of patients with
mCRC responsive to bevacizumab/FOLFOX treatment (AUC = 0.8015;
sensitivity, 82%; specificity, 64%). Notably, Boisen et al
(12) did not identify any of the
predictive miRNAs of the present study, and observed only
miR-664-3p and miR-455-5p to be associated with overall survival in
patients with mCRC treated with capecitabine, oxaliplatin and
bevacizumab.
As the course of the survival curves appears similar
for all significant miRNAs identified by the present study, it is
hypothesized that the miRNAs are associated with the same
biological feature, for example high angiogenic density,
responsible for sensitivity to anti-angiogenic treatment. In
addition, these miRNAs may share identical mechanism of secondary
resistance, represented by the significant increase in progression
rate following 20 months of therapy.
Association between miR-10a-5p expression and
clinical parameters in CRC has been described previously (20). The high expression of miR-10b-5p has
been associated with tumors located in the right colon relative to
the left colon and rectum, but no association to metastasis-free
survival has been observed (20). In
agreement with the observations of the present study, the
expression levels of miR-10a-5p in tumor FFPE samples were
demonstrated to be >2-fold higher in progession-free patients
compared with patients with progressive disease in bladder cancers
(21). The prognostic significance of
miR-10a-5p was also confirmed in acute myeloid leukemia, where the
serum expression levels of this miRNA were significantly higher in
comparison with a control group (22). In patients with recurrent
platinum-resistant ovarian cancer, higher expression levels of
miR-10a-5p were noted in peripheral blood plasma and associated
with a sensitivity to decitabine followed by carboplatin
chemotherapy (23).
Changes in miR-125a-5p expression levels have been
identified in several types of cancer. Gattolliat et al
(24) and Ozcan et al
(25) suggested that miR-125a-5p was
deregulated in CRC tissue samples. In addition, downregulated
miR-125a-5p was significantly associated with gastric cancer
metastases (26) and was
significantly downregulated in the serum of patients with lung
cancer (P<0.001) (27), indicating
its potential tumor-suppressive role in cancer. In the present
study, a higher level of miR-125a-5p were observed to be associated
with an improved outcome in patients with mCRC treated with
bevacizumab/FOLFOX. Understanding the processes accompanying
hypoxia is one of the main goals of studies investigating cancer,
as it is known that hypoxia serve a role in the resistance to
chemotherapy and radiotherapy (28).
Using the grade IV glioma U87MG cell line, it was demonstrated that
miR-92b-3p was downregulated by >2-fold under hypoxic conditions
(29), suggesting miR-92-3p
expression to be indicative of hypoxic environment. At present,
miR-3156-5p expression has not been described in association with
cancer.
There are certain potential limitations of the
present study, and a number of issues need to be addressed in order
to establish miRNAs as clinical diagnostic and predictive tools.
The validation cohort in the present study was not large enough to
support the clinical utility of the 4-miRNA predictive panel
identified. Additionally, all samples were enrolled from a single
center, and all patients were of the same ethnicity (European
descent). There is a need for a prospective, multi-centric study
involving large independent cohorts. As the levels of circulating
miR-126 have been observed to be associated with the response to
bevacizumab treatment (14), it would
be also of interest to evaluate the predictive value of the tissue
miRNAs identified in the predictive panel from the present study in
peripheral blood or blood serum/plasma samples, as a potentially
non-invasive diagnostic option.
In conclusion, 4 miRNAs (miR-92b-3p, miR-3156-5p,
miR-10a-5p and miR-125a-5p) were identified to exhibit higher
expression levels in the tumors of patients who responded to
bevacizumab/FOLFOX therapy compared with non-responders. Subsequent
to further independent validations, detection of these miRNA would
enable the identification of patients with mCRC who may potentially
benefit from this therapy. These miRNAs may also serve as novel
candidates for targeted therapeutic interventions in patients with
mCRC, even though the role of these miRNAs and the signaling
pathways these miRNAs regulate remain to be clarified.
Acknowledgements
The present study was financially supported by the
Czech Ministry of Health (grant no. 16-31765A), project MZ CR-RVO
(grant no. MOU, 00209805) and the Ministry of Education, Youth and
Sports of the Czech Republic under the project Central European
Institute of Technology 2020 (grant no. LQ1601).
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Can. 136:E359–E386. 2014.
View Article : Google Scholar
|
|
2
|
van Cutsem E, Nordlinger B and A; ESMO
Cervantes; Guidelines Working Group, : Advanced colorectal cancer:
ESMO Clinical Practice Guidelines for treatment. Ann Oncol. 21
Suppl 5:v93–v97. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Saltz LB, Clarke S, Díaz-Rubio E,
Scheithauer W, Figer A, Wong R, Koski S, Lichinitser M, Yang TS,
Rivera F, et al: Bevacizumab in combination with oxaliplatin-based
chemotherapy as first-line therapy in metastatic colorectal cancer:
A randomized phase III study. J Clin Oncol. 26:2013–2019. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bennouna J, Sastre J, Arnold D, Österlund
P, Greil R, van Cutsem E, von Moos R, Viéitez JM, Bouché O, Borg C,
et al: Continuation of bevacizumab after first progression in
metastatic colorectal cancer (ML18147): A randomised phase 3 trial.
Lancet Oncol. 14:29–37. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hurwitz HI, Tebbutt NC, Kabbinavar F,
Giantonio BJ, Guan ZZ, Mitchell L, Waterkamp D and Tabernero J:
Efficacy and safety of bevacizumab in metastatic colorectal cancer:
Pooled analysis from seven randomized controlled trials.
Oncologist. 18:1004–1012. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pai SG and Fuloria J: Novel therapeutic
agents in the treatment of metastatic colorectal cancer. World J
Gastrointest Oncol. 8:99–104. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Spiliopoulou P and Arkenau HT: Rationally
designed treatment for metastatic colorectal cancer: Current drug
development strategies. World J Gastroenterol. 20:10288–10295.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bhartiya D and Scaria V: Genomic
variations in non-coding RNAs: Structure, function and regulation.
Genomics. 107:59–68. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vychytilova-Faltejskova P, Radova L,
Sachlova M, Kosarova Z, Slaba K, Fabian P, Grolich T, Prochazka V,
Kala Z, Svoboda M, et al: Serum-based microRNA signatures in early
diagnosis and prognosis prediction of colon cancer. Carcinogenesis.
37:941–950. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mishra PJ: MicroRNAs as promising
biomarkers in cancer diagnostics. Biomark Res. 2:192014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cekaite L, Eide PW, Lind GE, Skotheim RI
and Lothe RA: MicroRNAs as growth regulators, their function and
biomarker status in colorectal cancer. Oncotarget. 7:6476–6505.
2016.PubMed/NCBI
|
|
12
|
Boisen MK, Dehlendorff C, Linnemann D,
Nielsen BS, Larsen JS, Osterlind K, Nielsen SE, Tarpgaard LS,
Qvortrup C, Pfeiffer P, et al: Tissue microRNAs as predictors of
outcome in patients with metastatic colorectal cancer treated with
first line capecitabine and oxaliplatin with or without
bevacizumab. PLoS One. 15:e1094302014. View Article : Google Scholar
|
|
13
|
Hansen TF, Carlsen AL, Heegaard NH,
Sørensen FB and Jakobsen A: Changes in circulating microRNA-126
during treatment with chemotherapy and bevacizumab predicts
treatment response in patients with metastatic colorectal cancer.
Br J Cancer. 112:624–629. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carvalho BS and Irizarry RA: A framework
for oligonucleotide microarray preprocessing. Bioinformatics.
26:2363–2367. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
R Development Core Team, . R: a language
and environment for statistical computing. R Foundation for
Statistical Computing; Vienna, Austria: http://www.r-project.org/March 31–2015
|
|
17
|
Smyth GK: Limma: linear models for
microarray dataBioinformatics and Computational Biology Solutions
using R and Bioconductor. Gentleman R, Carey V, Dudoit S, Irizarry
R and Huber W: Springer; New York, NY: pp. 397–420. 2005,
View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vandesompele J, De Preter K, Pattyn F,
Poppe B, van Roy N, De Paepe A and Speleman F: Accurate
normalization of real-time quantitative RT-PCR data by geometric
averaging of multiple internal control genes. Genome Biol.
3:RESEARCH0034. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schee K, Lorenz S, Worren MM, Günther CC,
Holden M, Hovig E, Fodstad O, Meza-Zepeda LA and Flatmark K: Deep
sequencing the microRNA transcriptome in colorectal cancer. PLoS
One. 8:e661652013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Segersten U, Spector Y, Goren Y, Tabak S
and Malmström PU: The role of microRNA profiling in prognosticating
progression in Ta and T1 urinary bladder cancer. Urol Oncol.
32:613–618. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhi Y, Xie X, Wang R, Wang B, Gu W, Ling
Y, Dong W, Zhi F and Liu Y: Serum level of miR-10-5p as a
prognostic biomarker for acute myeloid leukemia. Int J Hematol.
102:296–303. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Benson EA, Skaar TC, Liu Y, Nephew KP and
Matei D: Carboplatin with decitabine therapy, in recurrent platinum
resistant ovarian cancer, alters circulating mirnas concentrations:
A pilot study. PLoS One. 10:e01412792015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gattolliat CH, Uguen A, Pesson M, Trillet
K, Simon B, Doucet L, Robaszkiewicz M and Corcos L: MicroRNA and
targeted mRNA expression profiling analysis in human colorectal
adenomas and adenocarcinomas. Eur J Cancer. 51:409–420. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ozcan O, Kara M, Yumrutas O, Bozgeyik E,
Bozgeyik I and Celik OI: MTUS1 and its targeting miRNAs in
colorectal carcinoma: Significant associations. Tumour Biol.
37:6637–6645. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu Y, Huang Z and Liu Y: Reduced
miR-125a-5p expression is associated with gastric carcinogenesis
through the targeting of E2F3. Mol Med Rep. 10:2601–2608.
2014.PubMed/NCBI
|
|
27
|
Ferracin M, Lupini L, Salamon I, Saccenti
E, Zanzi MV, Rocchi A, da Ros L, Zagatti B, Musa G, Bassi C, et al:
Absolute quantification of cell-free microRNAs in cancer patients.
Oncotarget. 6:14545–14555. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Muz B, de la Puente P, Azab F and Azab AK:
The role of hypoxia in cancer progression, angiogenesis,
metastasis, and resistance to therapy. Hypoxia (Auckl). 3:83–92.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Agrawal R, Pandey P, Jha P, Dwivedi V,
Sarkar C and Kulshreshtha R: Hypoxic signature of microRNAs in
glioblastoma: Insights from small RNA deep sequencing. BMC
Genomics. 15:6862014. View Article : Google Scholar : PubMed/NCBI
|