Introduction
MicroRNAs (miRNAs or miRs) are small non-coding RNAs
(~22 nucleotides) that serve important roles in the regulation of
gene expression at the post-transcriptional level. The regulation
process can occur by degradation or inhibition of translation of
their target messenger RNAs through base pairing at 3′ or 5′
untranslated regions (1,2). miRNAs are involved in important
biological processes such as cell differentiation, proliferation
and death (3). Furthermore,
dysregulation of miRNA expression is associated with multiple
diseases, including several types of cancer such as lymphoma,
glioblastoma, and colorectal, lung and breast cancer (BC) (4).
The importance of miRNAs in cancer is supported by
the fact that ~50% of miRNAs are located at fragile sites or
genomic regions associated with diseases (5–7), as well
as by their ability to act as tumor suppressors or oncogenes
(8–10). Additionally, miRNA expression
signatures can be used to clearly categorize different solid tumors
(11). In BC, miRNA signatures can
help to discriminate between normal and cancer tissues (12). Furthermore, miRNAs are involved in
cancer initiation and progression, consequently regulating the
processes of invasion and metastasis in several types of cancer
(13,14). miR-10b and miR-335 were the first
miRNAs to be described as promoters or inhibitors of metastasis,
respectively (15). In BC, certain
miRNAs have been associated with the invasion-metastasis cascade,
including let-7, miR-10b, miR-21, miR-34, miR-200 and miR-335
(16).
In a previous study, our group demonstrated that
miR-183 and miR-494 are potential biomarkers of metastasis in BC
(17). miR-183 is located on
chromosome 7q32 and is dysregulated in several types of cancer,
including BC, hepatic, colorectal and lung cancer, as well as
leukemia and osteosarcoma (18). A
recent study reported that miR-183 is downregulated in
retinoblastoma tumor tissues and cell lines compared with its
expression in their normal counterparts (19). Furthermore, ectopic expression of
miR-183 inhibited cell migration and invasion in vitro
(19). Similarly, miR-494 was
observed to be downregulated in human gastric carcinoma tissues and
cell lines, in which the restoration of miR-494 expression
decreased the cell proliferation rate (20). These findings indicate that both
miRNAs are implicated in the regulation of biological processes
that are important in cancer progression, thus demonstrating the
requirement for further studies to evaluate their involvement in
other cancer types. Therefore, the present study evaluated the
expression and functional role of miR-183 and miR-494 in BC cell
lines, uncovering new evidence of the involvement of both miRNAs in
BC onset and progression.
Materials and methods
Cell culture
Seven BC cell lines (MCF-7, MCF-7/AZ, T47D, BT-20,
Hs578T, MDA-MB-231 and MDA-MB-468) were evaluated in the present
study. The cell lines, which were a kind donated by Dr Rui M. Reis
(Barretos Cancer Hospital, Barretos, Brazil) in August 2012, were
cultured in Dulbecco's modified Eagle's medium (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) supplemented with 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
1% penicillin-streptomycin solution (Sigma-Aldrich; Merck KGaA).
Cells were maintained at 37°C in a humidified 5% CO2
atmosphere.
Authentication of all BC-derived cell lines was
performed by short tandem repeat DNA typing according to the
International Reference Standard for Authentication of Human Cell
Lines, as previously reported (21).
All the cell lines had their genotype confirmed.
Total RNA isolation
Total RNA was isolated using TRIzol reagent (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
RNA quantification was performed using a NanoDrop ND-1000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc., Wilmington, DE, USA), and RNA quality was assessed using RNA
6000 Nano chips (Agilent Technologies, Inc., Santa Clara, CA, USA)
in a 2100 Bioanalyzer (Agilent Technologies, Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
miRNA expression was assessed by RT-qPCR using
TaqMan® MicroRNA Assays (Thermo Fisher Scientific,
Inc.). Reverse transcription of primers to the sequence of miR-183
(5′-UAUGGCACUGGUAGAAUUCACU-3′) and miR-494,
(5′-AGGUUGUCCGUGUUGUCUUCUCU-3′) and the complementary DNA synthesis
was performed with total RNA (10 ng) using TaqMan® Small
RNA Assays (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol.
The PCR with a final volume of 10 µl was performed
at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and
60°C for 1 min. All reactions were performed in triplicate a 7900HT
Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The analyses were performed using the R
statistical computing environment (https://www.r-project.org) according to the
2−ΔΔCq method (22). The
small non-coding nucleolar RNA RNU48 provided in the
TaqMan® Control MicroRNA Assay kit (Thermo Fisher
Scientific, Inc.) was used as a control for the analysis.
miRNA transfection
Homo sapiens (hsa)-miR-183 and hsa-miR-494
Pre-miR™ miRNA Precursors (Ambion; Thermo Fisher Scientific, Inc.)
were transfected into MDA-MB-231 and MDA-MB-468 BC cells using
siPORT™ Amine Transfection Agent (Thermo Fisher Scientific, Inc.)
at a final concentration of 20 nM, according to the manufacturer's
protocol. Cells transfected with the Pre-miR™ miRNA Precursor
negative control (Ambion; Thermo Fisher Scientific, Inc.) were used
as a control for the transfection. The efficiency of overexpression
of miR-183 and miR-494 was evaluated at 24, 48 and 72 h after
transfection by RT-qPCR.
Proliferation and migration
assays
Proliferation and migration assays were carried out
in the xCELLigence RTCA DP Instrument (Roche Applied Science,
Rotkreuz, Switzerland), using the E-Plate for measuring
proliferation and the CIM-Plate for measuring migration (both from
Roche Applied Science). The xCELLigence system transforms
automatically the impedance of electron flow caused by cells in a
cell index (CI) value according to the formula CI = (impedance at
time point n-impedance in the absence of cells) / nominal impedance
value (23). The experiments were
performed according to the manufacturer's protocol. For the
proliferation assay, concentrations of 6×103 and
8×103 MDA-MB-231 and MDA-MB-468 cells, respectively,
were used. For the migration assay, 2×104 cells were
used for both BC cell lines. The CI value was monitored for 72 h in
the proliferation assay and for 24 h in the migration assay.
In silico target prediction of miR-183
and miR-494
Target prediction was performed using the
miRWalk2.0, Micro T4, miRMap, PITA and RNAhybrid algorithms in the
comparative platform of miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/micrornapredictedtarget.html).
The criteria for miRNA target gene selection was identification in
the above algorithms and presence in both miRNAs. The targets were
then filtered by Breast Neoplasm (code C2910) National Cancer
Institute disease term using the cancer gene index of Reactome FI
Cytoscape Plugin 4 (http://wiki.reactome.org/index.php/Reactome_FI_Cytoscape_Plugin).
Finally, targets were enriched by biological process using BiNGO
Cytoscape plugin using a hypergeometric test with false discovery
rate correction (P≤0.05) with the GoSlim Gene Ontology dataset
(24).
Western blot assay
Cells transfected with each miRNA were lysed
directly on ice using lysis buffer (50 mM Tris pH 7.6–8.0, 150 mM
NaCl, 5 mM EDTA, 1 mM Na3VO4, 10 mM NaF, 10
mM sodium pyrophosphate and 1% NP-40) supplemented with a protease
inhibitor cocktail (1 mM dithiothreitol, 1 µg/ml leupeptin
hemisulfate, 1 µg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride
and 1 mM EDTA). The lysates were centrifuged at 10,000 × g for 15
min at 5°C and the supernatant was stored at −80°C. The soluble
proteins were quantitated using Bradford Reagent (Sigma-Aldrich;
Merck KGaA) according to the manufacturer's protocol. Total protein
(20 µg) was separated by SDS-PAGE (10% gel) and transferred onto a
Hybond-C-supported nitrocellulose membrane (GE Healthcare Life
Sciences, Chalfont, UK) using a Mini-PROTEAN Trans-Blot®
Turbo Transfer System (Bio-Rad Laboratories, Inc., Hercules, CA,
USA) at 100 V for 30 min. Membranes were blocked using TBS with
0.1% Tween-20 and 5% powdered milk at room temperature for 1 h, and
then incubated at 5°C overnight in a 1:500-dilution of anti-RB1
mouse polyclonal antibody (#sc-102, Santa Cruz Biotechnology, Inc.
MA, USA) in blocking buffer. Upon washing with TBS-T (0.1%), the
membranes were probed using an anti-mouse anti-rabbit secondary
antibody (#7076, Cell Signaling Technology, Inc., Danvers, MA, USA)
at dilution 1:5,000 for 2 h at room temperature. Subsequently,
specific binding was detected with enhanced chemiluminescence (GE
Healthcare Life Sciences). The detection of the chemiluminescent
signal was performed in the gel documentation system ImageQuant LAS
4000 Mini (GE Healthcare Life Sciences). Subsequently, the labeled
bands were analyzed and quantified using ImageJ software (version
1.49v; NIH-Scion Corporation, Bethesda, MD, USA) and bar graphs
represent densitometric analysis of each band that were plotted
using the gplots R package (https://cran.r-project.org/web/packages/gplots/index.html)
(25,26). Normalization was performed using
β-tubulin as the endogenous control (#3873, Cell Signaling
Technology, Inc.) at dilution 1:5,000 for 2 h at room
temperature.
Statistical analysis
Statistical analysis was performed using the R
statistical computing environment (https://www.r-project.org). Data are displayed as mean
values ± standard error of ≥3 independent experiments. Differences
were evaluated with the Student's t-test. P<0.05 was considered
to indicate a significant difference.
Results
Overexpression and functional analysis
of mir-183 in BC cell lines
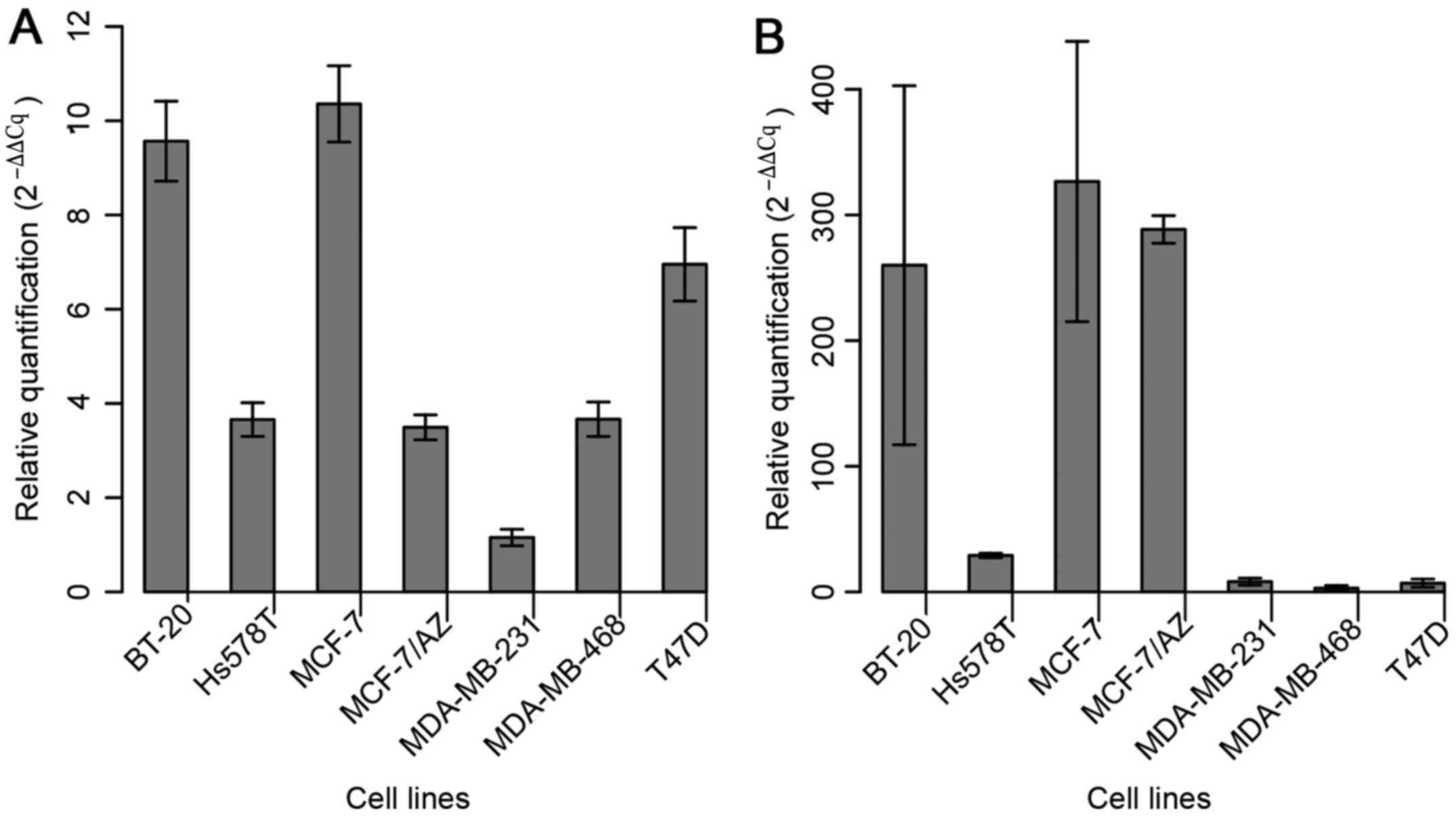
The expression of miR-183 was assessed in the
aforementioned seven BC cell lines in order to select those with
the lowest levels of miR-183 to further evaluate its role in
gain-of-function assays (Fig. 1A).
MDA-MB-231 cells exhibited the lowest basal level of miR-183;
considering that this cell line represents a model of the
basal-like molecular subtype (more aggressive) of BC, it was
selected for the functional approach. Among the cell lines that
displayed the second lowest level of expression (Hs578T, MCF-7/AZ
and MDA-MB-468), MDA-MB-468 was selected due to its low expression
level of miR-494 (Fig. 1B).
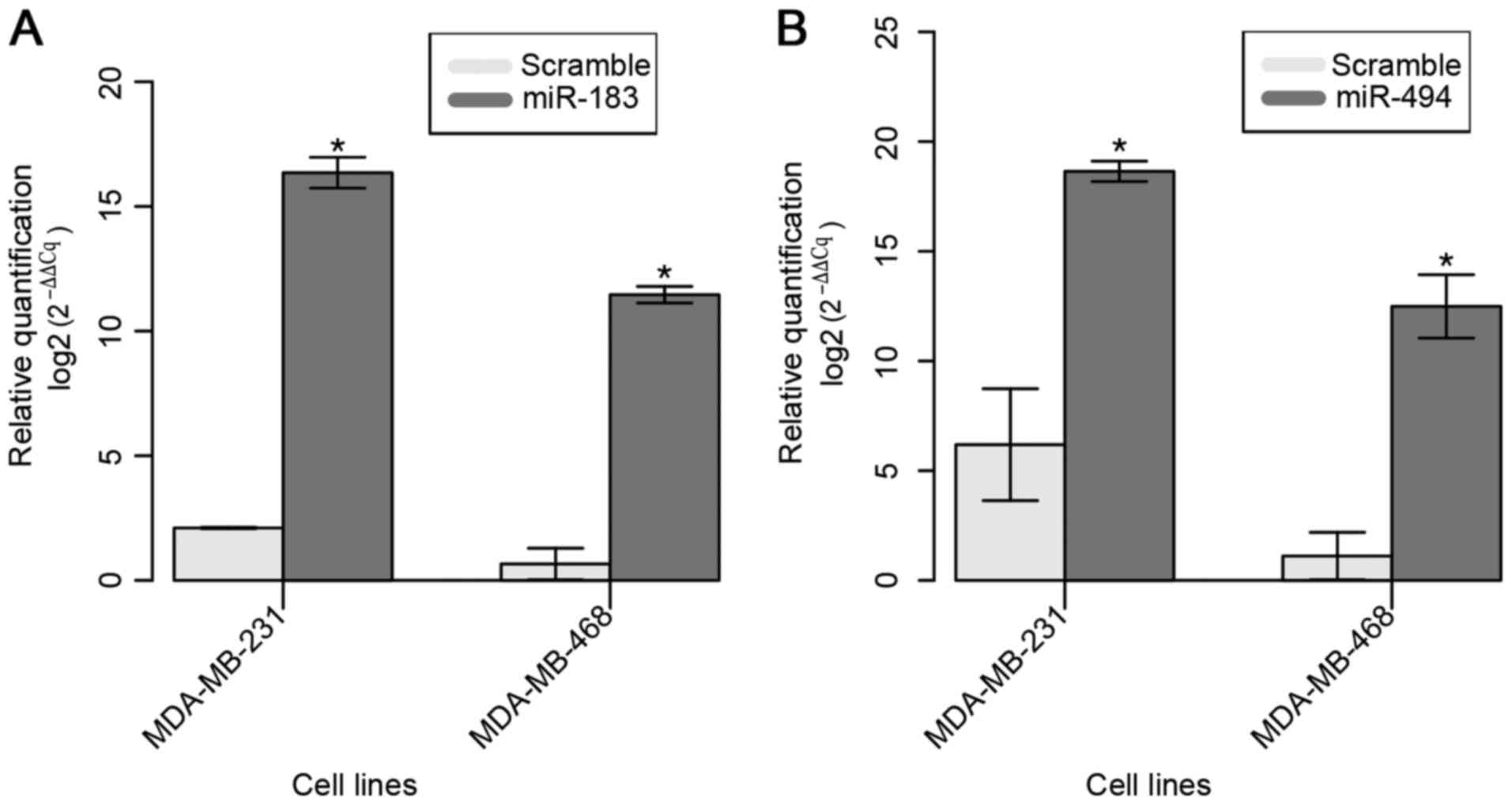
The restoration of miR-183 expression in the
selected two BC cell lines was confirmed 24 h after Pre-miR™ miRNA
Precursor transfection (Fig. 2A). A
mean 15-fold increase in the expression of miR-183 was observed in
transfected cell lines compared with that in parental cell
lines.
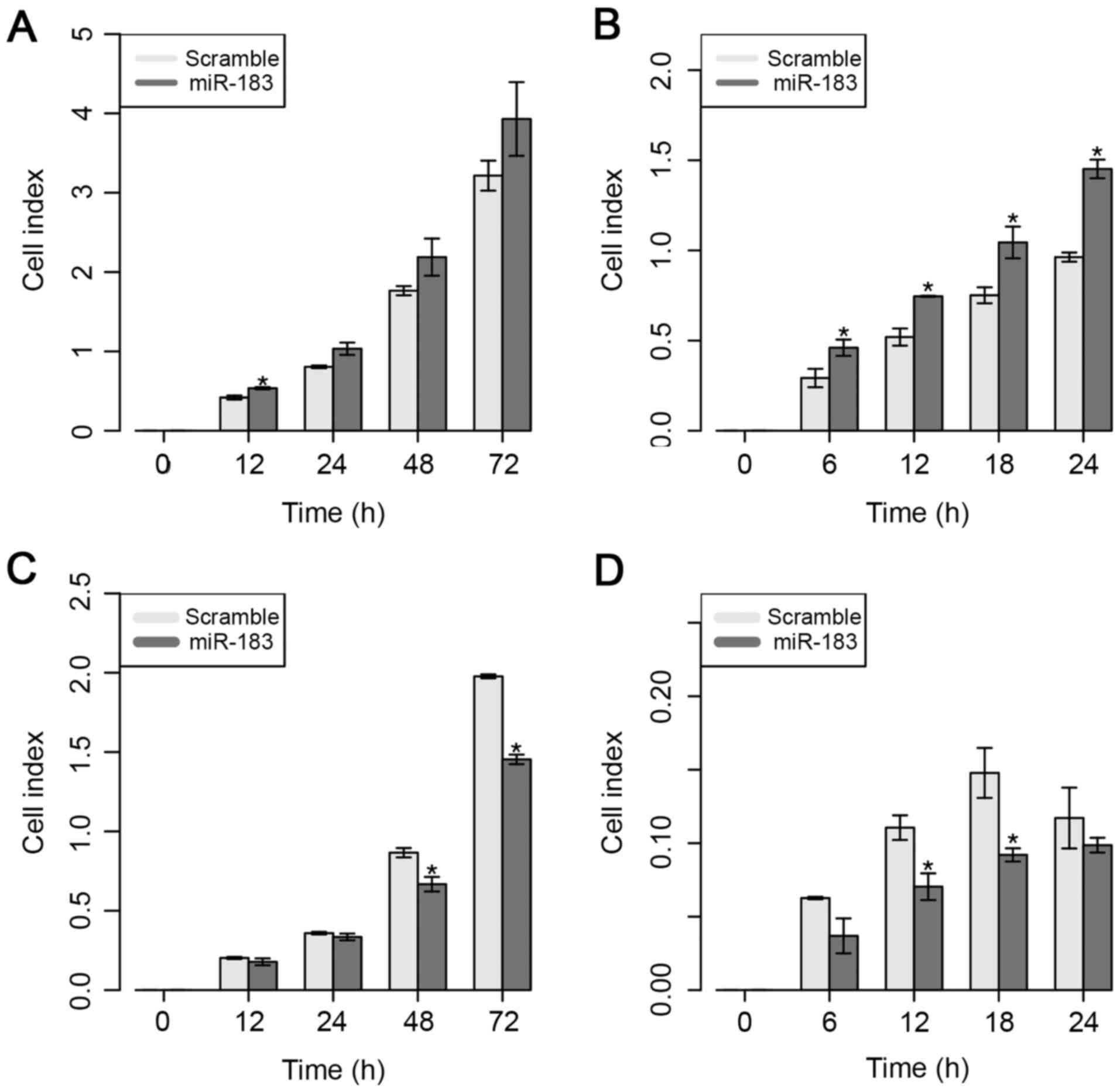
Next, the effect of ectopic expression of miR-183 on
the proliferative and migratory capacities of MDA-MB-231 cells was
evaluated, as shown in Fig. 3.
Overexpression of miR-183 induced a significant increase in
proliferative capacity in vitro at 12 h; however, a higher
proliferative rate at 24 and 48 h was also observed, even though
these findings were not significant (Fig.
3A). Regarding the migratory ability in vitro, the
present results demonstrated that the overexpression of miR-183
significantly increased the migratory ability from 6 to 24 h
post-transfection (Fig. 3B).
By contrast, overexpression of miR-183 significantly
reduced cell proliferation at 48 and 72 h in the MDA-MB-468 cell
line (Fig. 3C). Similarly, the
present results demonstrated that overexpression of miR-183 also
significantly inhibited cell migration at 12 and 18 h
post-transfection (Fig. 3D). These
results suggest that the overexpression of miR-183 significantly
increased the proliferative and migratory abilities of both cell
lines, which suggests the potential role of this miRNA in BC
cellular behavior.
Overexpression and functional analysis
of miR-494 in the MDA-MB-231 BC-derived cell line
The aforementioned seven BC cell lines were used to
evaluate the basal expression of miR-494. The results indicated
that the MDA-MB-231 and MDA-MB-468 cell lines had the lowest basal
levels of miR-494; therefore, these lines were selected for the
functional evaluation of this miRNA (Fig.
1B).
The restoration of miR-494 expression in both cell
lines was confirmed at 24 h after Pre-miR™ miRNA Precursor
transfection (Fig. 2B). A mean
increase of ~10-fold in miR-183 expression was observed compared
with that in the parental cell lines.
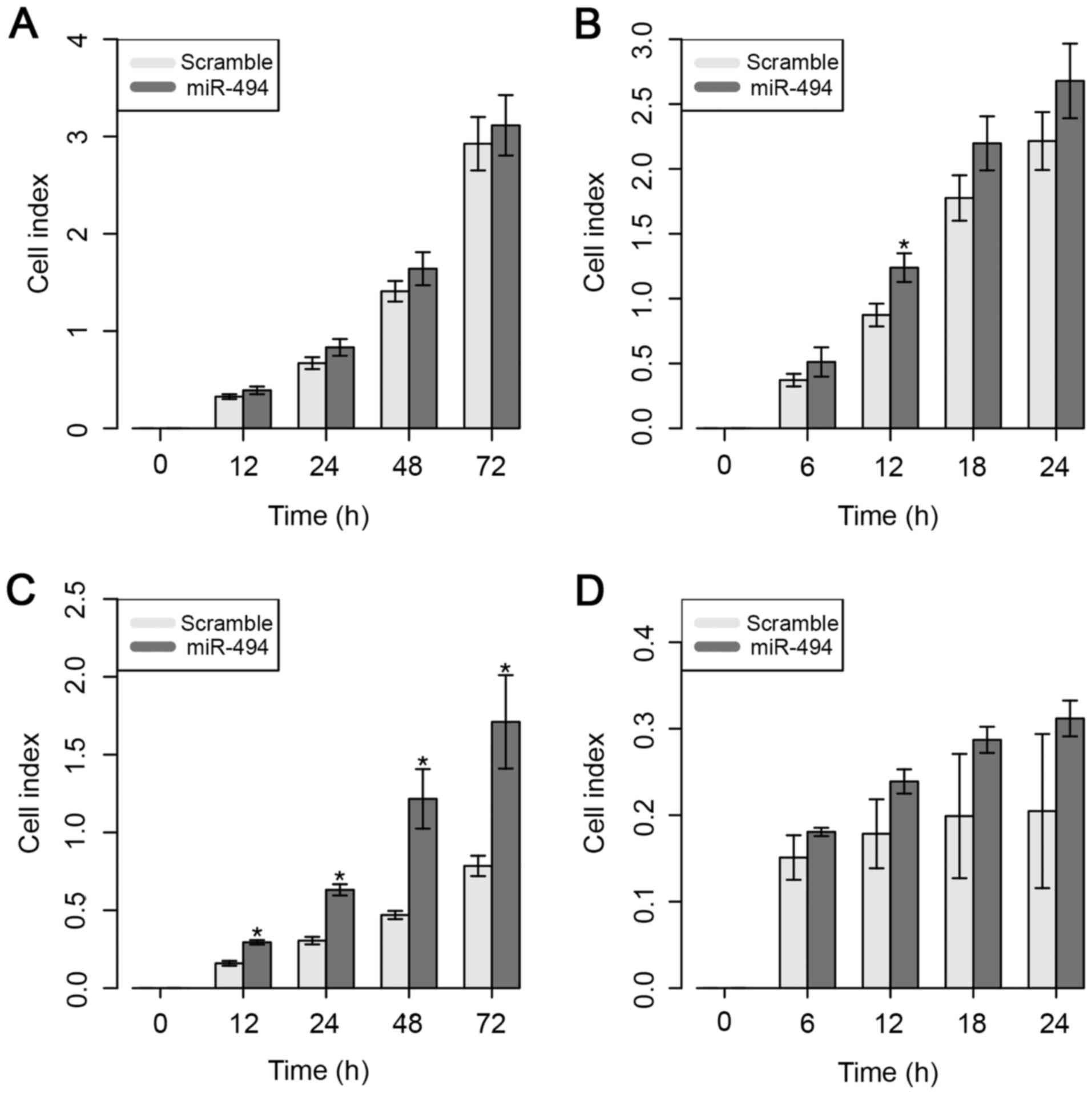
Despite the higher CI value, the ectopic expression
of miR-494 did not result in a significant difference in the cell
proliferative ability of MDA-MB-231 cells (Fig. 4A). However, overexpression of miR-494
significantly increased the cell migratory capacity of MDA-MB-231
cells, but only at 12 h after in vitro transfection
(Fig. 4B).
The results for the MDA-MB-468 cell line
demonstrated that cell proliferation was significantly increased 12
h after overexpression of miR-494 (Fig.
4C). However, no significant impact on cell migratory ability
was observed in this cell line (Fig.
4D).
These results suggest that the overexpression of
miR-494 does not necessarily control the ability of the cells to
migrate, but appears to be involved in the significant inhibition
of cell proliferative ability in the MDA-MB-231 cell line.
Identification and evaluation of RB1
protein expression by western blotting
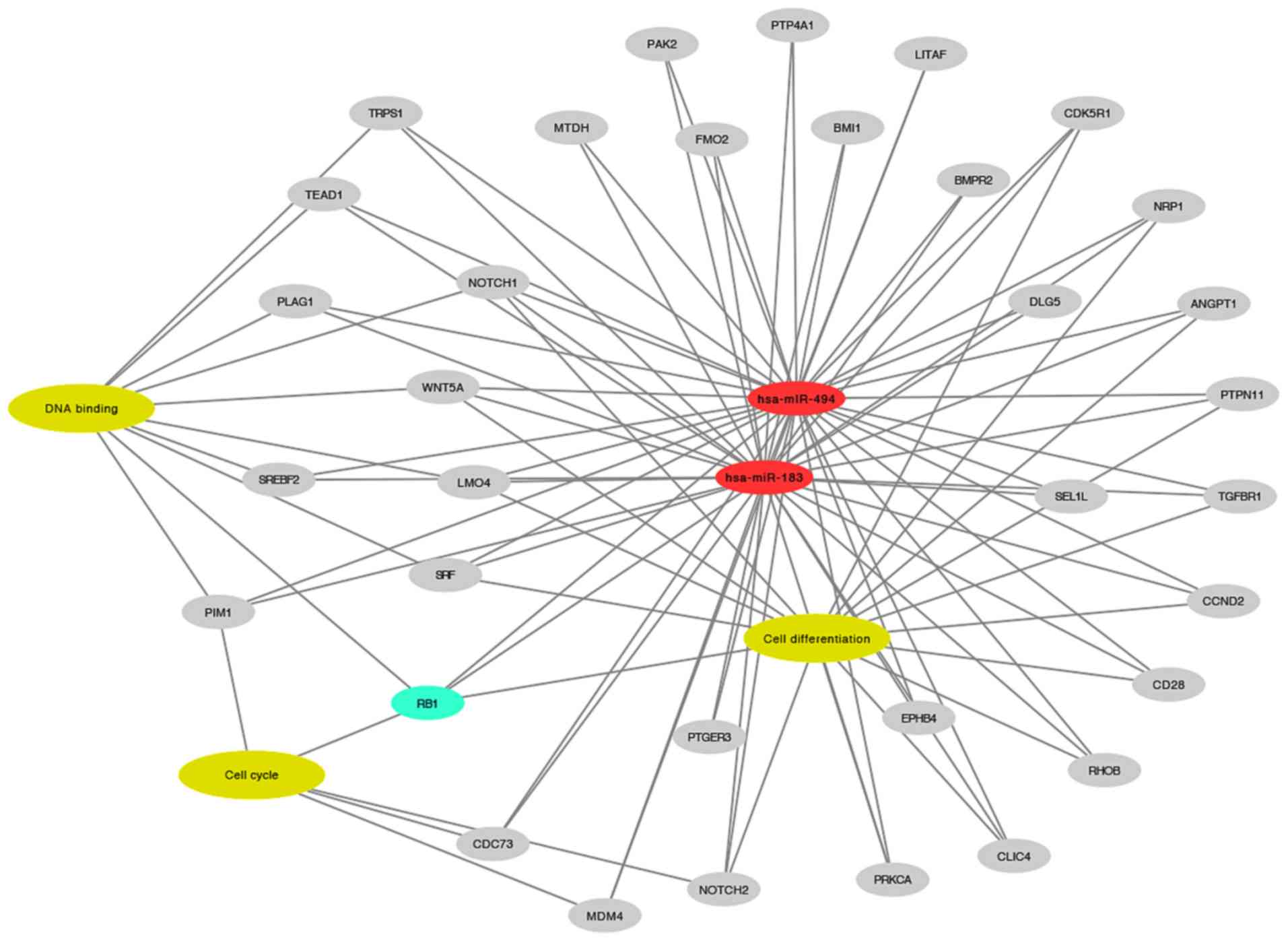
In silico analysis highlighted several
targets of miR-183 and miR-494, as shown in Fig. 5. RB1 was selected for western blot
validation due to its involvement in enriched biological processes
associated with BC, including cell differentiation
(P=4.09×10−7), cell cycle (P=3.58×10−3) and
DNA binding (P=2.65×10−2). Among these three processes,
RB1 was the only target identified to be involved in cell
differentiation and cell cycle progression.
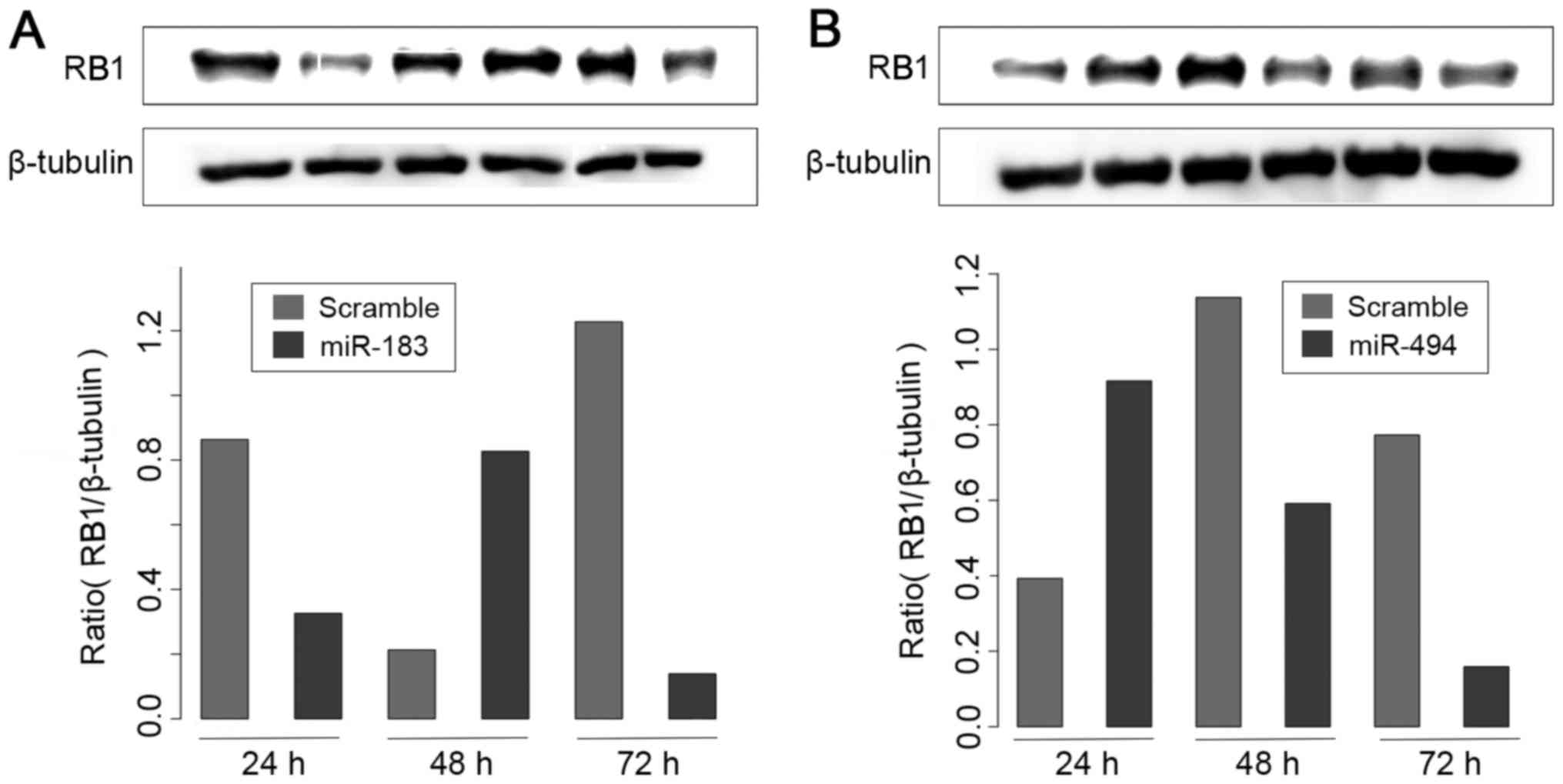
It was observed that transfection of miR-183
resulted in a significant reduction in RB1 protein levels in
MDA-MB-231 cells (Fig. 6A). By
contrast, miR-494 transfection did not produce an effect on RB1
protein levels (Fig. 6B). These
results suggest that miR-183 may act as a potential regulator of
RB1 protein, at least in a subset of BC cell lines.
Discussion
Several miRNAs have been associated with BC
progression (12). However, the
mechanism involved in miRNA-controlled metastasis is not completely
understood yet (16). Evidence from a
previous study by our group (17) and
from studies about the roles of miR-183 and miR-494 in other cancer
types (18–20) suggests that these miRNAs are
dysregulated in BC.
Considering the expression of these two miRNAs in a
panel of BC cell lines, MDA-MB-231 and MDA-MB-468 were selected for
functional ectopic overexpression assays due to the low levels of
expression of both miRNAs in these two cell lines. According to
Subik et al (27), the
MDA-MB-231 cell line is characterized as a triple-negative/basal B
mammary carcinoma. Haga and Phinney (28) evaluated the expression of miR-494 in
MDA-MB-231 cells by qPCR and revealed a low expression level, thus
corroborating the findings of the present study. Additionally,
Lowery et al (29) also
performed in vitro overexpression of miR-183 but in the T47D
cell line. According to the present findings, the T47D cell line
was not the best model for the study, compared with the other BC
cell lines tested. Indeed, the authors observed that overexpression
of miR-183 in the T47-D cell line resulted in an increase in
migration, but without a significant difference when compared with
that exhibited by the negative control (29).
For the functional assays, the xCELLigence System
was selected in the present study. Limame et al (30) performed a comparative analysis between
the xCELLigence System and conventional functional cellular assays,
showing a strong correlation (>90%) between the two methods. The
present results demonstrated that the ectopic overexpression of
miR-183 affected the behavior of the MDA-MB-231 and MDA-MB-468 cell
lines in a contradictory manner. Overexpression of miR-183 resulted
in an increase in the proliferative and migratory capacities of the
cells over time (proliferation was significantly increased only at
12 h). By contrast, in MDA-MB-468 cells, the overexpression of
miR-183 inhibited cell proliferation after 48 and 72 h, and cell
migration after 12 and 18 h compared with the control (scramble
miRNA). Similarly, Zhao et al showed that the overexpression
of miR-183 in an osteosarcoma cell line with a high invasive
capacity (F5M2) led to the inhibition of cell migration and
invasion, which is partly in agreement with the results obtained in
MDA-MB-468 cells (18).
Overexpression of miR-494 had no significant effect on cell
proliferation in the MDA-MB-231 cell line; however, an increase in
cell migratory ability was observed after 12 h of overexpression
in vitro. The overexpression of miR-494 increased the cell
proliferative capacity of MDA-MB-468 cells after 12 h, but did not
affect their migration. Accordingly, the results obtained with
overexpression of miR-494 in the MDA-MB-231 cells were similar to
those of a recent study, which noticed that the ectopic expression
of this miRNA inhibited cell proliferation in a lung cancer cell
line (A549) (31).
The differences in proliferative and migratory
behavior between the two cell lines evaluated in the present study
can be explained on the basis of their molecular subtype, since the
MDA-MB-231 cell line is an invasive ductal carcinoma, while the
MDA-MB-468 cell line is a metastatic basal-like adenocarcinoma
(32). Consequently, these cell lines
have different genotypes with specific mutations in genes that
control the mechanisms of cell proliferation and apoptosis,
including cyclin-dependent kinase (CDK) inhibitor 2A, BRAF, KRAS,
tumor protein P53, phosphatase and tensin homolog, neurofibromin 2,
RB1 and SMAD family member 4 (33,34). It
has been widely reported that various MDA-MB-468 clones have a
homozygous deletion of RB1, which can explain such differences in
cellular behavior (35) but this
finding has no influence on the results of the present study.
Among the genes evaluated in the present study,
miR-183 and miR-494 possibly regulated the expression of RB1, a
tumor suppressor that negatively regulates the cell cycle. Cell
cycle control by RB1 involves other factors, including
CDKs-cyclins, which phosphorylate the RB1 protein, thus rendering
it inactive. Inactive RB1 subsequently releases E2 factor, a
transcription factor that activates the transcription of several
genes that promote the cell cycle transition from G1 to S phase or
activate cell proliferation (36). In
the present study, RB1 protein expression was negatively regulated
at 72 h after overexpression of miR-183 and miR-494 in the
MDA-MB-231 cell line. Thus, the inhibition of proliferation induced
by miR-183 in MDA-MB-231 cells may be modulated via the
downregulation of RB1.
In conclusion, the present study provided evidence
that miR-183 and miR-494 may be associated with the progression of
BC. Furthermore, the present results suggest a new perspective for
future research on the potential role of these miRNAs in the
regulation of key metastatic biological processes during BC
progression.
Acknowledgements
The authors thank Dr Rene A.C. Vieira (Barretos
Cancer Hospital, Brazil) for his suggestions. The present study was
supported by Fundação de Amparo à Pesquisa do Estado de São
Paulo-FAPESP, São Paulo, Brazil (grant nos. 2010/16796-0 and
2012/17111-7) and partially supported by FINEP (grant no.
MCTI/FINEP/MS/SCTIE/DECIT-01/2013-FPXII-BIOPLAT).
References
|
1
|
Munker R and Calin GA: MicroRNA profiling
in cancer. Clin Sci (Lond). 121:141–158. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Da Sacco L and Masotti A: Recent insights
and novel bioinformatics tools to understand the role of microRNAs
binding to 5′untranslated region. Int J Mol Sci. 14:480–495. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Meltzer PS: Cancer genomics: Small RNAs
with big impacts. Nature. 435:745–746. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gaur A, Jewell DA, Liang Y, Ridzon D,
Moore JH, Chen C, Ambros VR and Israel MA: Characterization of
microRNA expression levels and their biological correlates in human
cancer cell lines. Cancer Res. 67:2456–2468. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:pp. 2999–3004. 2004; View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro- RNA genes miR15
and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad
Sci USA. 99:pp. 15524–15529. 2002; View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chang TC, Wentzel EA, Kent OA, et al:
Transactivation of miR-34a by p53 broadly influences gene
expression and promotes apoptosis. Mol Cell. 26:745–752. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mendell JT: miRiad roles for the miR-17-92
cluster in development and disease. Cell. 133:217–222. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:pp. 2257–2261.
2006; View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Galasso M, Sandhu SK and Volinia S:
MicroRNA expression signatures in solid malignancies. Cancer J.
18:238–243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sassen S, Miska EA and Caldas C: MicroRNA:
Implications for cancer. Virchows Arch. 452:1–10. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Negrini M and Calin GA: Breast cancer
metastasis: A microRNA story. Breast Cancer Res. 10:2032008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Valastyan S: Roles of microRNAs and other
non-coding RNAs in breast cancer metastasis. J Mammary Gland Biol
Neoplasia. 17:23–32. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Marino AL, Evangelista AF, Macedo T,
Silveira HC, Kerr LM, Vieira RA, Longatto AF and Silveira MM:
Differential expression profile of microRNAs associated with human
breast cancer progression. BMC Proc. 7 Suppl 2:pp. P92013;
View Article : Google Scholar
|
|
18
|
Zhao H, Guo M, Zhao G, Ma Q, Ma B, Qiu X
and Fan Q: miR-183 inhibits the metastasis of osteosarcoma via
downregulation of the expression of Ezrin in F5M2 cells. Int J Mol
Med. 30:1013–1020. 2012.PubMed/NCBI
|
|
19
|
Wang J, Wang X, Li Z, Liu H and Teng Y:
MicroRNA-183 suppresses retinoblastoma cell growth, invasion and
migration by targeting LRP6. FEBS J. 281:1355–1365. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
He W, Li Y, Chen X, Lu L, Tang B, Wang Z,
Pan Y, Cai S, He Y and Ke Z: miR-494 acts as an anti-oncogene in
gastric carcinoma by targeting c-myc. J Gastroenterol Hepatol.
29:1427–1434. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Silva-Oliveira RJ, Silva VA, Martinho O,
Cruvinel-Carloni A, Melendez ME, Rosa MN, De Paula FE, de Souza
Viana L, Carvalho AL and Reis RM: Cytotoxicity of allitinib, an
irreversible anti-EGFR agent, in a large panel of human
cancer-derived cell lines: KRAS mutation status as a predictive
biomarker. Cell Oncol (Dordr). 39:253–263. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kammermann M, Denelavas A, Imbach A,
Grether U, Dehmlow H, Apfel CM and Hertel C: Impedance measurement:
A new method to detect ligand-biased receptor signaling. Biochem
Biophys Res Commun. 412:419–424. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maere S, Heymans K and Kuiper M: BiNGO: A
Cytoscape plugin to assess overrepresentation of gene ontology
categories in biological networks. Bioinformatics. 21:3448–3449.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schneider CA, Rasband WS and Eliceiri KW:
NIH Image to ImageJ: 25 years of image analysis. Nat Methods.
9:671–675. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Girish V and Vijayalakshmi A: Affordable
image analysis using NIH Image/ImageJ. Indian J Cancer.
41:472004.PubMed/NCBI
|
|
27
|
Subik K, Lee JF, Baxter L, Strzepek T,
Costello D, Crowley P, Xing L, Hung MC, Bonfiglio T, Hicks DG and
Tang P: The expression patterns of ER PR, HER2, CK5/6, EGFR, Ki-67
and AR by immunohistochemical analysis in breast cancer cell lines.
Breast Cancer (Auckl). 4:35–41. 2010.PubMed/NCBI
|
|
28
|
Haga CL and Phinney DG: MicroRNAs in the
imprinted DLK1-DIO3 region repress the epithelial-to-mesenchymal
transition by targeting the TWIST1 protein signaling network. J
Biol Chem. 287:42695–42707. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lowery AJ, Miller N, Dwyer RM and Kerin
MJ: Dysregulated miR-183 inhibits migration in breast cancer cells.
BMC Cancer. 10:5022010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Limame R, Wouters A, Pauwels B, Fransen E,
Peeters M, Lardon F, De Wever O and Pauwels P: Comparative analysis
of dynamic cell viability, migration and invasion assessments by
novel real-time technology and classic endpoint assays. PLoS One.
7:e465362012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ohdaira H, Sekiguchi M, Miyata K and
Yoshida K: MicroRNA-494 suppresses cell proliferation and induces
senescence in A549 lung cancer cells. Cell Prolif. 45:32–38. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Holliday DL and Speirs V: Choosing the
right cell line for breast cancer research. Breast Cancer Res.
13:2152011. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lacroix M and Leclercq G: Relevance of
breast cancer cell lines as models for breast tumours: An update.
Breast Cancer Res Treat. 83:249–289. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Neve RM, Chin K, Fridlyand J, Yeh J,
Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al: A
collection of breast cancer cell lines for the study of
functionally distinct cancer subtypes. Cancer Cell. 10:515–527.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mizuarai S, Machida T, Kobayashi T,
Komatani H, Itadani H and Kotani H: Expression ratio of CCND1 to
CDKN2A mRNA predicts RB1 status of cultured cancer cell lines and
clinical tumor samples. Mol Cancer. 10:312011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chinnam M and Goodrich DW: RB1,
development, and cancer. Curr Top Dev Biol. 94:129–169. 2011.
View Article : Google Scholar : PubMed/NCBI
|