Introduction
Cervical cancer (CC) is the second major cause of
female cancer-associated mortalities worldwide, which accounts for
~12% of all female mortalities due to cancer (1,2). A total
of ~49,000 incident cases of CC were diagnosed in 2011, which
occurred primarily in developing countries (3). At present, surgery, radiotherapy and
chemotherapy remain the standard treatment for patients with CC.
Clinical outcomes vary greatly between patients and are difficult
to predict (4,5). In addition, the clinical stage of
disease is important regarding the prognosis for patients with CC,
and the 5-year survival rate for all stages combined is ~70%
(6). Therefore, it is urgently
required to identify novel and effective biomarkers for early stage
diagnosis and for potential targets for CC.
The human genome contains >20,000 protein-coding
genes according to high-throughput transcriptome analysis, which
represents ~2% of the whole genome (7,8). In
addition, the rest of human genome may be transcribed into various
short or long non-coding RNAs (lncRNAs) (9). lncRNAs are a subgroup of RNAs that are
>200 nucleotides in length and may be implicated in various
types of gene regulation, including transcriptional,
post-transcriptional or epigenetic regulation (10). Additionally, these types of regulation
implicated by lncRNAs may induce the progression of cancer or other
diseases (11). Compared with short
non-coding RNAs, like miRNAs, the biological roles of lncRNAs have
largely been underestimated. However, there have already been
studies characterizing the regulatory roles of lncRNAs, including
cell proliferation, apoptosis and invasion, and parental imprinting
(12–14). In addition, results from emerging
studies have reported associations between the dysregulation of
lncRNAs to multiple human diseases, including cancer (15–24). These
studies have demonstrated that a large number of lncRNAs serve
crucial roles in the progression of colorectal, breast, prostate
and liver cancer, and other human tumors (25–28).
In the present study, the lncRNAs and mRNA
expression profiles in CC tissues were compared with adjacent
non-cancerous tissues. The differently expressed lncRNAs were
additionally studied with potential gene targets through gene
ontology (GO) and pathway analysis, and these results were
confirmed in 40 CC and adjacent non-cancerous tissues using reverse
transcription quantitative polymerase chain reaction (RT-qPCR).
Materials and methods
Ethics statement
The present study was approved for the use of human
biopsy samples by the Institution Review Board of Wenzhou Medical
University (Wenzhou, China). The written consent was received from
all participants in the present study at the time of surgery.
Clinical samples
A total of one CC tissue and one adjacent
non-cancerous tissue from 3 patients were included in the
microarray assay. Tissue samples from 43 patients (age range 20–65
years; average age 47.8 years) with cervical cancer between March
2011 and December 2013 were collected (Table I). All tissue samples were collected
during surgical resection at the First Affiliated Hospital of
Wenzhou Medical University and stored at −80°C in the tissue bank
for further use.
 | Table I.Clinicopathological characteristics
in 43 patient samples of cervical cancer. |
Table I.
Clinicopathological characteristics
in 43 patient samples of cervical cancer.
| Clinical
parameter | Number of cases
(%) |
|---|
| Age (years) |
|
|
<30 | 3 (7.0) |
|
30–55 | 29 (67.4) |
|
>55 | 11 (25.6) |
| Age at first birth
(years) |
|
|
<18 | 2 (4.7) |
|
18–24 | 10 (23.2) |
|
>24 | 23 (53.5) |
|
Nulliparous | 8 (18.6) |
| Caesarean
section |
|
|
Never | 34 (79.1) |
|
Ever | 9 (20.9) |
| Tumor types |
|
|
Endophytic type | 6 (14.0) |
|
Ulcerative type | 9 (20.9) |
|
Endocervical type | 11 (25.6) |
|
Exophytic type | 17(39.5) |
RNA extraction
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) was used to extract the total RNAs from tissues.
The mirVana miRNA Isolation kit (Ambion; Thermo Fisher Scientific,
Inc.) was used to purify small RNAs in accordance with the
manufacture's protocol. The concentration and purity of RNAs were
determined by OD260/280 readings using a spectrophotometer
(NanoDrop ND-2000; NanoDrop Technologies; Thermo Fisher Scientific,
Inc., Pittsburgh, PA, USA). By using the RNA 6000 Nano
Lab-on-a-Chip kit and the Bioanalyzer 2100 (Agilent Technologies,
Inc., Santa Clara, CA, USA), RNA integrity was determined by
capillary electrophoresis.
DNA microarray
Microarray assays were performed by Kangcheng
Biotechnology Co. (Shanghai, China). Arraystar human lncRNA and
mRNA Array version 3.0 (Qiagen GmbH, Hilden, Germany) was used in
the assay, which was designed with four identical arrays per slide
(4×180K format). Arraystar human lncRNA and mRNA Array contains
30,586 human lncRNAs probes and 26,109 human mRNAs probes, which
were collected from a number of sources, including GENCODE/ENSEMBL
(http://www.gencodegenes.org/data_ensembl.html), the
Human lncRNA Catalog (17), RefSeq
(https://www.ncbi.nlm.nih.gov/refseq/), USCS Genome
Browser (http://genome.ucsc.edu/cgi-bin/hgGateway), Non-coding
(nc) RNA Expression Database (http://nred.matticklab.com/cgi-bin/ncrnadb.pl),
Antisense ncRNA pipeline (http://research.imb.uq.edu.au/rnadb/rnadb2_archive.htm),
homeotic gene ncRNAs and the ultra-conserved regions. Each RNA was
detected by corresponding probes in duplicate experiments.
RNA amplification, labeling and
hybridization
By using Eberwine's linear RNA amplification method
and subsequent enzymatic reaction (29), fluorescent dye (Cy5 and Cy3) labeled
complimentary (c)DNAs were produced using the CapitalBio cRNA
Amplification and Labeling kit (CapitalBio Corporation, Beijing,
China), according to the manufacturer's instructions.
Microarray imaging and data
analysis
GeneSpring software (version 12.0; Agilent
Technologies, Inc.) was applied to analyze the lncRNAs and mRNA
array data. Threshold values of ≥2 and ≤-2-fold change were used to
identify the differentially expressed genes. The data was
log(2) transformed and median
centered by genes using the Adjust Data function of Multiexperiment
Viewer software (MeV 4.3.02) (Dana-Farber Cancer Institute, Boston,
MA, USA). The data were subsequently analyzed with hierarchical
clustering with average linkage. GO and pathway analysis were
performed on Gene-Cloud of Biotechnology Information (GCBI)
(https://www.gcbi.com.cn/gclib/html/index) according
the protocol of the manufacturer.
RT-qPCR analysis
Total RNA of 40 clinical samples was isolated using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The
purity and concentration of RNA samples were determined by a
spectrophotometer (NanoDrop ND-2000; Thermo Fisher Scientific,
Inc.). The samples with an optical density 260/280 ratio >1.8
were reversely transcribed using a GoScript™ Reverse Transcription
system kit (Promega Corporation, Madison, WI, USA) according to the
manufacturer's protocol. The expression of selected lncRNAs were
analyzed using qPCR with a GoTaq® qPCR Master Mix kit
(Promega Corporation) on the StepOne Plus PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). PCR reactions were
performed using standard cycling parameters (stage 1: 50°C for 2
min, Stage 2: 95°C for 10 min then 40 cycles of 95°C for 15 Sec and
60°C for 1 min). β-actin was used as an internal control for
normalization, and the 2−ΔΔCt method (30) was used to calculate the expression of
lncRNAs. Each reaction was performed 3 times. The sequences of the
primers for qPCR are listed in Table
II.
 | Table II.Primers for quantitative reverse
transcription polymerase chain reaction. |
Table II.
Primers for quantitative reverse
transcription polymerase chain reaction.
| Sequence name
(ID) | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| β-actin |
ATCGTGCGTGACATTAAGGAGAAG |
AGGAAGGAAGGCTGGAAGAGTG |
| TCONS_00027301
(hsaLB_LI_107288) |
GCCGACAAAGAGAAGGGAAGA |
GCAGATAGTGAAGGCATGGAAGT |
| TCONS_00029064
(hsaLB_LI_114731) |
TGCTGCCGGAAACGTGTG |
GGCTTCTGGGGAGAGTGGG |
| TCONS_00010587
(hsaLB_LI_38766) |
CACCACCAAGACCCCCTCAC |
GCGAACAAGCGTCCAGGTAA |
| TCONS_00003380
(hsaLB_LI_16629) |
GCGAACAAGCGTCCAGGTAA |
CGGCACAGAAGGAATCCAAC |
| TCONS_00026907
(hsaLB_LI_16629) |
TGGATTGTTGGGTATATTTTGGA |
TGTATGAAGAGGATGCTGAAGGC |
Statistical analysis
All data are expressed as the mean ± standard
deviation. P<0.05 was considered to indicate a statistically
significant difference. Comparison between groups was analyzed by a
Student's t-test. All the statistical analyses were conducted using
GraphPad Prism (version 5; GraphPad Software, Inc., La Jolla, CA,
USA).
Results
Overview of the lncRNA and mRNA
profiles in CC tissues and adjacent non-cancerous tissues
In the present study, a commercial human lncRNA
microarray (Kangcheng Biotechnology Co.) was used to investigate
the characteristic expression profiles of CC tissues and adjacent
non-cancerous tissues by using total RNA isolated from different
patients with CC. The lncRNA profiles of CC tissues were analyzed
by comparing CC tissues with adjacent non-cancerous tissues.
Hierarchical clustering analysis was used to
investigate the expression level of the lncRNAs (Fig. 1 and Table
III). The results of the lncRNA expression profiles are
summarized in Table III. The
alternation of lncRNA expressions were evaluated by fold change. As
demonstrated in Table III, 32,829
lncRNAs on the microarray exhibited expression above background
levels, and 10.22% (3,356/32,829) of the lncRNAs were significantly
differentially expressed between the CC sample and corresponding
adjacent non-cancerous sample (absolute fold-change ≥2). A total of
55.33% (1,857/3,356) of the significantly differentially expressed
lncRNAs were upregulated in the CC tissue. By contrast, 8.25%
(1,987/24,063) of the mRNAs demonstrated significantly different
expression between cancer and corresponding adjacent non-cancerous
tissues (absolute fold-change ≥2), whilst 56.72% (1,127/1,987) of
the mRNAs were significantly upregulated.
 | Table III.Number of differentially expressed
lncRNAs and mRNAs in cervical cancer and corresponding adjacent
non-cancerous tissues. |
Table III.
Number of differentially expressed
lncRNAs and mRNAs in cervical cancer and corresponding adjacent
non-cancerous tissues.
| Parameter | n (%) | Type of change | Fold change | n (%) |
|---|
| Total lncRNAs on
the microarray | 32,829 |
|
|
|
|
Differentially expressed
lncRNAs | 3,356
(10.22) | Up | ≥2 | 1,857 (55.33) |
|
|
| Down | ≥2 | 1,499 (44.67) |
| Total mRNAs | 24,063 |
|
|
|
|
Differentially expressed
mRNAs | 1,987 (8.25) | Up | ≥2 | 1,127 (56.72) |
|
|
| Down | ≥2 |
860 (43.28) |
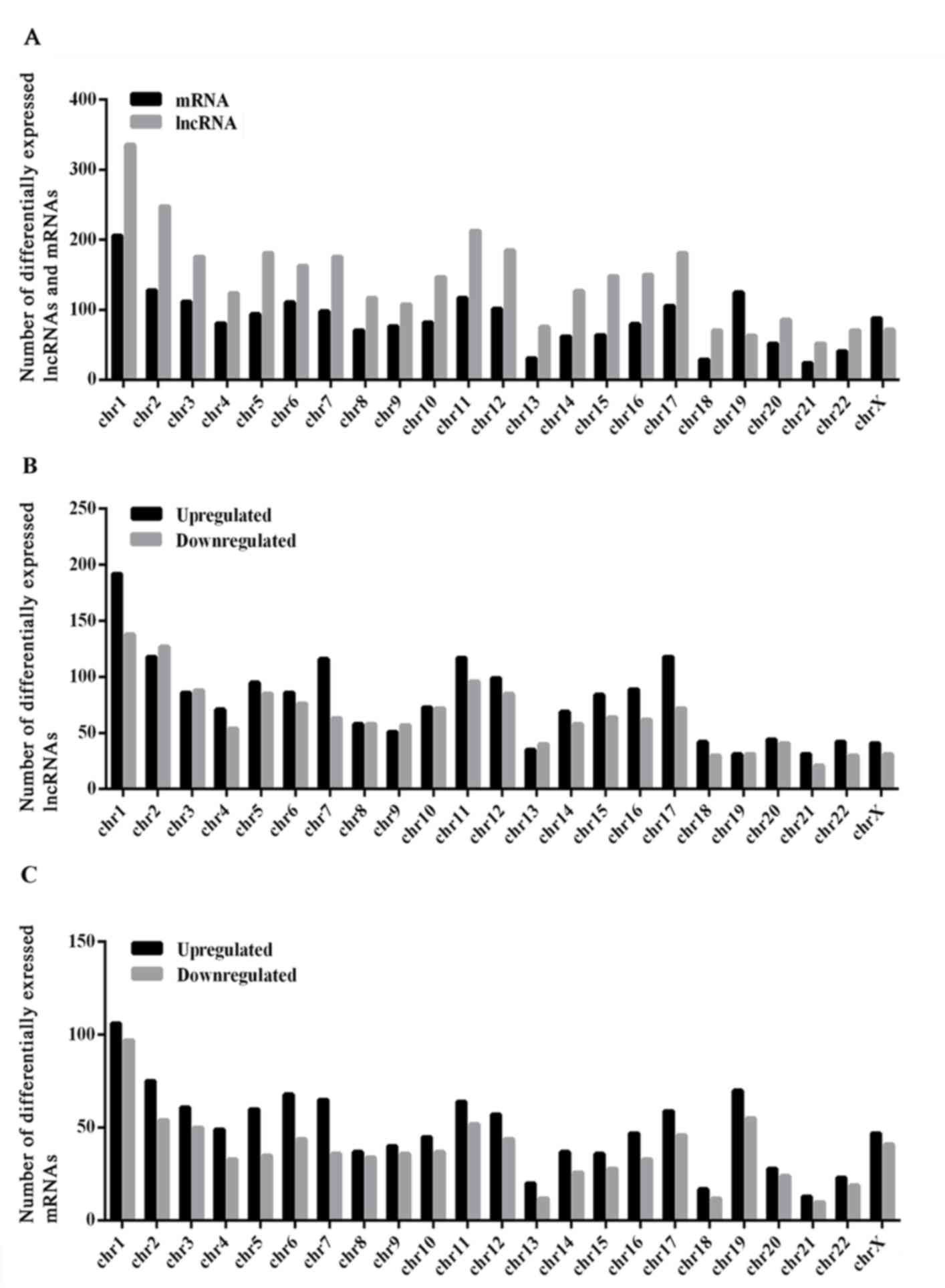
Analysis of the distribution of differentially
expressed lncRNAs and protein-coding mRNAs reveals that the lncRNAs
and mRNAs were not distributed equally on each chromosome (Fig. 2). The analysis reveals that chromosome
1 had the highest number of differentially expressed altered
lncRNAs and protein-coding mRNAs (Fig.
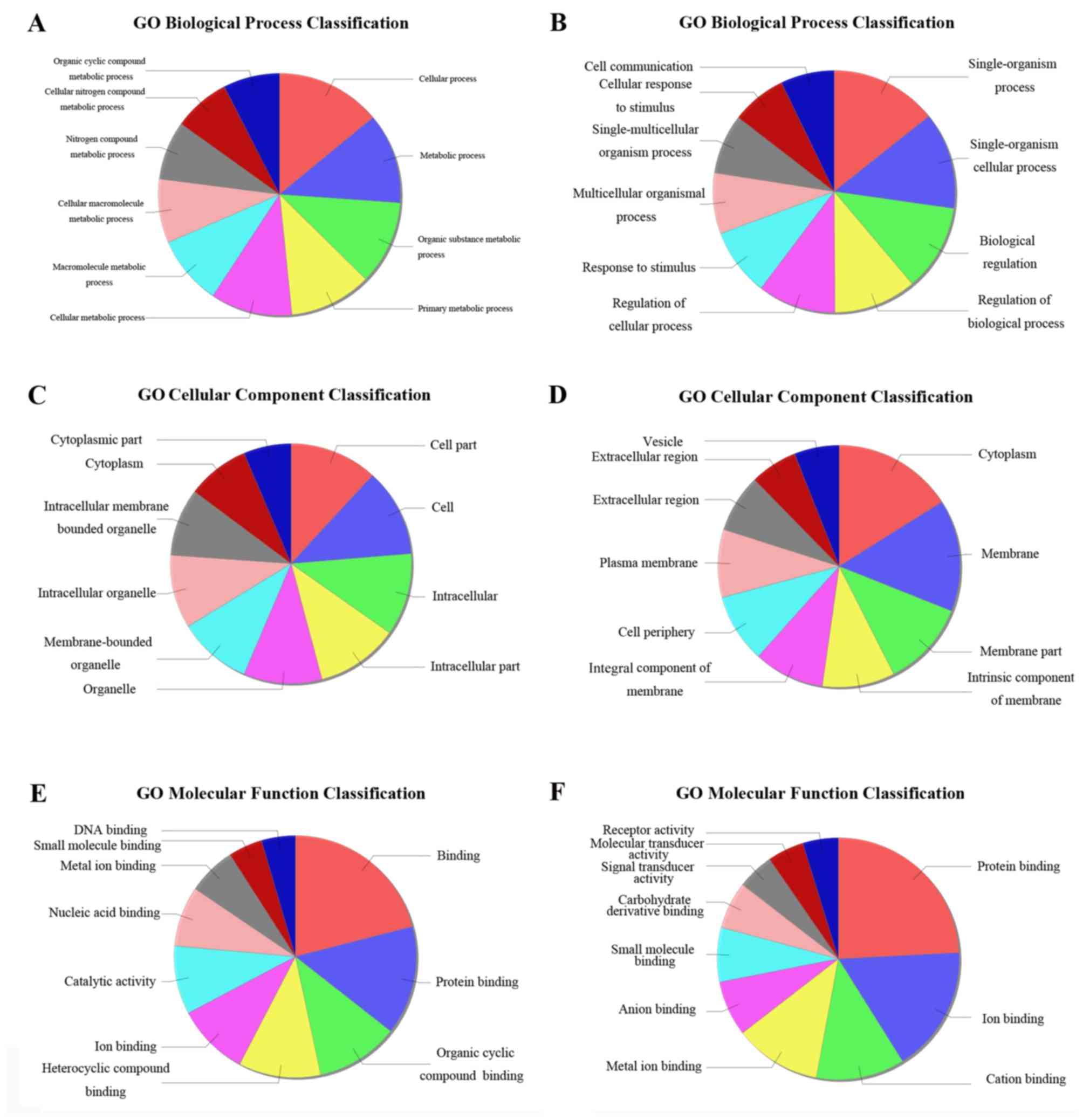
2). All differentially expressed protein-coding genes were
submitted for GO term enrichment analysis. A total of three domains
were studied, including biological process, cellular component and
molecular function (Fig. 3).
Classification and subgroup analysis
of lncRNAs
lncRNAs may be classified into five subgroups,
including sense lncRNAs, antisense lncRNAs, intronic lncRNAs,
intergenic lncRNAs and bidirectional lncRNAs based on different
transcription forms. Sense lncRNAs exhibit the same transcriptional
direction with exons of protein-coding genes, and a previous study
identified that certain sense lncRNAs may be viewed as non-coding
transcript variants of genes (31).
These non-coding transcript variants may regulate gene expression
(32). The present study identified
331 upregulated and 175 downregulated sense lncRNAs in CC tissues
compared with adjacent non-cancerous tissues (Table IV).
 | Table IV.Types of differentially expressed
lncRNAs and mRNAs in cervical cancer tissues compared with
corresponding adjacent non-cancerous samples. |
Table IV.
Types of differentially expressed
lncRNAs and mRNAs in cervical cancer tissues compared with
corresponding adjacent non-cancerous samples.
| Type of
lncRNAs | Type of change | Fold change | Number of
lncRNAs |
|---|
| Sense | Up | ≥2 | 331 |
|
| Down | ≥2 | 175 |
| Antisense | Up | ≥2 | 355 |
|
| Down | ≥2 | 263 |
| Intronic | Up | ≥2 | 415 |
|
| Down | ≥2 | 345 |
| Intergenic | Up | ≥2 | 613 |
|
| Down | ≥2 | 808 |
| Bidirectional | Up | ≥2 | 140 |
|
| Down | ≥2 | 63 |
There were 355 antisense lncRNAs significantly
upregulated in CC tissues, and 263 antisense lncRNAs that were
downregulated. Anti-sense lncRNAs are transcribed against
overlapping genes, regulate their protein-coding counterparts via
multiple mechanisms, including chromatin remodeling, alternative
splicing, translational interference and promoter targeting
(33).
Intronic lncRNAs have been demonstrated to regulate
the expression of neighbourhood genes or other genes through
alternative splicing, miRNA, RNA interference, transcriptional
disrution and chromatin modification in previous studies (34,35). The
present study identified 415 upregulated and 345 downregulated
intronic lncRNAs in CC tissues, as illustrated in Table IV.
Bidirectional lncRNAs may regulate the expression of
their neighboring genes through epigenetic modification (36). Bidirectional non-coding genes share
paired transcriptional initiation sites with separate transcripts,
which are in opposite orientations, but with close proximity
(37). Among the significantly
changed lncRNAs in the present study, 140 bidirectional lncRNAs
were upregulated and 63 were downregulated.
Intergenic non-coding RNAs are able to regulate the
expression of target genes with a distance >10 kb through
recruiting histone-modifying enzymes to the chromatin. Therefore,
the target genes of intergenic non-coding RNAs may be located
across the genome. The present study identified 613 upregulated and
808 downregulated intergenic lncRNAs in CC tissues compared with
adjacent non-cancerous tissues.
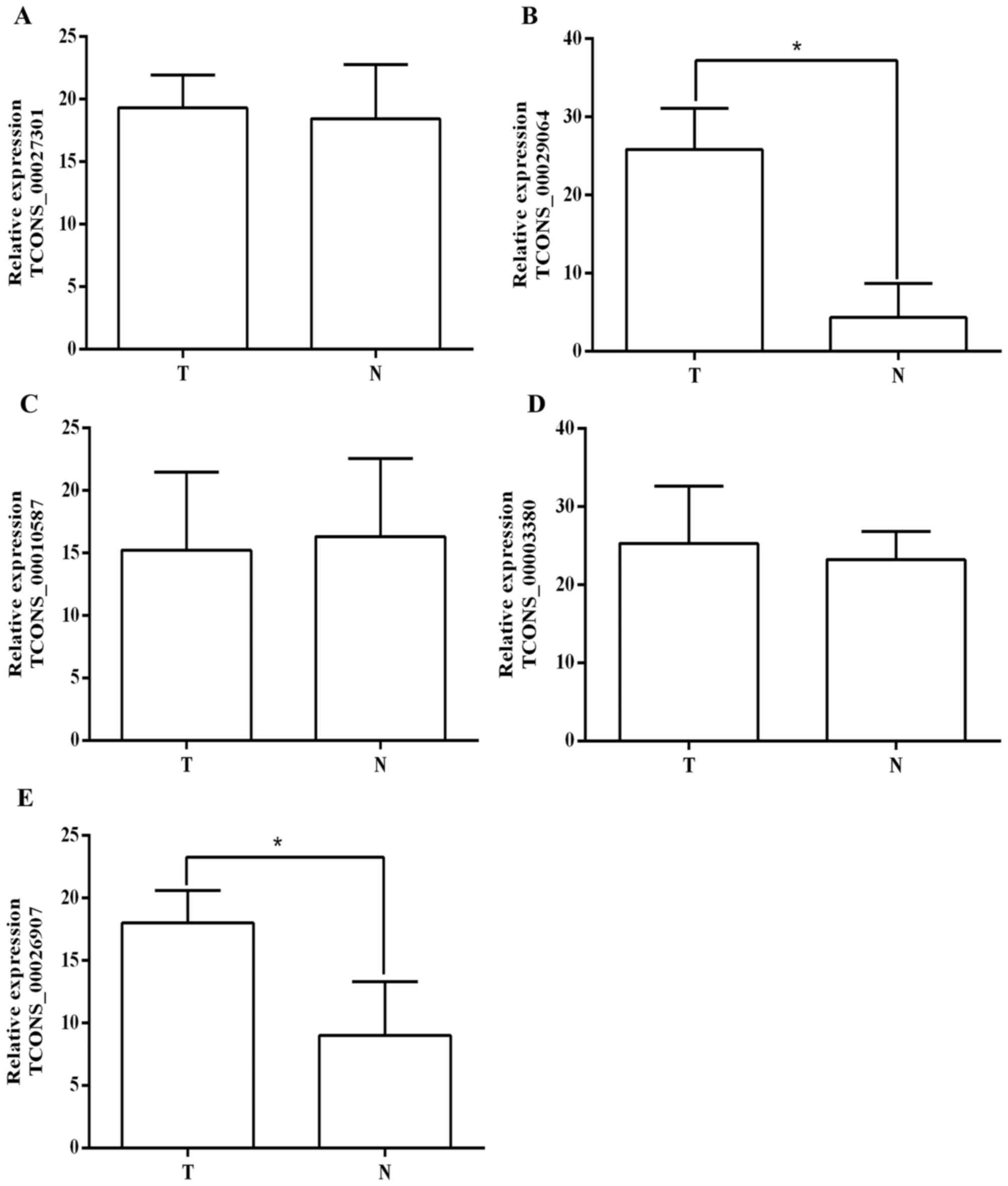
Validation of the significantly
changed lncRNAs by RT-qPCR
The expression level of differentially expressed
lncRNAs was confirmed using RT-qPCR, (Fig. 4) and a total of 5 lncRNAs that were
particularly markedly upregulated were selected. The result
demonstrates that 3 of the selected lncRNAs did not exhibit an
altered expression pattern. By contrast, 2 lncRNAs (TCONS_00029064
and TCONS_00026907) were significantly upregulated in CC tissues
compared with adjacent non-cancerous tissues (Fig. 4), which was consistent with the
microarray result (Fig. 1). RT-qPCR
analysis indicates that TCONS_00029064 and TCONS_00026907 may serve
important roles in CC tumorigenesis and other associated biological
process.
Discussion
CC remains an enormous challenge and worldwide
public health problem (38). Despite
the development of advanced therapeutic strategies, the prognosis
in patients with CC varies and is difficult to predict. Therefore,
novel molecular mechanisms are needed in order to develop effective
therapeutic strategies. In previous studies protein-coding genes
and non-coding RNAs have been reported to be involved in the
molecular mechanism of carcinogenesis (39,40).
Compared to the knowledge of coding genes and short non-coding
RNAs, such as miRNAs, general understanding of lncRNAs remains
limited. An increasing number of studies demonstrate that certain
lncRNAs serve crucial roles in cancer and associated biological
functions, including cell migration/invasion (41) and cell-cycle regulation (42). Non-coding RNAs, including SPRY4-IT1,
have been reported to have a key role in cell growth and
differentiation in melanoma cell lines (43). Additionally, the altered expression of
a number of non-coding RNAs have been associated with cancer
progression (44). Despite the
previous findings, the associations between lncRNA and CC remain
unknown.
In the present study, the overall lncRNAs expression
profile of CC tissues was established by comparing the expression
of lncRNAs in CC samples with the expression in corresponding
adjacent non-cancerous tissues. A total of 3,356 differentially
expressed lncRNAs, and a total of 1,987 differentially expressed
mRNAs were revealed. Based on the association of the lncRNAs with
coding genes, the lncRNAs may be classified into five subgroups
that include sense, antisense, intronic, bidirectional and
intergenic lncRNAs (45). The results
of the present study indicated that the different subtypes of
differentially expressed lncRNAs are unequally distributed across
the genome, which is consistent with a previous study (46). Although all the five subgroups of
lncRNAs were detected, intergenic lncRNAs constituted a major
portion. This result indicates that among differentially expressed
lncRNAs, intergenic lncRNAs are more abundant compared with others,
which suggests that intergenic lncRNAs may serve important roles in
CC.
Overall, the present study characterized the
expression profile of lncRNAs and mRNAs in CC tissues and
corresponding adjacent non-cancerous tissues using microarray and
identified a large portion of differentially expressed lncRNAs and
mRNAs. These results remain limited due to sample quantities.
Further studies using greater numbers of CC samples may provide
more accurate expression information for lncRNAs and mRNAs.
Meanwhile, the data of the present study will facilitate other
groups to better understand the function of lncRNAs in CC
development and metastasis.
Acknowledgements
The present study was supported by grants from the
Medical and Health Project of Zhejiang Province (grant no.
2016KYA137 to Dr H.Z), Wenzhou Public Welfare Science and
Technology Project (grant no. Y20140707 to Dr H.Z) and the
Incubation Project of the First Affiliated Hospital of Wenzhou
Medical University (grant no. FHY2014009 to Dr H.Z).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ellenson LH and Wu TC: Focus on
endometrial and cervical cancer. Cancer Cell. 5:533–538. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schiffman M, Wentzensen N, Wacholder S,
Kinney W, Gage JC and Castle PE: Human papillomavirus testing in
the prevention of cervical cancer. J Natl Cancer Inst. 103:368–383.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Waggoner SE: Cervical cancer. Lancet.
361:2217–2225. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Biewenga P, van der Velden J, Mol BW,
Stalpers LJ, Schilthuis MS, van der Steeg JW, Burger MP and Buist
MR: Prognostic model for survival in patients with early stage
cervical cancer. Cancer. 117:768–776. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shingleton HM, Jones WB, Russell A,
Fremgen A, Chmiel JS, Ocwieja K, Winchester DP and Clive R:
Hysterectomy in invasive cervical cancer: A national patterns of
care study of the American College of Surgeons. J Am Coll Surg.
183:393–400. 1996.PubMed/NCBI
|
|
7
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
ENCODE Project Consortium, ; Birney E,
Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH,
Weng Z, Snyder M, Dermitzakis ET, et al: Identification and
analysis of functional elements in 1% of the human genome by the
ENCODE pilot project. Nature. 447:799–816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tsai MC, Spitale RC and Chang HY: Long
intergenic noncoding RNAs: New links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Maruyama R, Shipitsin M, Choudhury S, Wu
Z, Protopopov A, Yao J, Lo PK, Bessarabova M, Ishkin A, Nikolsky Y,
et al: Altered antisense-to-sense transcript ratios in breast
cancer. Proc Natl Acad Sci USA. 109:pp. 2820–2824. 2012; View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Loewer S, Cabili MN, Guttman M, Loh YH,
Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al:
Large intergenic non-coding RNA-RoR modulates reprogramming of
human induced pluripotent stem cells. Nat Genet. 42:1113–1117.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Villegas VE, Rahman MF, Fernandez-Barrena
MG, Diao Y, Liapi E, Sonkoly E, Ståhle M, Pivarcsi A, Annaratone L,
Sapino A, et al: Identification of novel non-coding RNA-based
negative feedback regulating the expression of the oncogenic
transcription factor GLI1. Mol Oncol. 8:912–926. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu XH, Liu ZL, Sun M, Liu J, Wang ZX and
De W: The long non-coding RNA HOTAIR indicates a poor prognosis and
promotes metastasis in non-small cell lung cancer. BMC Cancer.
13:4642013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu X, Fang Y, Wang Z, Xie J, Zhan Q, Deng
X, Chen H, Jin J, Peng C, Li H and Shen B: Downregulation of gas5
increases pancreatic cancer cell proliferation by regulating CDK6.
Cell Tissue Res. 354:891–896. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kong GY, Zhang JP, Zhang S, Shan CL, Ye LH
and Zhang XD: Hepatitis B virus X protein promotes hepatoma cell
proliferation via upregulation of MEKK2. Acta Pharmacol Sin.
32:1173–1180. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yuan SX, Yang F, Yang Y, Tao QF, Zhang J,
Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al: Long noncoding RNA
associated with microvascular invasion in hepatocellular carcinoma
promotes angiogenesis and serves as a predictor for hepatocellular
carcinoma patients' poor recurrence-free survival after
hepatectomy. Hepatology. 56:2231–2241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu
WQ, Xie WP and Hou YY: Long non-coding RNA MEG3 inhibits NSCLC
cells proliferation and induces apoptosis by affecting p53
expression. BMC Cancer. 13:4612013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schmidt LH, Spieker T, Koschmieder S,
Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D,
Marra A, et al: The long noncoding MALAT-1 RNA indicates a poor
prognosis in non-small cell lung cancer and induces migration and
tumor growth. J Thorac Oncol. 6:1984–1992. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Matouk IJ, DeGroot N, Mezan S, Ayesh S,
Abu-lail R, Hochberg A and Galun E: The H19 non-coding RNA is
essential for human tumor growth. PLoS One. 2:e8452007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW,
Li MQ, Chen YC, Qian XP, Lu TJ, Yu LZ, et al: Rapid identification
of UCA1 as a very sensitive and specific unique marker for human
bladder carcinoma. Clin Cancer Res. 12:4851–4858. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
de Kok JB, Verhaegh GW, Roelofs RW,
Hessels D, Kiemeney LA, Aalders TW, Swinkels DW and Schalken JA:
DD3(PCA3), a very sensitive and specific marker to detect prostate
tumors. Cancer Res. 62:2695–2698. 2002.PubMed/NCBI
|
|
25
|
Gao Y, Chen G, Zeng Y, Zeng J, Lin M, Liu
X and Liu J: Invasion and metastasis-related long noncoding RNA
expression profiles in hepatocellular carcinoma. Tumour Biol.
36:7409–7422. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ren S, Peng Z, Mao JH, Yu Y, Yin C, Gao X,
Cui Z, Zhang J, Yi K, Xu W, et al: RNA-seq analysis of prostate
cancer in the Chinese population identifies recurrent gene fusions,
cancer-associated long noncoding RNAs and aberrant alternative
splicings. Cell Res. 22:806–821. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY,
Zhang F and Zheng SS: Overexpression of long non-coding RNA HOTAIR
predicts tumor recurrence in hepatocellular carcinoma patients
following liver transplantation. Ann Surg Oncol. 18:1243–1250.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Phillips J and Eberwine JH: Antisense RNA
Amplification: A Linear amplification method for analyzing the mRNA
population from single living cells. Methods. 10:283–288. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Carninci P, Kasukawa T, Katayama S, Gough
J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al:
The transcriptional landscape of the mammalian genome. Science.
309:1559–1563. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang X, Arai S, Song X, Reichart D, Du K,
Pascual G, Tempst P, Rosenfeld MG, Glass CK and Kurokawa R: Induced
ncRNAs allosterically modify RNA-binding proteins in cis to inhibit
transcription. Nature. 454:126–130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Faghihi MA and Wahlestedt C: Regulatory
roles of natural antisense transcripts. Nat Rev Mol Cell Biol.
10:637–643. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Louro R, El-Jundi T, Nakaya HI, Reis EM
and Verjovski-Almeida S: Conserved tissue expression signatures of
intronic noncoding RNAs transcribed from human and mouse loci.
Genomics. 92:18–25. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang L, Lin C, Jin C, Yang JC, Tanasa B,
Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al:
lncRNA-dependent mechanisms of androgen-receptor-regulated gene
activation programs. Nature. 500:598–602. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Louro R, Smirnova AS and Verjovski-Almeida
S: Long intronic noncoding RNA transcription: Expression noise or
expression choice? Genomics. 93:291–298. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Trinklein ND, Aldred SF, Hartman SJ,
Schroeder DI, Otillar RP and Myers RM: An abundance of
bidirectional promoters in the human genome. Genome Res. 14:62–66.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schiffman M, Castle PE, Jeronimo J,
Rodriguez AC and Wacholder S: Human papillomavirus and cervical
cancer. Lancet. 370:890–907. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jin G, Sun J, Isaacs SD, Wiley KE, Kim ST,
Chu LW, Zhang Z, Zhao H, Zheng SL, Isaacs WB and Xu J: Human
polymorphisms at long non-coding RNAs (lncRNAs) and association
with prostate cancer risk. Carcinogenesis. 32:1655–1659. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao
X, Chen WS and Li B: LncRNA-UCA1 exerts oncogenic functions in
non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett.
371:99–106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim T, Cui R, Jeon YJ, Fadda P, Alder H
and Croce CM: MYC-repressed long noncoding RNAs antagonize
MYC-induced cell proliferation and cell cycle progression.
Oncotarget. 6:18780–18789. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Khaitan D, Dinger ME, Mazar J, Crawford J,
Smith MA, Mattick JS and Perera RJ: The melanoma-upregulated long
noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer
Res. 71:3852–3862. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lujambio A, Portela A, Liz J, Melo SA,
Rossi S, Spizzo R, Croce CM, Calin GA and Esteller M: CpG island
hypermethylation-associated silencing of non-coding RNAs
transcribed from ultraconserved regions in human cancer. Oncogene.
29:6390–63401. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Derrien T, Johnson R, Bussotti G, Tanzer
A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG,
et al: The GENCODE v7 catalog of human long noncoding RNAs:
Analysis of their gene structure, evolution, and expression. Genome
Res. 22:1775–1789. 2012. View Article : Google Scholar : PubMed/NCBI
|