Introduction
Esophageal cancer is the eighth most common
malignancy and the sixth leading cause of cancer-associated
mortality worldwide (1–3). Even subsequent to combined multimodality
treatment, clinical outcomes remain extremely poor (4,5). More
effective treatments based on novel mechanisms are required. DNA
repair pathways play a vitally important role for maintaining
genomic integrity. Failure of these pathways may lead to unrepaired
DNA lesions, and the accumulation of such lesions is associated
with genomic instability (6). In
recent years, therefore, the strategy of inhibiting proteins
associated with DNA repair has shown promise for new treatments of
various malignancies.
Poly (ADP-ribose) polymerase-1 (PARP1) is a 113-kDa
nuclear polymerase that modifies substrates by poly
ADP-ribosylation, and can conjugate ADP from NAD+ to
target proteins, such as histones (7,8). At
present, it has been shown that PARP1 plays a role in the repair of
DNA damage and is activated by DNA strand breaks, particularly
those of single-stranded DNA (9–11).
Numerous studies have reported that PARP1 inhibitors are effective
in patients with breast and ovarian cancer, since the BRCA gene,
which is another DNA repair gene, is frequently mutated (12–14); PARP1
expression is upregulated to compensate for the impaired DNA repair
(12,15). Previously, PARP1 inhibitors have
received attention in patients with malignancies other than breast
and ovarian cancer (7,16). However, there are few studies
investigating PARP1 in esophageal cancer, particularly esophageal
squamous cell carcinoma (ESCC) (17).
The present study aimed to investigate the
association between PARP1 expression and prognosis in patients with
ESCC, as well as the effect of inhibiting PARP1 expression on the
proliferation of ESCC cells.
Materials and methods
Clinical tissue samples
Between January 1998 and December 2011, 86 tissue
samples were collected from patients who had undergone radical
esophagectomy, without preoperative therapies such as chemotherapy
or chemoradiotherapy, for primary ESCC at the Department of
Gastroenterological Surgery, Osaka University Hospital (Osaka,
Japan). Pathological tumor stage was evaluated using the seventh
edition of the TNM classification established by the Union for
International Cancer Control (18).
The present study was approved by the Ethics Committee of Osaka
University Hospital (Suita, Japan) and written consent was obtained
from all the patients in the present study.
Antibodies
The primary antibodies used for western blot
analysis were obtained from Cell Signaling Technology, Inc.
(Danvers, MA, USA) and were as follows: Rabbit anti-checkpoint
kinase 2 (Chk2) (catalog no. 2662S; dilution, 1:1,000); rabbit
anti-phospho-Chk2 (Thr68) (catalog no. 2661S; dilution, 1:1,000);
rabbit anti-cell division control (cdc) 25c (catalog no. 4688S;
dilution, 1:1,000); rabbit anti-phospho-cdc25c (Thr48) (catalog no.
9527S; dilution, 1:1,000); mouse anti-cdc2 (catalog no. 9116S;
dilution, 1:1,000); rabbit anti-phospho-cdc2 (Tyr15) (catalog no.
9111S; dilution, 1:1,000); and rabbit anti-cyclin B1 (catalog no.
4138S; dilution, 1:1,000). Mouse anti-PARP1 (catalog no. sc-8007;
immunohistochemistry dilution, 1:50; western blotting dilution,
1:1,000) was obtained from Santa Cruz Biotechnology, Inc. (Dallas,
TX, USA). Secondary antibodies used for western blot analysis were
obtained from GE Healthcare Life Sciences (Little Chalfont, UK) and
were as follows: Horseradish peroxidase (HRP)-conjugated sheep
anti-mouse IgG (catalog no. NA931; dilution, 1:100,000) and
HRP-conjugated donkey anti-rabbit IgG (catalog no. NA934; dilution,
1:100,000).
Immunohistochemical (IHC)
staining
A mouse anti-PARP1 antibody (catalog no. sc-8007;
dilution, 1:50; Santa Cruz Biotechnology, Inc.) was used. In brief,
4-µm-thick sections of 10% formalin-fixed and paraffin-embedded
blocks were used for immunohistochemistry. These sections were
deparaffinized in xylene, dehydrated in graded ethanol, and heated
in 10 mM citrate buffer (pH 6.0) for 40 min at 95°C for antigen
retrieval by autoclave. Endogenous peroxidase activity was blocked
with 0.3% H2O2 in methanol. Nonspecific
binding was blocked with horse serum for 20 min using the
VECTASTAIN Elite ABC kit (catalog no. PK-6102, Vector Laboratories,
Burlingame, CA, USA). The sections were incubated overnight with
mouse anti-PARP1 antibody (catalog no. sc-8007; dilution, 1:50,
Santa Cruz Biotechnology, Inc.) at 4°C in a moist chamber. Antibody
staining was visualized with the VECTASTAIN Elite ABC kit, followed
by 3,3′-diaminobenzidine tetrahydrochloride plus
H2O2 for 2 min 30 sec. All sections were
counterstained with Mayer's hematoxylin (catalog no. 131-09665;
Wako, Osaka, Japan).
PARP1 expression was evaluated by the intensity of
stained cancer samples, particularly nuclei, as previously reported
(6). Tonsil tissues collected from
patients who had undergone tonsillectomy between January and
December 2011 at the Department of Otorhinolaryngology-Head and
Neck Surgery, Osaka University Hospital (Osaka, Japan), were used
as a positive control. The intensity was scored from 0 to 3 (0,
none; 1, weak; 2, moderate; 3, strong). Expression was considered
to be low when scores were 0 or 1 and high when scores were 2 or 3.
Evaluation was performed by two double-blinded independent
observers, who were unaware of the clinicopathological data and
outcome. When a discrepant evaluation between the two independent
observers was found, the evaluation was rechecked and
discussed.
Cell lines and culture conditions
Human ESCC TE1, TE4, TE5, TE6, TE8, TE9, TE10 and
TE11 cell lines were obtained from the Riken Bioresource Center
Cell Bank (Tsukuba, Japan). All cell lines were cultured in
RPMI-1640 (Nacalai Tesque, Inc., Kyoto, Japan), supplemented with
10% fetal bovine serum (Thermo Fisher Scientific, Inc., Waltham,
MA, USA) and 1% penicillin/streptomycin (Thermo Fisher Scientific,
Inc.). These cell lines were incubated in 5% CO2 at
37°C.
Small interfering RNA (siRNA)
design
siRNA against PARP1 (siPARP1;catalog nos.,
sc-29437A-C) and nontargeting siRNA (negative control siRNA;
catalog no., sc-37007) were purchased from Santa Cruz
Biotechnology, Inc. The sequence of siPARP1 was designed as
follows: sc-29437A sense, 5′-GAGUCAAGAGUGAAGGAAATT-3′ and
antisense, 5′-UUUCCUUCACUCUUGACUCTT-3′; sc-29437B sense,
5′-GGUAUCAACAAAUCUGAAATT-3′ and antisense,
5′-UUUCAGAUUUGUUGAUACCTT-3′; sc-29437C sense,
5′-GCAACAAACUGGAACAGAUTT-3′ and antisense,
5′-AUCUGUUCCAGUUUGUUGCTT-3′. Cells were cultured to 50–60%
confluency. The siRNA oligonucleotides (20 nM) in Opti-MEM (Thermo
Fisher Scientific, Inc.) were transfected into cells using
Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific, Inc.) and
incubated in Opti-MEM, according to the manufacturer's protocol.
Cells were incubated for 48 h (RT-qPCR and cell proliferation
assay) and 72 h (for western blotting and cell cycle assay)
subsequent to transfection. The transfection efficiency was
confirmed by RT-qPCR and western blot analysis.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total cellular RNA was extracted from cell pellets
with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. Relevant complementary
DNA was amplified by PCR with the Reverse Transcription System
A3500 (Promega Corporation, Madison, WI, USA). Reverse
transcription was performed at 42°C for 60 min followed by heating
at 95°C for 5 min.
The primer sequences were customized by
Sigma-Aldrich (Merck KGaA, Darmstadt, Germany) as follows: PARP1
(accession no. NM_001618) forward, 5′-GACGAGCTAAAGAAAGTGTGTTCAA-3′
and reverse, 5′-GGTCCAAGATCGCCGACTC-3′; GAPDH (accession nos.
NM_002046, NM_0012,56799, NM_001289745 and NM_001289746) forward,
5′-CAACTACATGGTTTACATGTTC-3′ and reverse, 5′-AAATGAGCCCCAGCCTTC-3′,
which was used as an internal control.
RT-qPCR was performed using the FastStart DNA Master
SYBR Green I (Roche Diagnostics, Basel, Switzerland) and
LightCycler System (Roche Diagnostics). The cycling conditions were
as follows: 1 cycle at 95°C for 10 min followed by 45 cycles at
95°C for 10 sec, 54°C for 10 sec, and 72°C for 10 sec.
Western blot analysis
Cells were resuspended in ice-cold
radioimmunoprecipitation assay buffer (Thermo Fisher Scientific,
Inc.) supplemented with a cocktail of protease/phosphatase
inhibitors (Thermo Fisher Scientific, Inc.). Cells were further
incubated on ice for 10 min and centrifuged at 14,000 × g at 4°C
for 20 min. Subsequent to determination of the protein
concentration using Bio-Rad Protein Assay (catalog no., 5000006JA;
Bio-Rad Laboratories, Inc., Hercules, CA, USA), the proteins in
each sample were resolved on SDS-PAGE (10% gel; Bio-Rad
Laboratories, Inc.) and transferred onto polyvinylidene difluoride
membranes (Merck KGaA). The membranes were washed with
Tris-buffered saline containing 0.1% Tween-20 (TBST) and blocked
with Blocking One-P (Nacalai Tesque, Inc.) at room temperature. The
membranes were incubated with respective primary antibodies against
different targets at 4°C overnight. Subsequent to incubation with
primary antibodies, secondary antibodies were added and incubated
for 1 h at room temperature. Subsequent to incubation with
secondary antibodies, signals were detected with ECL Prime Western
Blotting Detection Reagent (GE Healthcare Life Sciences, Little
Chalfont, UK).
Cell proliferation assay
Cells were seeded into 96-well plates at a density
of 5×103 cells/100 µl/well (Costar; Corning Inc.,
Corning, NY, USA) for 24 h, and then transfected with siPARP
(catalog nos. sc-29437A-C) or negative control siRNA (catalog no.
sc-37007). Cell viability was quantified subsequent to 0, 24, 48
and 72 h of transfection by the WST-8 assay using the Cell Counting
Kit-8 (catalog no. 343-07623; Dojindo Laboratories, Kumamoto,
Japan), according to the manufacturer's protocol. Absorbance was
measured at 450 nm with the Model 680XR Microplate Reader (Bio-Rad
Laboratories, Inc.).
Cell cycle assay
The cell cycle was assessed by flow cytometry.
First, 96 h prior to analysis, cells were seeded into 6-well plates
at a density of 5×105 cells/2 ml/well. A total of 72 h
prior to analysis, cells were transfected with siPARP1 or negative
control. Subsequent to treatment with siRNA, cells were harvested,
washed twice with PBS, and suspended with 70% cold ethanol (500 µl)
for 30 min on ice. Cells were washed with PBS, then resuspended
with 100 µg/ml RNase A (Qiagen, Hilden, Germany) for 20 min at 37°C
and 25 µg/ml propidium iodide (PI; Dojindo Laboratories) for 20 min
on ice. Subsequent to treatment, PI fluorescence was analyzed using
FACS Canto II (BD Biosciences, Franklin Lakes, NJ, USA). Data from
at least 10,000 cells were analyzed using BD FACSDIVA 7.0 (BD
Biosciences). Cell cycle distribution was calculated with FlowJo
software version 8.8.7 (Tree Star, Inc., San Carlos, CA, USA).
Statistical analysis
Associations between PARP1 expression and
clinicopathological factors were analyzed using the χ2
test for categorical variables and the Mann-Whitney U test for
continuous variables. Overall survival (OS) was defined as the
period from the date of surgery to the date of mortality from any
cause. Survival was estimated using the Kaplan-Meier method and
compared using the log-rank test. The hazard ratio (HR) for
recurrence or mortality in the group with high expression of PARP1
was estimated using a Cox proportional hazards model. Multivariate
Cox regression analysis was performed to adjust for potential
confounding factors. A statistically significant difference was
indicated by P<0.05. All reported P-values were two-tailed. All
statistical analyses were performed with JMP Pro 11.2.1 software
(SAS Institute, Cary, NC, USA).
Results
Patient characteristics and expression
of PARP1 in ESCC tissues
The present study examined the expression of PARP1
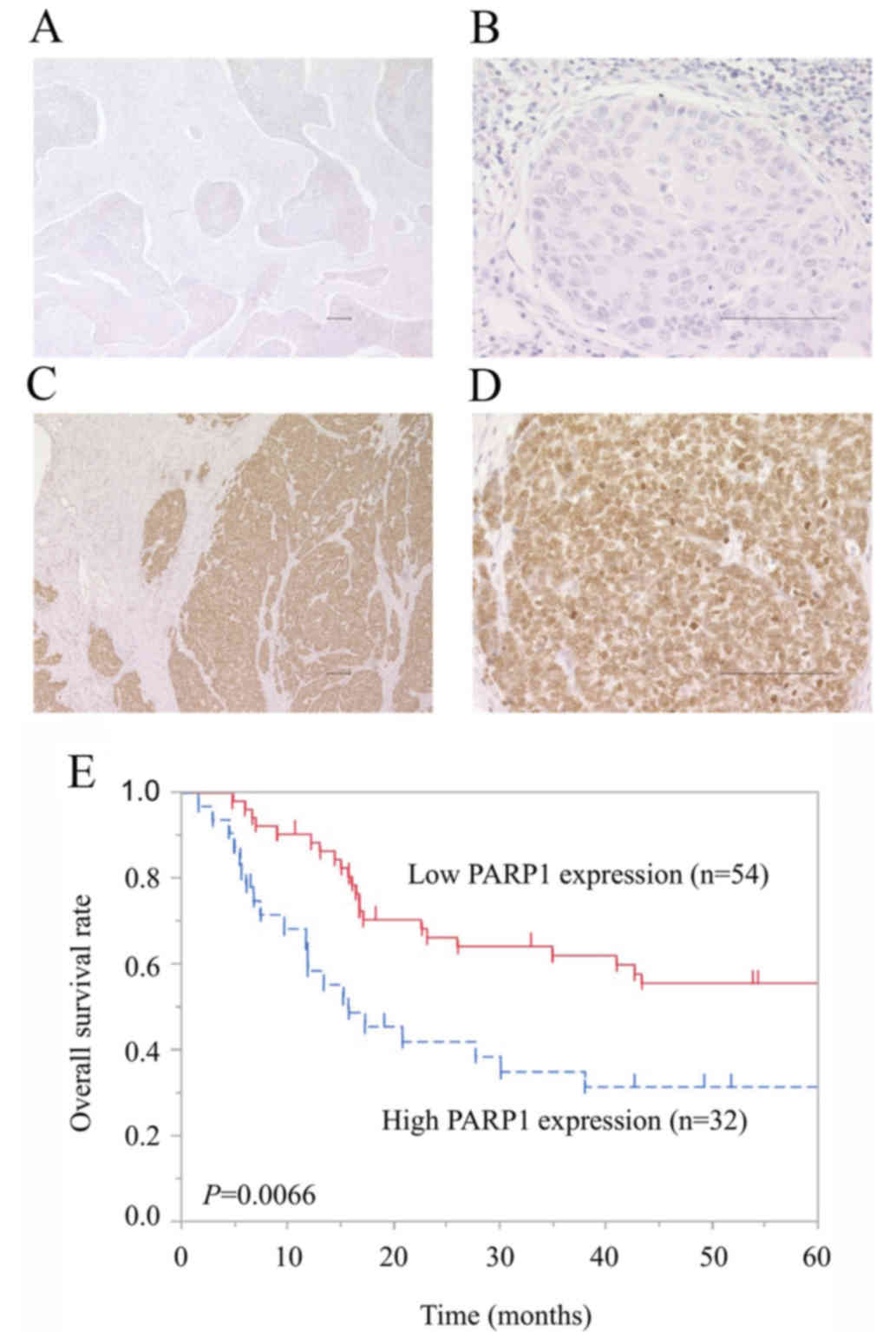
in ESCC tissues by IHC staining. The expression PARP1 in cancer
cells was observed in the cytoplasm and nucleus, but predominantly
in the nucleus (Fig. 1A-D). Among the
86 patients with ESCC, low expression of PARP1 was observed in 54
patients (62.8%) and high expression was observed in 32 patients
(37.2%). No significant difference in clinicopathological features
was observed between the patients with low and the patients with
high expression of PARP1 (Table
I).
 | Table I.Association between PARP1 expression
and clinicopathological features in 86 patients with ESCC. |
Table I.
Association between PARP1 expression
and clinicopathological features in 86 patients with ESCC.
|
| PARP1 expression,
n |
|
|
|---|
|
|
|
|
|
|---|
| Characteristic | Low | High | Total, n | P-value |
|---|
| Total | 54 | 32 | 86 |
|
| Age, years |
|
|
| 0.7295 |
|
<65 | 24 | 13 | 37 |
|
| ≥65 | 30 | 19 | 49 |
|
| Sex |
|
|
| 0.4653 |
| Male | 46 | 29 | 75 |
|
|
Female | 8 | 3 | 11 |
|
| Esophageal
location |
|
|
| 0.2792 |
|
Upper | 7 | 7 | 14 |
|
|
Middle/lower | 47 | 25 | 72 |
|
| Histology of SCC |
|
|
| 0.6725 |
|
Well/moderately
differentiated | 40 | 25 | 65 |
|
| Poorly
differentiated | 14 | 7 | 21 |
|
| Venous invasion |
|
|
| 0.8681 |
| No | 26 | 16 | 42 |
|
| Yes | 28 | 16 | 44 |
|
| Lymphatic
invasion |
|
|
| 0.6351 |
| No | 5 | 4 | 9 |
|
| Yes | 49 | 28 | 77 |
|
| Depth of tumor
invasion |
|
|
| 0.9500 |
|
pT1-2 | 24 | 14 | 38 |
|
|
pT3-4 | 30 | 18 | 48 |
|
| Lymph node
metastasis |
|
|
| 0.3370 |
| pN0 | 17 | 7 | 24 |
|
|
pN1-3 | 37 | 25 | 62 |
|
Association between prognosis and
expression of PARP1
The mean follow-up time for all patients in the
present study was 45.3±41.8 months. The group with high PARP1
expression had a significantly worse OS compared with patients with
low expression [HR 2.25; 95% confidence interval (CI), 1.23–4.11;
P=0.0092; Fig. 1E]. The 5-year OS
rate was 31.6% in the high-expression group and 55.7% in the
low-expression group. Multivariate Cox regression analysis revealed
that high expression of PARP1 was a statistically significant
independent prognostic factor of poor OS, along with pT3 and 4 and
pN1-3 disease (Table II). The
adjusted HR for OS in the group with high PARP1 expression was 2.39
(95% CI, 1.29–4.44; P=0.0051).
 | Table II.Univariate and multivariate Cox
regression analyses for overall survival in 86 patients with
ESCC. |
Table II.
Univariate and multivariate Cox
regression analyses for overall survival in 86 patients with
ESCC.
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Characteristic | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age, ≥65 vs. <65
years | 1.24 | 0.68–2.36 | 0.4846 |
|
|
|
| Sex, male vs.
female | 4.02 | 1.23–24.67 | 0.0169 | 3.29 | 0.97–20.55 | 0.0578 |
| Location,
middle/lower vs. upper | 1.31 | 0.69–2.41 | 0.3949 |
|
|
|
| Histology of SCC,
poorly vs. well/moderately differentiated | 1.48 | 0.73–2.82 | 0.2631 |
|
|
|
| Venous invasion,
positive vs. negative | 1.47 | 0.80–2.71 | 0.2112 |
|
|
|
| Lymphatic invasion,
positive vs. negative | 2.57 | 0.79–15.75 | 0.1315 |
|
|
|
| pT, 3–4 vs.
1–2 | 2.68 | 1.42–5.35 | 0.0021 | 2.10 | 1.06–4.42 | 0.0335 |
| pN, 1–3 vs. 0 | 3.36 | 1.52–8.88 | 0.0018 | 2.66 | 1.14–7.29 | 0.0219 |
| PARP1 expression,
high vs. low | 2.25 | 1.23–4.11 | 0.0092 | 2.39 | 1.29–4.44 | 0.0061 |
PARP1 mRNA expression in ESCC cell
lines
RT-qPCR analysis was performed to compare the mRNA
expression of PARP1 in the ESCC TE1, TE4, TE5, TE6, TE8, TE9, TE10
and TE11 cell lines. The expression of PARP1 mRNA was the highest
in TE9 cells and the second highest in TE6 cells (Fig. 2A). Accordingly, TE6 and TE9 cells were
selected for subsequent analysis in a PARP1 inhibition assay.
RT-qPCR and western blot analysis showed that siPARP1 significantly
reduced the expression of PARP1 mRNA and protein compared with
negative control cells (Fig. 2B).
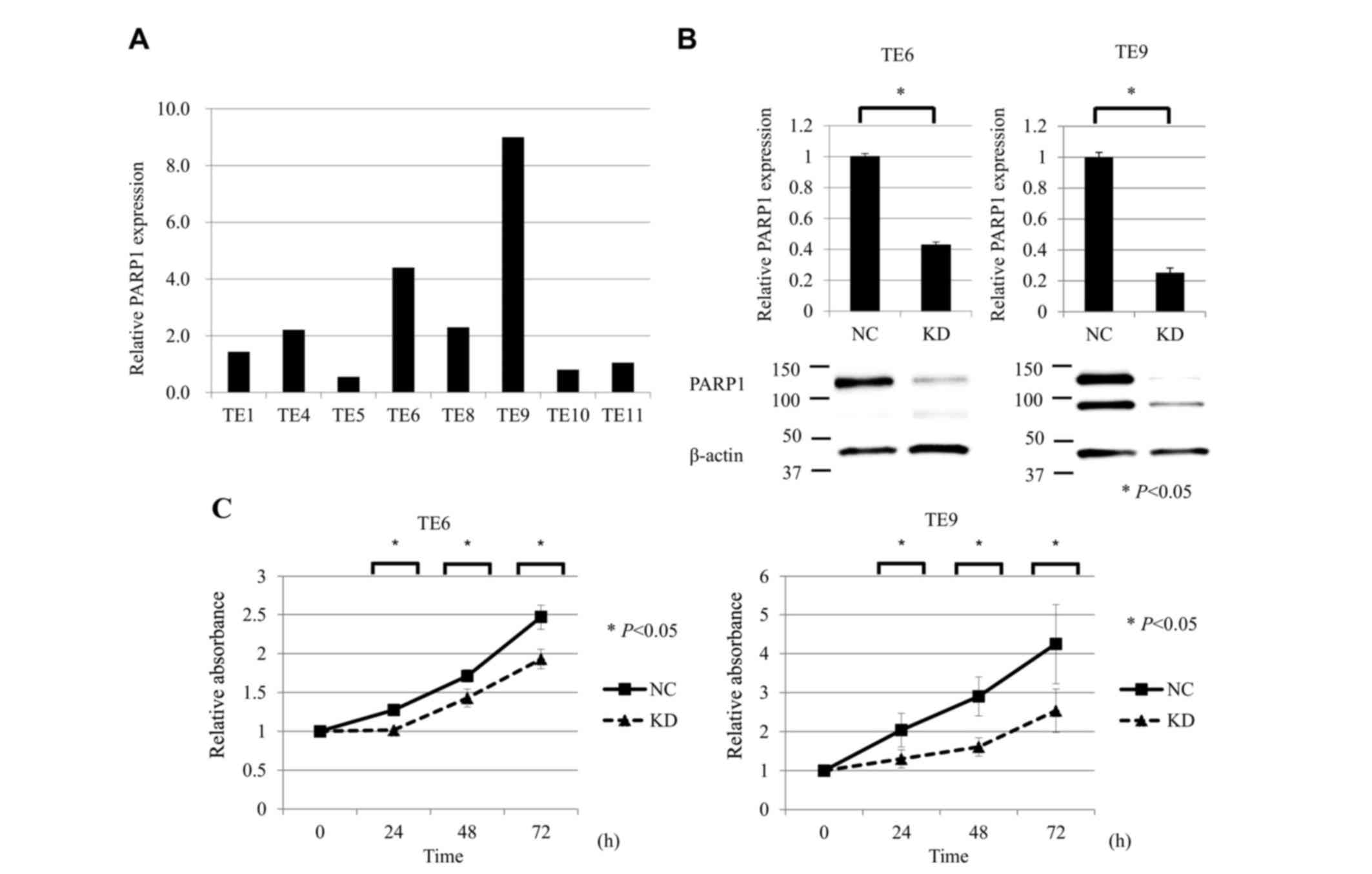 | Figure 2.Suppression of PARP1 inhibited ESCC
cell growth. (A) RT-qPCR analysis of PAPR1 mRNA expression in the
ESCC TE1, TE4, TE5, TE6, TE8, TE9, TE10 and TE11 cell lines. The
expression of PARP1 mRNA was the highest in TE9 cells and the
second highest in TE6 cells. (B) RT-qPCR and western blot analysis
showed that siPARP1 significantly reduced the expression of PARP1
mRNA and protein compared with the NC in TE6 and TE9 cells. (C)
Cell proliferation assay of ESCC cells (TE6 and TE9) comparing NC
and KD of PARP1. ESCC cells transfected with siPARP1 showed
significantly decreased proliferation compared with NC (at 24, 48
and 72 h). For all experiments, n=5, and KD was compared with NC at
each time-point (0, 24, 48, and 72 h). *P<0.05. PARP, poly
(ADP-ribose) polymerase-1; ESCC, esophageal squamous cell
carcinoma; RT-qPCR, reverse transcription-quantitative polymerase
chain reaction; siPARP1, small interfering RNA against PARP1; NC,
negative control; KD, knock-down. |
Inhibition of PARP1 reduces the
proliferative activity of ESCC cell lines
The effects of PARP1 inhibition on the proliferative
activity of ESCC TE6 and TE9 cell lines were examined. siPARP1
significantly inhibited cell growth compared with the negative
control siRNA at 24, 48 and 72 h after transfection (Fig. 2C).
Inhibition of PARP1 induces cell cycle
arrest in ESCC cell lines
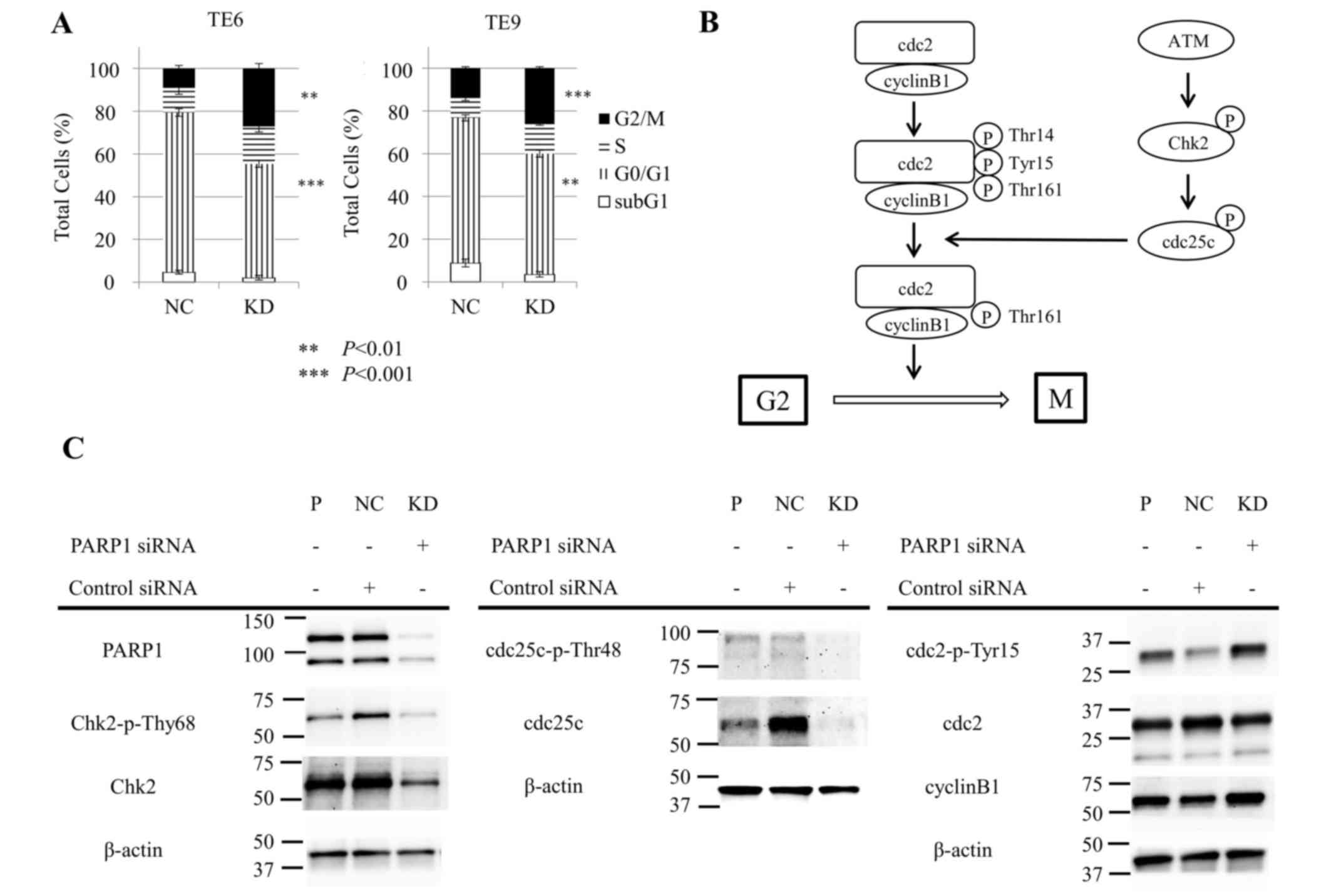
Subsequently, the effect of siPARP1 on the cell
cycle in ESCC TE6 and TE9 cells was examined with flow cytometry.
siPARP1 significantly increased the ratio of cells in the G2/M
phase compared with negative control siRNA (TE6, P<0.01; TE9,
P<0.001) and significantly decreased the ratio of cells in the
G0/G1 phase (TE6, P<0.001; TE9, P<0.01) (Fig. 3A). These results indicate that siPARP1
affected the G2/M checkpoint. Fig. 3B
shows the schema of the G2/M checkpoint. siPARP1-induced G2/M
arrest by was investigated by western blotting with monoclonal
antibodies specific for several key regulators, consisting of Chk2,
phosphorylated (p-)Chk2 (Thr68), cdc25c, p-cdc25c (Thr48), cdc2,
p-cdc2 (Tyr15) and cyclin B1. Cells treated with siPARP1 showed a
notable decrease in p-Chk2 (Thr68) and p-cdc25c (Thr48) expression,
and a notable increase in p-cdc2 (Tyr15) expression compared with
parental and negative control cells (Fig.
3C).
Discussion
In the present study, IHC staining was used to
demonstrate that the expression level of PARP1 was useful in
predicting clinical outcomes in patients with ESCC. Furthermore, it
was demonstrated that inhibition of PARP1 with siPARP1 reduced the
proliferative activity of ESCC cells, and the effect of PARP1
inhibition induced cell cycle arrest at the G2/M phase. At present,
although various anticancer drugs have been widely used in patients
with ESCC, molecular targeted drugs have not been established as
the treatment for patients with ESCC. The present study also
suggests that new molecular targeted treatment for ESCC using PARP
inhibitors may have potential in a clinical setting.
Previous studies have reported that PARP1 plays a
critical role in responding to DNA damage by activating DNA repair
pathways responsible for cellular survival (11,19). In
breast and ovarian cancers in particular, PARP inhibitors are the
most recent treatment and are highly regarded (12). Additionally, previous studies have
indicated an association between PARP1 and the cell cycle. Jelinic
and Levine (16) reported that PARP
inhibitors did not affect homology-directed DNA damage, but did
affect cell cycle arrest at the G2 phase in a human osteosarcoma
cell line. Park et al (7)
reported that PARP1 inhibition significantly attenuated growth and
colony formation, and induced G2/M arrest in gastric cancer cells.
Overall, it was hypothesized that PARP1 inhibition suppressed
proliferation and regulated the cell cycle at the G2/M checkpoint
in ESCC. The present study supported this hypothesis by analyzing
experimental data from proliferation and cell cycle assays. Flow
cytometry showed that PARP1 inhibition induced cell cycle arrest at
the G2/M phase. By contrast, no significant difference in apoptosis
was observed between the negative control group and the
siPARP1-treated group (data not shown). These results were
supported by a previous study (7).
In addition, western blotting was used to examine
the detailed mechanisms of G2/M arrest induced by PARP1 inhibition.
This analysis showed that PARP1 inhibition inhibited the
phosphorylation of Chk2 and cdc25c, the latter of which is
responsible for removal of phosphates at Thr14 and Tyr15 and the
subsequent activation of cdc2 (20,21).
Therefore, these results revealed that siPARP1 induced cell cycle
arrest at the G2/M phase through the ataxia telangiectasia mutated
(ATM)-Chk2-cdc25c pathway, suggesting that PARP1 may interact with
the ATM-Chk2 pathway. PARP1 inhibition has potential in ESCC
therapy by acting via the induction of cell cycle arrest at the
G2/M phase, through the ATM-Chk2-cdc25c pathway.
There are several limitations to the present study.
One is the relatively small number of tissue samples from patients
with ESCC, thus restricting the IHC analysis. Additional
multicenter studies involving more patients are required. Another
limitation is that the present study was conducted strictly in
vitro. Additional studies focusing on the effect of PARP
inhibition in vivo are required to investigate the potential
clinical application of the present findings in patients with
ESCC.
In conclusion, the present IHC analysis showed that
PARP1 may be an independent prognostic marker in ESCC, and
experiments using ESCC cells demonstrated that PARP1 inhibition
could induce cell cycle arrest at the G2/M phase through the
ATM-Chk2-cdc25c pathway. With respect to personalized treatments,
PARP inhibitors may be of use in patients with ESCC that show high
PARP1 expression in the future.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nakamura M, Iwahashi M, Nakamori M, Ojima
T, Katsuda M, Iida T, Hayata K, Kato T and Yamaue H: New prognostic
score for the survival of patients with esophageal squamous cell
carcinoma. Surg Today. 44:875–883. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Okumura H, Uchikado Y, Setoyama T,
Matsumoto M, Owaki T, Ishigami S and Natsugoe S: Biomarkers for
predicting the response of esophageal squamous cell carcinoma to
neoadjuvant chemoradiation therapy. Surg Today. 44:421–428. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Borghesi S, Hawkins MA and Tait D:
Oesophagectomy after definitive chemoradiation in patients with
locally advanced oesophageal cancer. Clin Oncol (R Coll radiol).
20:221–226. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Makino T, Yamasaki M, Miyata H, Tanaka K,
Takahashi T, Kurokawa Y, Nakajima K, Takiguchi S, Mori M and Doki
Y: Solitary lymph node recurrence of esophageal squamous cell
carcinoma: Surgical failure or systemic disease? Ann Surg Oncol.
23:2087–2093. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Alexander BM, Wang XZ, Niemierko A, Weaver
DT, Mak RH, Roof KS, Fidias P, Wain J and Choi NC: DNA repair
biomarkers predict response to neoadjuvant chemoradiotherapy in
esophageal cancer. Int J tadiat Oncol Biol Phys. 83:164–171. 2012.
View Article : Google Scholar
|
|
7
|
Park SH, Jang KY, Kim MJ, Yoon S, Jo Y,
Kwon SM, Kim KM, Kwon KS, Kim CY and Woo HG: Tumor suppressive
effect of PARP1 and FOXO3A in gastric cancers and its clinical
implications. Oncotarget. 6:44819–44831. 2015.PubMed/NCBI
|
|
8
|
Hoeijmakers JH: Genome maintenance
mechanisms for preventing cancer. Nature. 411:366–374. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Juarez-Salinas H, Sims JL and Jacobson MK:
Poly(ADP-ribose) levels in carcinogen-treated cells. Nature.
282:740–741. 1979. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Durkacz BW, Omidiji O, Gray DA and Shall
S: (ADP-ribose)n participates in DNA excision repair. Nature.
283:593–596. 1980. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rouleau M, Patel A, Hendzel MJ, Kaufmann
SH and Poirier GG: PARP inhibition: PARP1 and beyond. Nat Rev
Cancer. 10:293–301. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hoeijmakers JH: DNA damage, aging and
cancer. N Engl J Med. 361:1475–1485. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Helleday T, Petermann E, Lundin C, Hodgson
B and Sharma RA: DNA repair pathways as targets for cancer therapy.
Nat Rev Cancer. 8:193–204. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rottenberg S, Jaspers JE, Kersbergen A,
van der Burg E, Nygren AO, Zander SA, Derksen PW, de Bruin M,
Zevenhoven J, Lau A, et al: High sensitivity of BRCA1-deficient
mammary tumors to the PARP inhibitor AZD2281 alone and in
combination with platinum drugs. Proc Natl Acad Sci USA. 105:pp.
17079–1784. 2008; View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bryant HE, Schultz N, Thomas HD, Parker
KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ and Helleday T:
Specific killing of BRCA2-deficient tumours with inhibitors of
poly(ADP-ribose) polymerase. Nature. 434:913–917. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jelinic P and Levine DA: New insights into
PARP inhibitors' effect on cell cycle and homology-directed DNA
damage repair. Mol Cancer Ther. 13:1645–1654. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sakogawa K, Aoki Y, Misumi K, Hamai Y, Emi
M, Hihara J, Shi L, Kono K, Horikoshi Y, Sun J, et al: Involvement
of homologous recombination in the synergism between cisplatin and
poly (ADP-ribose) polymerase inhibition. Cancer Sci. 104:1593–1599.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sobin LH and Compton CC: TNM seventh
edition: what's new, what's changed: Communication from the
international union against cancer and the American Joint committee
on cancer. Cancer. 116:5336–5339. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gradwohl G, Ménissier de Murcia JM,
Molinete M, Simonin F, Koken M, Hoeijmakers JH and de Murcia G: The
second zinc-finger domain of poly(ADP-ribose) polymerase determines
specificity for single-stranded breaks in DNA. Proc Natl Acad Sci
USA. 87:pp. 2990–2994. 1990; View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Atherton-Fessler S, Liu F, Gabrielli B,
Lee MS, Peng CY and Piwnica-Worms H: Cell cycle regulation of the
p34cdc2 inhibitory kinases. Mol Biol Cell. 5:989–1001. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hunter T: Protein kinases and
phosphatases: The yin and yang of protein phosphorylationand
signaling. Cell. 80:225–236. 1995. View Article : Google Scholar : PubMed/NCBI
|