Introduction
Liver cancer is the most common primary malignant
tumor in the liver and ranks 5th among malignant tumors and 3rd in
the mortality rate (1). Various
factors can cause liver cancer, including excessive alcohol
consumption, viral hepatitis, and chronic liver inflammation caused
by non-alcoholic hepatic steatosis (2–4). Chronic
inflammation in the liver can lead to recurrent injuries and
proliferation of hepatic cells, further activating oncogenes as
well as liver cancer-associated signal pathways and deactivating
the tumor suppressor genes (5,6).
MicroRNAs (miRNAs) are non-coding single-stranded
small RNAs 18–25 nucleotides in length. miRNAs can exert gene
regulatory functions at the translational level, and play key roles
in development, proliferation, differentiation, apoptosis, and
carcinogenesis (7). Over 1,000 miRNAs
are encoded in the human genome, which have the ability to regulate
the expression of 60% of protein-encoding genes (8). miRNAs can act on the 3′-untranslated
region (UTR) of target mRNAs, resulting in the abnormal
downregulation of target genes (9).
Reports have described the abnormal expressions of
multiple miRNAs in hepatic cells and peripheral serum of liver
cancer patients (10). Among these
miRNAs, miR-375, which is located on the genetic regions of
cryba2 and Ccdc108 on 2q35, can inhibit the transcription and
translation of the oncogene astrocyte elevated gene-1 (AEG-1),
exerting the anti-carcinoma function in liver cancer cells
(11). miR-221, which is
located on the P11.3 region of the X chromosome, is closely
correlated with tumors because the expression of miR-221 is
significantly elevated in many malignant tumor cells (12). miR-221 can regulate the cell
cycle, differentiation and apoptosis, as well as participate in the
occurrence and development of tumor by adjusting the expression of
p27, p53 upregulated modulator of apoptosis (PUMA), Bcl-2 modifying
factor (BMF), c-kit, and DNA-damage-inducible transcript 4 protein
(DDIT4) (13).
In a previous study, the differences in miRNA
expression in liver cancer cells and normal liver cells were
compared, and the results confirmed that upregulated expression of
miR-221 and downregulated expression of miR-375 are
involved in the occurrence and development of liver cancer by
regulating the cell proliferation, cycle, apoptosis, migration and
invasion, as well as clone formation (14,15).
However, to the best of our knowledge, currently, there are no
reports on the expressions of miR-375 and miR-221
directly in liver cancer tissues along with the pathological
parameters and prognosis of liver cancer.
In the present study, we used quantitative RT-(q)PCR
to determine the expression levels of miR-375 and
miR-221 in liver tumor and tumor-adjacent normal tissue. We
analyzed the correlation of miR-375 and miR-221
expressionwith clinicopathological parameters and prognosis of
liver cancer in combination with clinical data.
Materials and methods
Human tumor tissue
We collected frozen tumors and tumor-adjacent normal
tissue from 70 patients with liver cancer who were admitted to the
Department of General Surgery of Zhejiang Hospital for treatment
between January 2008 and December 2010. These tumors were diagnosed
as liver cancer through pathological examinations. All 70 patients
received surgical treatment for the first time and had no
chemotherapy history. This cohort had 38 males and 32 females, and
the age range was 25–78 years with a median age of 45 years. The
acquisition of samples was approved by the Clinical Ethics
Committee of Zhejiang Hospital and all enrolled patients or their
family signed the written informed consent. The 70 patients
received postoperative follow-up for 5 years and the follow-up rate
reached 100%. Recording of the survival time started from the 1st
day after operation, and ended on the date of death of the patient
or the last day of follow-up. Statistical analysis was carried out
with the month as the unit.
Quantitative RT-PCR
Two samples (~50 mg each) were used from each frozen
tumor and tumor-adjacent normal tissue. Total RNA was extracted
according to the instructions of the RNA extraction kit (Invitrogen
Life Technologies, Carlsbad, CA, USA). Concentration and
purification of total RNA were detected using ultraviolet-visible
spectrophotometer (Hitachi High-Technologies Corp., Tokyo, Japan),
and extracted RNA was classified as qualified when the ratio of
A260/A280 was between 1.8 and 2.0. Then, cDNA was generated by
reverse transcription according to the instructions in the
reverse-transcription kit. With the cDNA as template, the
expression of miR-375 and miR-221 was detected
according to the method given in the instructions of the RT-PCR kit
with U6 RNA as internal reference. Synthesis of primer,
reverse-transcription kit, and real-time fluorescent quantitative
PCR kit (Takara Bio, Dalian, China). Primer sequences of
miR-375, miR-221 and U6 are shown in Table I, and reaction conditions were: 95°C
for 10 min, 95°C for 15 sec, 60°C for 1 min; 40 cycles of
amplification. Ct value was obtained, and the relative expression
was calculated using the method of 2−∆∆Ct. Calculation
was carried out according to the formula: ∆Ct (target gene) = Ct
(target gene) - Ct (reference gene).
 | Table I.Primer sequences used for qRT-PCR. |
Table I.
Primer sequences used for qRT-PCR.
| Gene | Primer sequences |
|---|
| miR-375 | F:
5-GGCTCTAGAGGGGACGAAGC-3 |
|
| R:
5-GGCAAGCTTTTTCCACACCTCAGCCTTG-3 |
| miR-221 | F:
5-CAAGGAATCATGTATGCTGTAG-3 |
|
| R:
5-AGGATGACATTACACCTTATCTC-3 |
| U6 | F:
5-GCTTCGGCAGCACATATACTAAAAT-3 |
|
| R:
5-CGCTTCACGAATTTGCGTGTCAT-3 |
Statistical analysis
Data processing was performed using SPSS 17.0
software (International Business Machines Corporation, Armonk, NY,
USA). Measurement data are presented as mean ± standard deviation
and t-test was used for intergroup comparison. For countable data,
Chi-square test was used for intergroup comparison. Single-factor
survival analysis was carried out using the Kaplan-Meier method,
the log-rank method was used to identify the difference in survival
curve, and multivariate survival analysis was carried out using the
Cox proportional hazards model. P≤0.05 indicates that the
difference has statistical significance.
Results
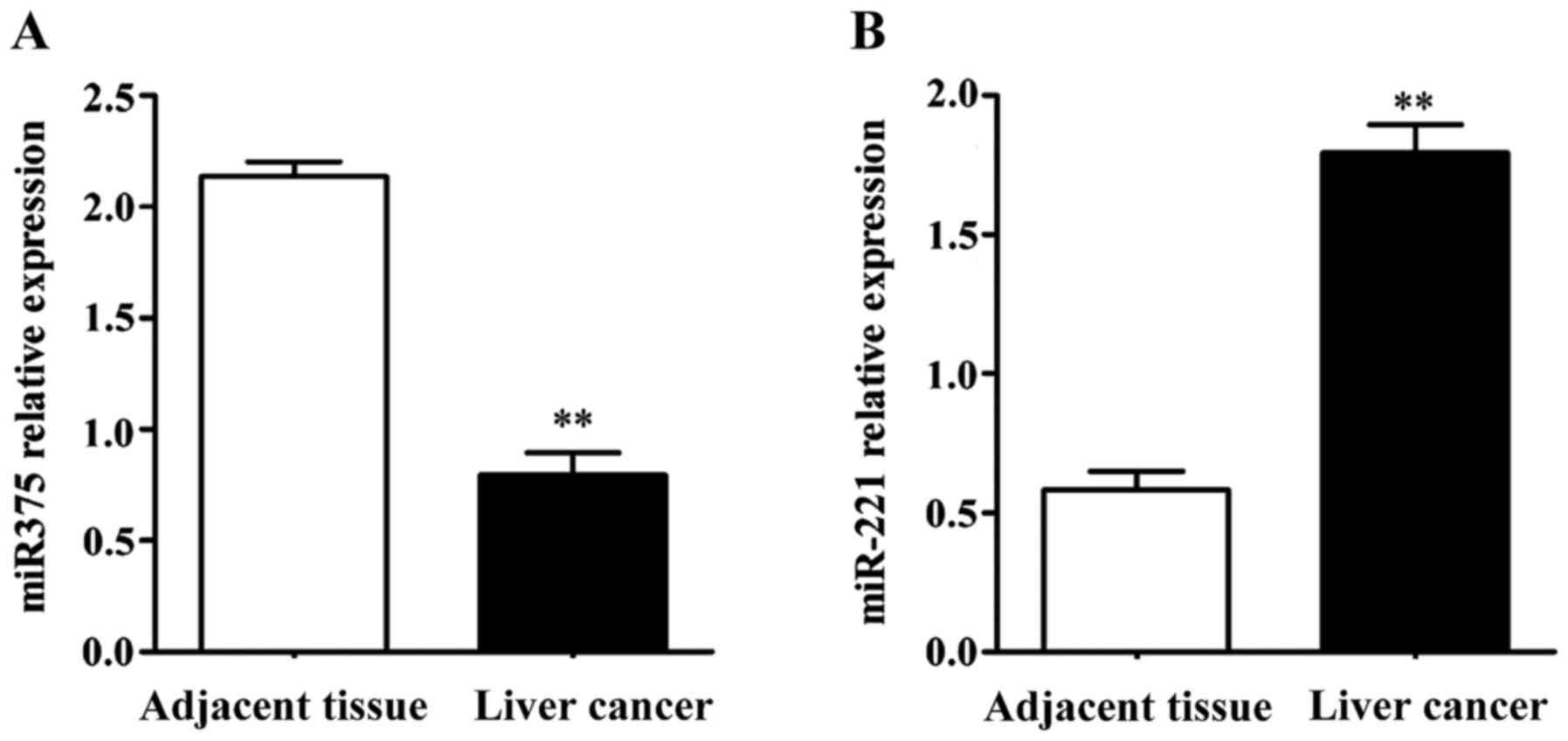
Expression of miR-375 and miR-221 by
RT-qPCR
We extracted RNA from the liver tumor and normal
adjacent tissues and examined the expression of miR-375 and
miR-221. Compared with the tumor-adjacent normal tissues,
the expression of miR-375 was significantly decreased in
liver cancer (Fig. 1). By contrast,
the expression of miR-221 was significantly elevated in
liver cancer (Fig. 1).
Correlation of miR-375 and miR-221
expression with pathological parameters of liver cancer
Based on the expression levels of miR-375 and
miR-221 in the 70 liver cancer tissues, the samples were
divided into the miR-375 high-expression (≥2.135),
miR-375 low-expression (<2.135), miR-221
high-expression (≥1.795), and miR-221 low-expression
(<1.795) groups. No statistically significant differences were
identified in the comparisons of factors such as age, sex and
smoking history between the two groups (Table II). According to the clinical
materials, we analyzed the correlations between the expression of
miR-375 and miR-221 and the pathological parameters.
Chi-square test showed that the abnormal expression of
miR-375 and miR-221 correlated with the occurrence of
metastasis and TNM staging (tumor-node-metastasis), but was not
correlated with sex, age and tumor size (Table II).
 | Table II.Correlation of miR-375 and
miR-221 expression with pathological parameters. |
Table II.
Correlation of miR-375 and
miR-221 expression with pathological parameters.
|
|
| miR-375 | miR-221 |
|---|
|
|
|
|
|
|---|
| Clinical data | Case | High expression
(case, %) | Low expression
(case, %) | P-value | High expression
(case, %) | Low expression
(case, %) | P-value |
|---|
| Male | 38 | 12 (31.6) | 26 (68.4) | >0.05 | 23 (60.5) | 15 (39.5) | >0.05 |
| Female | 32 | 11 (34.4) | 21 (65.6) |
| 22 (68.8) | 10 (31.2) |
|
| Age ≥50 years | 41 | 14 (34.1) | 27 (65.9) | >0.05 | 25 (60.9) | 16 (39.1) | >0.05 |
| Age <50
years | 29 | 9 (31.0) | 20 (69.0) |
| 20 (68.9) | 9 (31.1) |
|
| Tumor size ≥5
cm | 44 | 15 (34.1) | 29 (63.6) | >0.05 | 26 (59.1) | 18 (40.9) | >0.05 |
| Tumor size <5
cm | 26 | 8 (30.8) | 18 (69.2) |
| 19 (73.1) | 7 (26.9) |
|
| Invasion and
metastasis | 48 | 12 (25.0) | 36 (75.0) | <0.05 | 37 (77.1) | 11 (22.9) | <0.05 |
| No invasion or
metastasis | 22 | 11 (50.0) | 11 (50.0) |
| 8 (36.4) | 14 (63.6) |
|
| Stage (I–II) | 42 | 18 (42.9) | 24 (57.1) | <0.05 | 23 (54.8) | 19 (45.2) | <0.05 |
| Stage (III–IV) | 28 | 5 (17.9) | 23 (82.1) |
| 22 (78.6) | 6 (21.4) |
|
Survival of patients with liver
cancer
The 70 liver cancer patients were followed-up for 5
years. At that point, there were 9 patients alive and 61 patients
dead due to further progression of liver cancer. The overall 5-year
survival rate was 12.9% (9/70) and the mortality rate was 87.1%
(61/70).
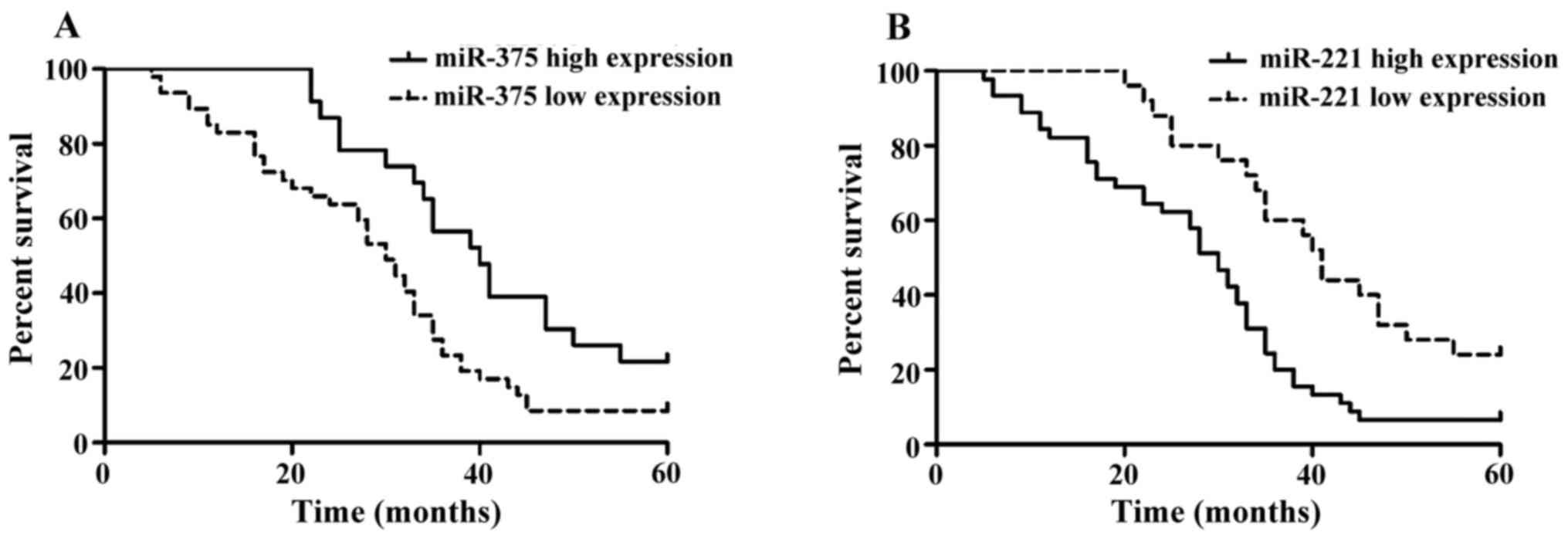
Single-factor analysis of the patient
prognosis
We next analyzed the Kaplan-Meier survival curves of
70 liver cancer patients with expression of miR-375 and
miR-221 (Fig. 2). Patients
with high miR-375 expression and low miR-221
expression had a better survival prognosis. Differences in the
curves of overall survival rate were analyzed using the log-rank
test (Table III). According to the
single-factor survival analysis, statistical significance was
identified in the effects of miR-375 and miR-221 on
the overall survival rate of liver cancer (Table III).
 | Table III.Single-factor analysis of
miR-375 and miR-221 expression with overall
survival. |
Table III.
Single-factor analysis of
miR-375 and miR-221 expression with overall
survival.
| Group | Case | 5-year survival
cases | 5-year survival
rate (%) | Wald
(log-rank) | P-value |
|---|
| miR-375 |
|
|
| 7.033 | <0.05 |
| High
expression | 23 | 5 | 21.7% |
|
|
| Low
expression | 47 | 4 | 8.5% |
|
|
| miR-221 |
|
|
| 11.23 | <0.05 |
| High
expression | 45 | 3 | 6.7% |
|
|
| Low
expression | 25 | 6 | 24% |
|
|
Multivariate analysis of expression of
miR-375 and miR-221 with survival
Correlation of miR-375 and miR-221
expression with overall survival rate of liver cancer patients were
analyzed via multivariate survival analysis by Cox proportional
hazards model. Expression levels were all substituted into the
formula, in which the substitution level was set as 0.05 and the
deletion level was set as 0.1. In the factors affecting the
survival of patients with liver cancer, the regression coefficient
of miR-375 was negative, indicating a relatively long
survival time of liver cancer patients with a high expression of
miR-375 (Table IV). However,
the regression coefficient of miR-221 was positive,
indicating a relatively short survival time of liver cancer
patients with a high expression of miR-221 (Table IV).
 | Table IV.Multivariate survival analysis of
miR-375 and miR-221 expression with overall survival
rate by Cox proportional hazards model. |
Table IV.
Multivariate survival analysis of
miR-375 and miR-221 expression with overall survival
rate by Cox proportional hazards model.
| Variate | B | SE | Wald | P-value | RR (95% CI) |
|---|
| miR-375 | −1.010 | 0.827 | 4.755 | 0.014 | 0.153
(0.035–0.837) |
| miR-221 | 0.748 | 0.203 | 4.870 | 0.012 | 1.743
(1.004–3.772) |
Discussion
As a common malignant tumor, liver cancer is
characterized by a relatively high mortality rate due to the lack
of effective early diagnosis and treatments (16). Recent studies on the effect and action
mechanism of miRNAs have identified key roles in the occurrence and
progression of tumors, changes in the microenvironment of tumor and
tumor-associated domestication of immune cells. Thus, miRNAs can be
used, not only in the diagnosis and prognosis of tumors, but also
as a target in the treatment of the tumor (17).
At present, there are several studies on the
correlation between miR-375 expression and tumors. As
biomarkers of liver cancer, miR-25, miR-375 and let-7f can
distinguish liver cancer cells, particularly when miR-375
was used alone to detect liver cancer cells, with 96% specificity
and 100% sensitivity (18). An in
vitro study showed that miR-375 expression is
significantly decreased in a liver cancer cell strain. High
expression of miR-375 can suppress the proliferation and
migration of tumor cells and induce cell cycle arrest and apoptosis
(14). Furthermore, it was shown that
miR-375 can weaken the migration and invasion capability of
liver cancer cells via inhibiting the expression of AEG-1 (11). Recent studies have reported a high
expression of miR-221 in liver cancer cells, which can
promote the growth and proliferation of liver cancer cells by
regulating cell differentiation (19). miR-221 suppresses the
activities of p27 and p57 by acting on cell cycle-dependent kinase,
further increasing the number of cells in the S phase of cell cycle
and facilitating the progression of the cell cycle in liver cancer
cells (20). Additionally,
miR-221 can also interfere with the mTOR (mammalian target
of rapamycin) signaling pathway via suppressing the DDIT4, thus
inducing the occurrence and progression of tumors (21).
To investigate the expression of miR-375 and
miR-221 in liver cancer, we found that miR-375
expression was elevated in liver cancer, but miR-221
expression was decreased. Further studies in combination with the
clinicopathological characteristics of patients showed that the
expression of miR-375 and miR-221 were correlated
with the metastasis and TNM staging of patients, but not correlated
with the sex, age and tumor size. We also found that miR-375
and miR-221 significantly affected the overall survival time
of liver cancer patients. Analysis by multivariate Cox proportional
hazards model showed that miR-375 and miR-221
affected the survival time of liver cancer patients and they were
the independent indexes for estimating the prognosis of liver
cancer patients. High expression of miR-375 can serve as an
indicator for excellent prognosis, while a high expression of
miR-221 is an indicator for poor prognosis.
Many studies have shown that miR-375 and
miR-221 are closely correlated with multiple kinds of
tumors. Expression of miR-375 in non-small cell lung cancer
is positively correlated with its prognosis (22). In esophageal squamous carcinoma can
exert a tumor-suppressing effect by inhibiting the expression of
insulin-like growth factor 1 receptor (23). Expression of miR-221 is
significantly upregulated in liver cancer cells and can promote the
growth and metastasis of tumor cells through p27 and c-kit
(21). In vitro studies showed
that the upregulated expression of miR-221 and downregulated
expression of miR-375 are identified in liver cancer cells
(14,15). Mechanistic studies confirmed that
these miRs can regulate proliferation, cell cycle, apoptosis and
migration (14,15).
In conclusion, a low expression of miR-375
and high expression of miR-221 are closely correlated with
the occurrence and development of liver cancer, particularly the
lymphatic metastasis and TNM staging. Thus, miR-375 and
miR-221 can serve as reference biomarkers for guiding the
treatment of liver cancer and estimating the prognosis.
References
|
1
|
Yang JD and Roberts LR: Epidemiology and
management of hepatocellular carcinoma. Infect Dis Clin North Am.
24:899–919. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Duan XY, Zhang L, Fan JG and Qiao L: NAFLD
leads to liver cancer: Do we have sufficient evidence? Cancer Lett.
345:230–234. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Karagozian R, Derdák Z and Baffy G:
Obesity-associated mechanisms of hepatocarcinogenesis. Metabolism.
63:607–617. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nakagawa H, Umemura A, Taniguchi K,
Font-Burgada J, Dhar D, Ogata H, Zhong Z, Valasek MA, Seki E,
Hidalgo J, et al: ER stress cooperates with hypernutrition to
trigger TNF-dependent spontaneous HCC development. Cancer Cell.
26:331–343. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang DY and Friedman SL:
Fibrosis-dependent mechanisms of hepatocarcinogenesis. Hepatology.
56:769–775. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
He G and Karin M: NF-κB and STAT3 - key
players in liver inflammation and cancer. Cell Res. 21:159–168.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Siomi H and Siomi MC: On the road to
reading the RNA-interference code. Nature. 457:396–404. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Panera N, Gnani D, Crudele A, Ceccarelli
S, Nobili V and Alisi A: MicroRNAs as controlled systems and
controllers in non-alcoholic fatty liver disease. World J
Gastroenterol. 20:15079–15086. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: Tools for microRNA genomics. Nucleic Acids
Res. 36(Database): D154–D158. Nov 8–2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yoo BK, Emdad L, Su ZZ, Villanueva A,
Chiang DY, Mukhopadhyay ND, Mills AS, Waxman S, Fisher RA, Llovet
JM, et al: Astrocyte elevated gene-1 regulates hepatocellular
carcinoma development and progression. J Clin Invest. 119:465–477.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vigouroux C, Auclair M, Dubosclard E,
Pouchelet M, Capeau J, Courvalin JC and Buendia B: Nuclear envelope
disorganization in fibroblasts from lipodystrophic patients with
heterozygous R482Q/W mutations in the lamin A/C gene. J Cell Sci.
114:4459–4468. 2001.PubMed/NCBI
|
|
13
|
Goldman RD, Gruenbaum Y, Moir RD, Shumaker
DK and Spann TP: Nuclear lamins: Building blocks of nuclear
architecture. Genes Dev. 16:533–547. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He XX, Chang Y, Meng FY, Wang MY, Xie QH,
Tang F, Li PY, Song YH and Lin JS: MicroRNA-375 targets AEG-1 in
hepatocellular carcinoma and suppresses liver cancer cell growth in
vitro and in vivo. Oncogene. 31:3357–3369. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
He XX, Guo AY, Xu CR, Chang Y, Xiang GY,
Gong J, Dan ZL, Tian DA, Liao JZ and Lin JS: Bioinformatics
analysis identifies miR-221 as a core regulator in hepatocellular
carcinoma and its silencing suppresses tumor properties. Oncol Rep.
32:1200–1210. 2014.PubMed/NCBI
|
|
16
|
Tang W, Zhu J, Su S, Wu W, Liu Q, Su F and
Yu F: MiR-27 as a prognostic marker for breast cancer progression
and patient survival. PLoS One. 7:e517022012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu Z, Zhang X, Wang G and Zheng H: Role
of microRNAs in hepatocellular carcinoma. Hepat Mon. 14:e186722014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang YS, Dai Y, Yu XF, Bao SY, Yin YB,
Tang M and Hu CX: Microarray analysis of microRNA expression in
hepatocellular carcinoma and non-tumorous tissues without viral
hepatitis. J Gastroenterol Hepatol. 23:87–94. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cohen M, Lee KK, Wilson KL and Gruenbaum
Y: Transcriptional repression, apoptosis, human disease and the
functional evolution of the nuclear lamina. Trends Biochem Sci.
26:41–47. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yuan Q, Loya K, Rani B, Möbus S,
Balakrishnan A, Lamle J, Cathomen T, Vogel A, Manns MP, Ott M, et
al: MicroRNA-221 overexpression accelerates hepatocyte
proliferation during liver regeneration. Hepatology. 57:299–310.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pineau P, Volinia S, McJunkin K, Marchio
A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM and
Dejean A: miR-221 overexpression contributes to liver
tumorigenesis. Proc Natl Acad Sci USA. 107:pp. 264–269. 2010;
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, Jiang Q, Xia N, Yang H and Hu C:
Decreased expression of microRNA-375 in nonsmall cell lung cancer
and its clinical significance. J Int Med Res. 40:1662–1669. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kong KL, Kwong DL, Chan TH, Law SY, Chen
L, Li Y, Qin YR and Guan XY: MicroRNA-375 inhibits tumour growth
and metastasis in oesophageal squamous cell carcinoma through
repressing insulin-like growth factor 1 receptor. Gut. 61:33–42.
2012. View Article : Google Scholar : PubMed/NCBI
|