Introduction
It is well known that recurrence and metastasis are
important causes of mortality in nasopharyngeal carcinoma (NPC)
patients (1). Unfortunately, the
mechanisms underlying the metastasis of NPC remain unclear.
Therefore, there is an urgent need to investigate the mechanisms.
The CNE3 cell line was obtained from liver metastatic carcinoma of
primary NPC and established in 1992 (2). A molecular pathological study has
previously indicated that the histological type of the CNE3 cell
line transformed from an undifferentiated non-keratinizing
carcinoma with focal adenocarcinoma differentiation into a poorly
differentiated adenocarcinoma (3).
The results provided approaches for studying metastatic NPC.
Tumor heterogeneity is a subclonal process, where
clones of cancer cells may differ in characteristics, including
karyotype, invasiveness, growth rate, expression of cell surface
markers and sensitivity to therapeutics (4,5). According
to Paget's theory, metastasis is clonal in its nature (6). Therefore, heterogeneous characteristics
of clonal sublines are an important reason that malignant tumor is
inclined to recurrence and metastasis in the advanced stages
(7). Successful construction of tumor
sublines will provide an efficient tool for screening
metastasis-associated genes and investigating invasive and
metastatic mechanisms (8,9).
The pro-oncogene Zbtb7a is also named
Pokemon/FBI-1/OCZF/LRF, containing broad complex, tramtrack,
bric-a-brac/poxvirus and zinc finger (BTB/POZ) domain and
actin-binding repeats (10,11). Zbtb7a has a critical role in
oncogenesis (12). Zbtb7a is able to
repress transcription of Rb via its POZ domains in different cancer
cell lines (13), activate
transcription in fatty-acid synthase promoter and provide more
phospholipid membrane components required for rapid proliferation
of cancer cells (14). It has been
reported that Zbtb7a is closely associated with a number of
different cancer types, including breast cancer (15), prostate cancer (16), hepatocellular carcinoma (17), NPC (18)
and colorectal cancer (19). Early
studies by the present authors demonstrated that increasing
expression levels of Zbtb7a mRNA and protein were accompanied by
NPC progression in most cases (18).
Subsequently, short hairpin RNA (shRNA)2 plasmids were constructed
and screened for their ability to knock down Zbtb7a expression
(20). In the present study in order
to elucidate the characteristics of the CNE3 cell line from NPC
distant metastases, a number of sublines were established by single
cell cloning and stable passaging. Sublines with different
tumorigenicity were subsequently selected and heterogeneity in
cellular characteristics was investigated. Finally, the
associations between changes in Zbtb7a expression and heterogeneity
in cellular characteristics were analyzed.
Materials and methods
Cell culture
The human NPC epithelial cell line CNE3 was
originally obtained and preserved at the Research Center of Medical
Sciences, The People's Hospital of Guangxi Zhuang Autonomous Region
in Nanning (Guangxi, China) (2). The
CNE3 cells were grown in RPMI-1640 medium with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA).
Single cell cloning by serial
dilution
The single CNE3 cell clones were screened by serial
dilution. A total of 100 µl complete culture medium was added to 95
wells in the 96-well plate (Corning Incorporated, Corning, NY,
USA). A total of 200 µl CNE3 cell suspension (5×104/ml) was added
to one well (A1), and 100 µl suspension was transferred from well
A1 to B1 and mixed by gentle pipetting. The 1:2 dilutions were
repeated down the entire column, and 100 µl suspension was
discarded from well H1. A total of 100 µl additional medium was
added to each well in the first column, and 100 µl suspension was
transferred from the first column to the second column. The 1:2
dilutions were repeated across the entire plate, and 100 µl medium
was added to each well.
Soft agar colony formation
Thirteen CNE3 sublines were established by screening
single cell clones and stable passaging. The sublines were named
sequentially from CNE3-GX1 to CNE3-GX13. A base layer comprising 2
ml medium (0.5% agarose; Beijing Solarbio Science & Technology
Co., Ltd., Beijing, China) in a 6-well plate (Corning Incorporated)
was solidified at 4°C for 15 min. A cell layer containing 103 cells
in 1 ml medium (0.25% agarose) was solidified at 4°C for 15 min and
subsequently incubated at 37°C under 5% CO2. The number
of colonies (over 50 cells/colony) was counted after 14 days of
incubation using a CKX41 microscope (original magnification, ×40;
Olympus Corporation, Tokyo, Japan).
Xenograft mouse tumor model
Based on the results of soft agar colony formation
and cell morphology, 5 sublines and CNE3 were selected to establish
tumor model. A total of 60 BALB/c mice were obtained from the
Guangxi Medical University Laboratory Animal Centre. The mice were
female, 4-weeks old and weighed ~15 g, and were randomly divided
into six groups. Mice were raised in independent ventilation cages
(Tecniplast S.p.A, Buguggiate, Italy). The temperature was 20–25°C.
The atmosphere was 20–50 Pa. The light/dark cycle was 10/14 h. The
food and water were sterilized. A nude mouse was subcutaneously
injected with cells [107/200 µl phosphate-buffered saline (PBS)]
from different sublines. The end point of when the mice were
sacrificed was 8 weeks. Then they were anatomized and evaluated for
the probability of metastasis after 8 weeks. The weight and volume
of primary tumor were measured. The protocol for establishing the
transplanted tumor model of CNE3 was the same as the tumor model of
CNE3 sublines. Tumor volume was calculated by using the formula: V
= (π/6)(d1xd2)3/2. The present study was approved by the Ethics
Committee of the People's Hospital of Guangxi Zhuang Autonomous
Region (Nanning, China).
Histological evaluation and
immunohistochemistry (IHC)
Histological evaluation and IHC were performed as
previously described (3). The tissues
were obtained from nude mice transplanted with tumors of cells from
the CNE3 cell line and its sublines. The tissues were fixed for 24
h by 10% neutral formalin, at room temperature. Tissues were then
sliced into 4-µm thick sections. Tissues were stained with
hematoxylin and eosin (H&E); 0.5% hematoxylin stained for 5 min
and 0.5% eosin stained for 2 min at room temperature. A non-biotin
horseradish peroxidase ready-to-use two-step detection system
(ZSGB-BIO, Beijing, China) was used in the IHC analysis. There was
an increased abundance of positive staining observed in the brown
granules compared with the unspecific background staining. Staining
was primarily observed in the cell nucleus (p63) or cytoplasm
[cytokeratin (CK)5/6, CK7]. The positive cell rates and staining
intensities were analyzed in the intact slices by high power fields
(original magnification, ×200). The results of the positive cell
rates (<10%) and weak coloring were negative. The results of the
positive cell rates (>10%) and dark brown granules were
positive. Staining was determined as positive when >10% of the
cells were positively stained, and positive staining was observed
in the dark brown granules. H&E and IHC were analyzed using a
BX51 microscope (Olympus Corporation).
Quantitative reverse transcription
polymerase chain reaction (RT-qPCR) and western blotting
RT-qPCR and western blot analysis were performed as
previously described (18). For
RT-qPCR, total RNA from the CNE3-GX4, CNE3-GX6, CNE3-GX7, CNE3-GX10
and CNE3-GX11 sublines was extracted by TRIzol (Invitrogen; Thermo
Fisher Scientific, Inc.). RNA amount was detected by NanoDrop 2000
(Thermo Fisher Scientific, Inc.). Total RNA (4 µl) was analyzed by
1.2% agarose gel electrophoresis with 0.6 mol/l formaldehyde. The
gel was photographed by Bio Imaging System Gene Genius (Syngene,
Cambridge, UK). An equal amount (4 µg) of total RNA was synthesized
as a first-strand cDNA using the RevertAid™ First-Strand cDNA
Synthesis kit (Fermentas; Thermo Fisher Scientific, Inc.) according
to the manufacturer's protocol. The cDNA was the source of the
template. The Zbtb7a (NM_015898.2) primers were: Forward,
5′-GCTTGGGCCGGTTGAATGTA-3′ and reverse, 5′-GGCTGTGAAGTTACCGTCGG-3′.
GAPDH was used as an internal reference control. The GAPDH
(NM_002046.4) primers: forward, 5′-CATGAGAAGTATGACAACAGCC-3′ and
reverse, 5′-AGTCCTTCCACGATACCAAAGT-3′. All primer sequences were
designed and synthesized by Invitrogen (Thermo Fisher Scientific,
Inc.). The reaction mixture consisted of 10 µl Super Real PreMix
and 0.4 µl 50X ROX (both from Tiangen Biotech Co., Ltd., Beijing,
China), 0.5 µl template, 0.5 µl forward primer, 0.5 µl reverse
primer and 7.4 µl ddH2O. The thermocycling conditions
included an initial denaturation step at 95°C for 15 min,
denaturation at 95°C for 10 sec and annealing at 60°C for 32 sec
for 40 cycles, dissociation stage at 95°C for 15 sec, 60°C for 1
min, 95°C for 15 sec, 60°C for 15 sec. The reaction program was
executed by the 7500 Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The expression level of Zbtb7a
mRNA was calculated using the ΔΔCq method (21).
For western blotting, protein (20 µg) was subjected
to 10% SDS-PAGE and transferred onto 0.22 µm PVDF membranes
(Millipore, Billerica, MA, USA) using the Mini-Protean system
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). The membranes were
incubated with the primary antibodies against Zbtb7a (1:500
dilution; catalog no. ab70208; Abcam, Cambridge, UK) and β-actin
(1:1,200 dilution; catalog no. AA128; Beyotime Institute of
Biotechnology, Shanghai, China) overnight at 4°C. The membranes
were subsequently incubated with peroxidase-conjugated goat
anti-rabbit IgG (H+L; 1:8,000 dilution; catalog no. ZB-2301;
ZSGB-BIO) or horseradish peroxidase-labeled goat anti-mouse IgG
(H+L; 1:12,000 dilution; catalog no. A0216; Beyotime Institute of
Biotechnology) for 2 h at 37°C. The detection was performed using
the BeyoECL Plus kit (Beyotime Institute of Biotechnology)
according to the manufacturer's protocol. Fuji Medical X-ray Film
(Guangxi Yesstar Medical Systems Co., Ltd., Nanning, Guangxi,
China) was placed on top of the membrane and performed exposure.
The exposed X-ray film was scanned (Unisplendour Co., Ltd.,
Beijing, China).
Transient knockdown of Zbtb7a in CNE3
sublines
The sublines CNE3-GX6 and CNE3-GX11, with increased
tumorigenicity compared with other sublines, as demonstrated by
soft agar colony formation assay, animal experiments and analysis
of Zbtb7a expression levels, were selected. The pRNAT-U6.1/Neo
(GenScript, Piscataway, NJ, USA) vector was used for all
transfection experiments, and these were performed as previously
described (13). The cells were
plated in 6-well plates at a density of 105 cells/well, and
transient transfection was performed with 4 µg plasmid and 10 µl
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) when the cells covered >50% of the area. A
transfection efficiency of 70–80% was observed by IX71 fluorescence
microscopy (Olympus Corporation) after 48 h, and the transfected
cells were used for subsequent assays. The untreated cells and
cells transfected with the empty vector and shRNA were named as
blank control (BC), negative control (NC) and shRNA,
respectively.
MTT assay and colorimetric focus
forming assay
The role of Zbtb7a in growth control of CNE3
sublines was analyzed by MTT and focus forming assays. For MTT
assay, the cells were plated in 96-well plates at a density of
4,000 cells/well and incubated with 0.5% MTT (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) for 4 h at 37°C. Subsequently, the cells
were lysed using DMSO (Sigma-Aldrich; Merck KGaA), and the
absorbance was detected by using the GF-M3000 ELISA analyzer
(Shandong Gaomi Caihong Analytical Instruments Co., Ltd., Gaomi,
Shandong, China) at 490 nm. For colorimetric focus forming assay,
the NC and shRNA-transfected cells were plated in 6-well plates at
a density of 300 cells/well. Then the cells were fixed using 100%
methanol after 2 weeks of incubation at 37°C for 30 min. They were
subsequently stained by 5% crystal violet (Amresco, LLC, Solon, OH,
USA) in 100% methanol at 37°C for 10 min. The number of foci (over
50 cells/foci) were counted using a CKX41 microscope (original
magnification, ×40; Olympus Corporation).
Transwell migration and invasion
assay
Transwell migration and invasion assays were
performed in 6.5 mm Transwell chambers of 24-well plates (pore
size, 8 µm; Costar; Corning Incorporated). A total of 1×105 cells
(migration assay) and 4×105 cells (invasion assay) from the BC, NC
and shRNA-transfected groups were resuspended in 200 µl serum-free
RPMI-1640 medium in the upper chamber. A total of 800 µl medium
containing 10% FBS was added to the lower chamber. For invasion
assays, the upper surface was coated with 70 µl serum-free
RPMI-1640 medium (dilution, 1:3) and Matrigel (BD Biosciences, San
Jose, CA, USA) prior to the seeding of the cells. The cells were
incubated for 4 h at 37°C and 5% CO2. After 24 h
(migration assay) and 48 h (invasion assay) of incubation at 37°C,
non-migratory and non-invasive cells in the upper chamber were
completely removed by cotton swabs. The cells adhered to the lower
chamber were rinsed with PBS, fixed with 100% methanol and stained
with 1% crystal violet at 37°C for 20 min. The number of cells
migrated and invaded was quantified by counting using a BX51
microscope (original magnification, ×200). For each well, the mean
of five individual fields in the center of the filter was
obtained.
Statistical analysis
All assays were performed in three independent
experiments, and all data are expressed as the mean ± standard
deviation (SD). Data analysis and graphs were performed using Sigma
Plot software (version 12.5; SPSS Inc., Chicago, IL, USA).
Statistical differences between two groups were evaluated with
independent samples t-test, and one-way analysis of variance was
used for multiple comparisons. Least significance difference (LSD)
test and Student-Newman-Keuls (SNK) test were used as post hoc
tests (version 13.0; SPSS Inc.). P<0.05 was considered to
indicate a statistically significant difference.
Results
Soft agar colony forming and
morphology of CNE3 sublines
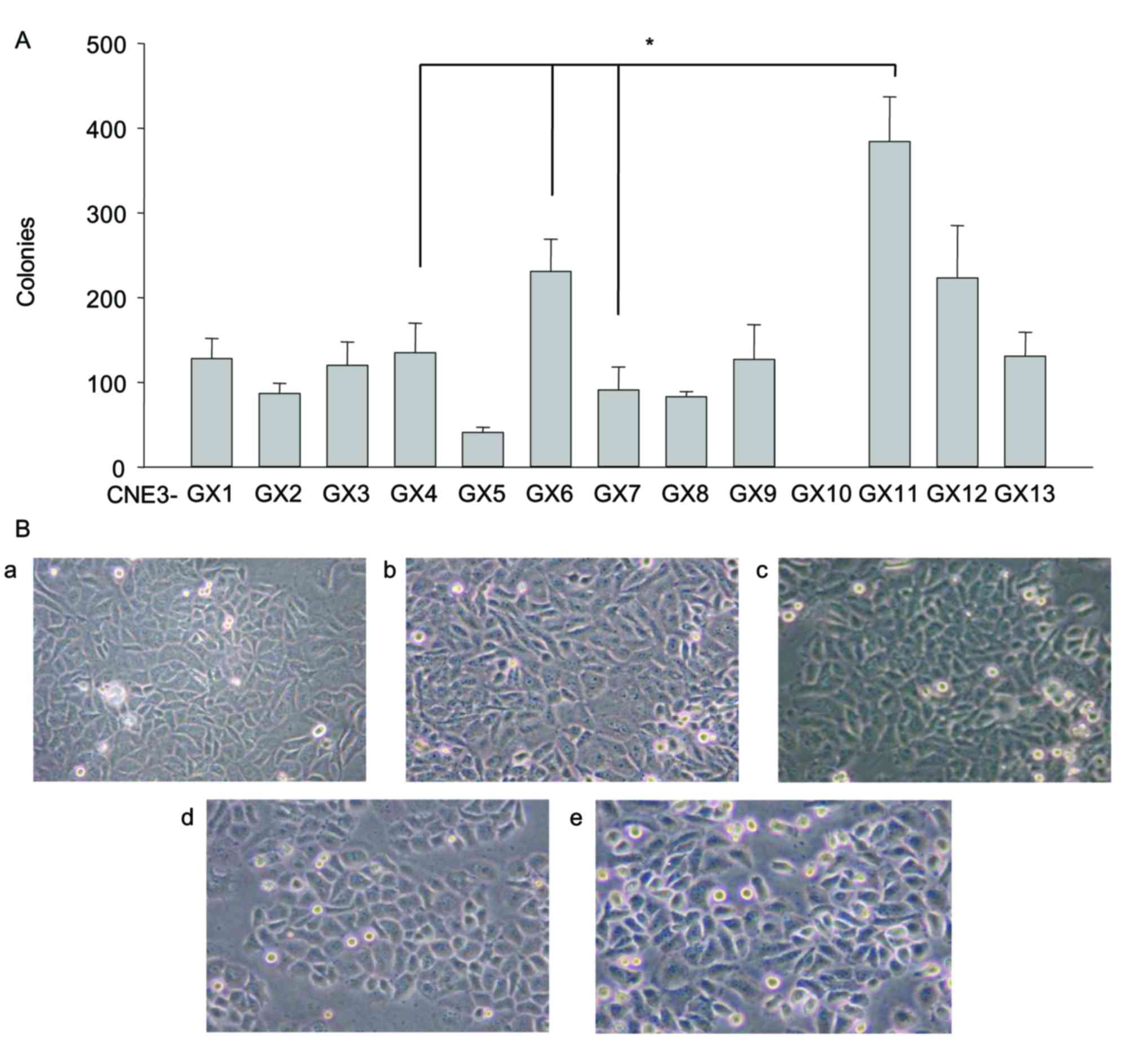
The number of colonies formed was markedly different
between the sublines. CNE3-GX10 cells were not able to form any
colonies. By contrast, CNE3-GX11 cells formed a significantly
increased number of colonies compared with cells from the other
sublines (Fig. 1A). These findings
suggested that proliferative capability of CNE3-GX10 cells was the
weakest and the proliferative capability of CNE3-GX11 cells was the
strongest. A total of 5 sublines (CNE3-GX4, CNE3-GX6, CNE3-GX7,
CNE3-GX10 and CNE3-GX11) were selected for the following
experiments according to the obvious differences of soft-agar
colony formation and cell morphology (Fig. 1B).
Xenograft mouse tumor model and
molecular pathological assays
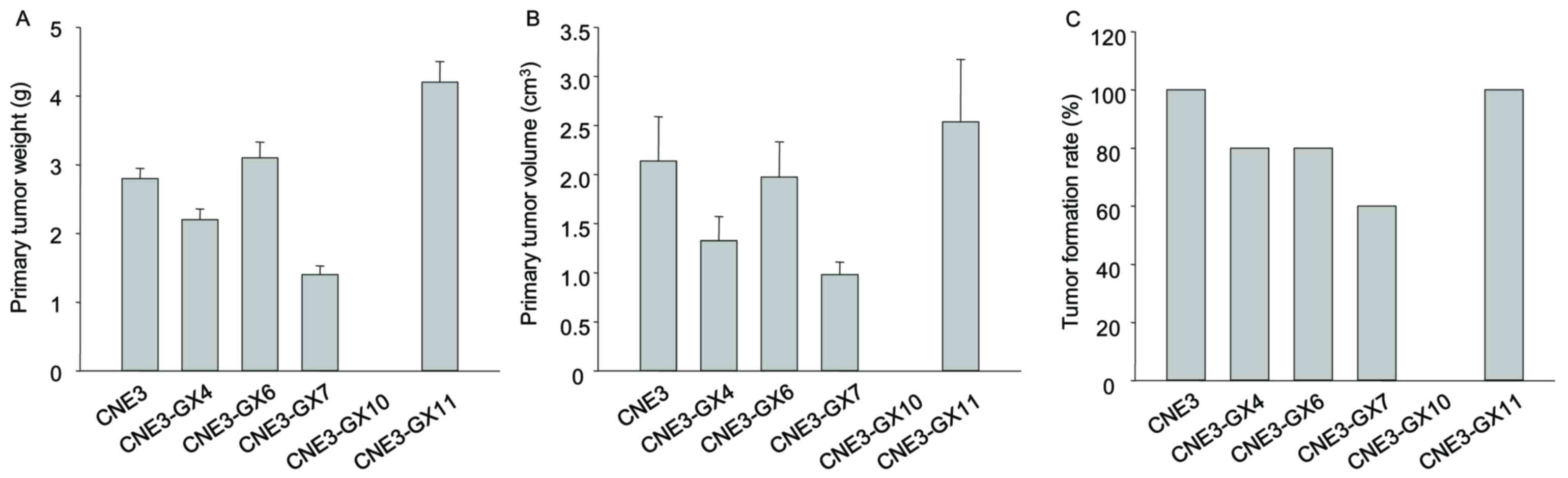
High metastatic potency was not observed in the
sublines CNE3-GX4, CNE3-GX6, CNE3-GX7, CNE3-GX10 and CNE3-GX11. All
BALB/c mice injected with CNE3-GX11 cells formed tumors (10/10),
while all mice injected CNE3-GX10 cells failed to form tumors
(0/10). The other sublines (CNE3-GX4, CNE3-GX6 and CNE3-GX7) partly
formed tumors. The weight of tumor and volume of tumors formed from
CNE3-GX11 cells was increased compared with other sublines
(Fig. 2). Findings from histological
examination and IHC analysis of tumor tissues formed from CNE3-GX4,
CNE3-GX6, CNE3-GX7 and CNE3-GX11 were similar with the results of
tumor tissues formed from CNE3 cells (3). H&E evaluation and IHC revealed that
CNE3 sublines had some features of poorly-differentiated
adenocarcinoma. CK5/6 and p63 expression was negative and CK7
expression was positive in the IHC assay (3).
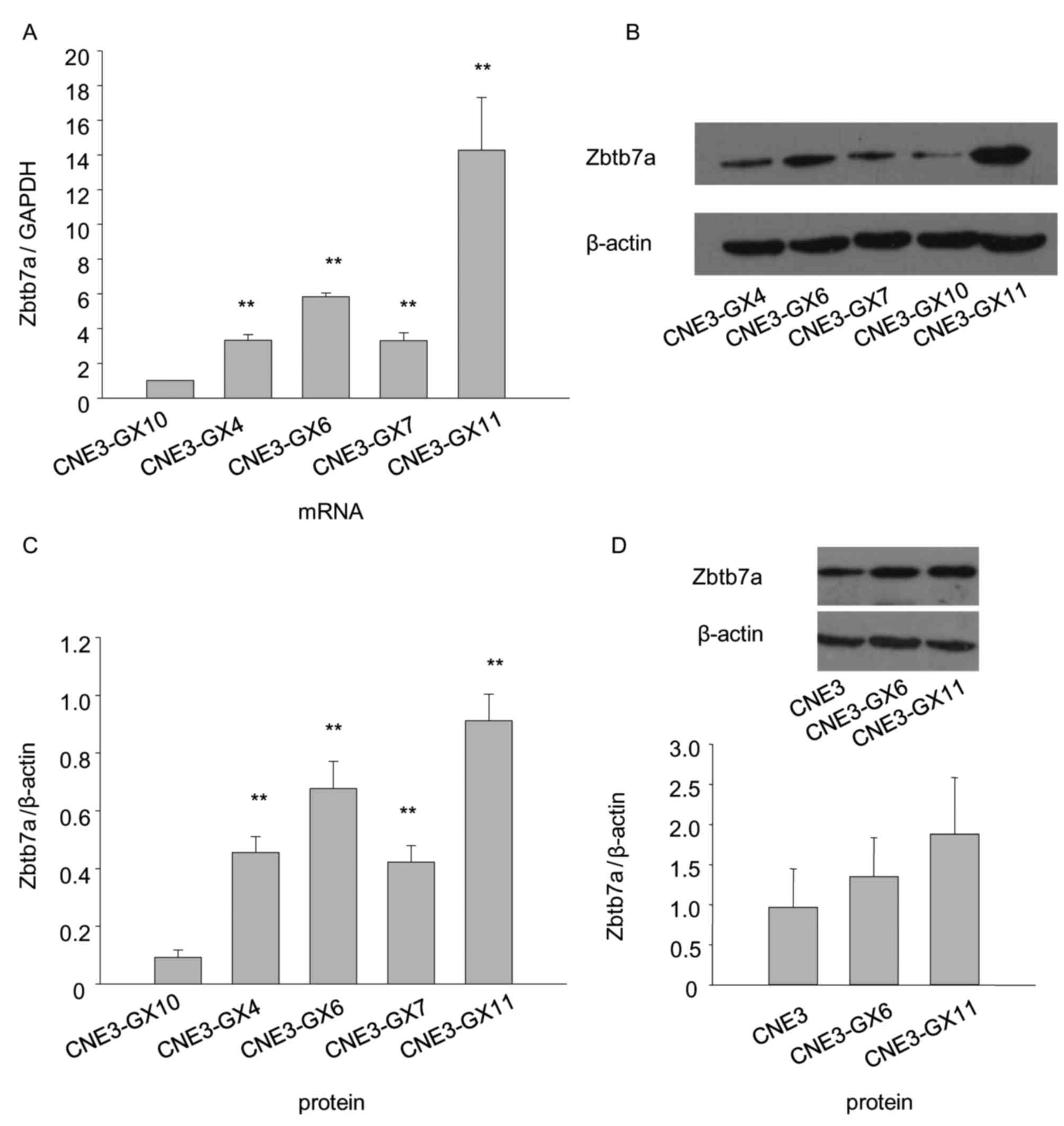
Zbtb7a expression levels of CNE3 and
CNE3 sublines
The results indicated that Zbtb7a expression in
CNE3-GX11 cells was the highest, while the expression in the
CNE3-GX10 cells was the lowest (Fig.
3). Due to their stronger tumorigenicity and higher expression
compared with other sublines, CNE3-GX6 and CNE3-GX11 were used for
the following assay.
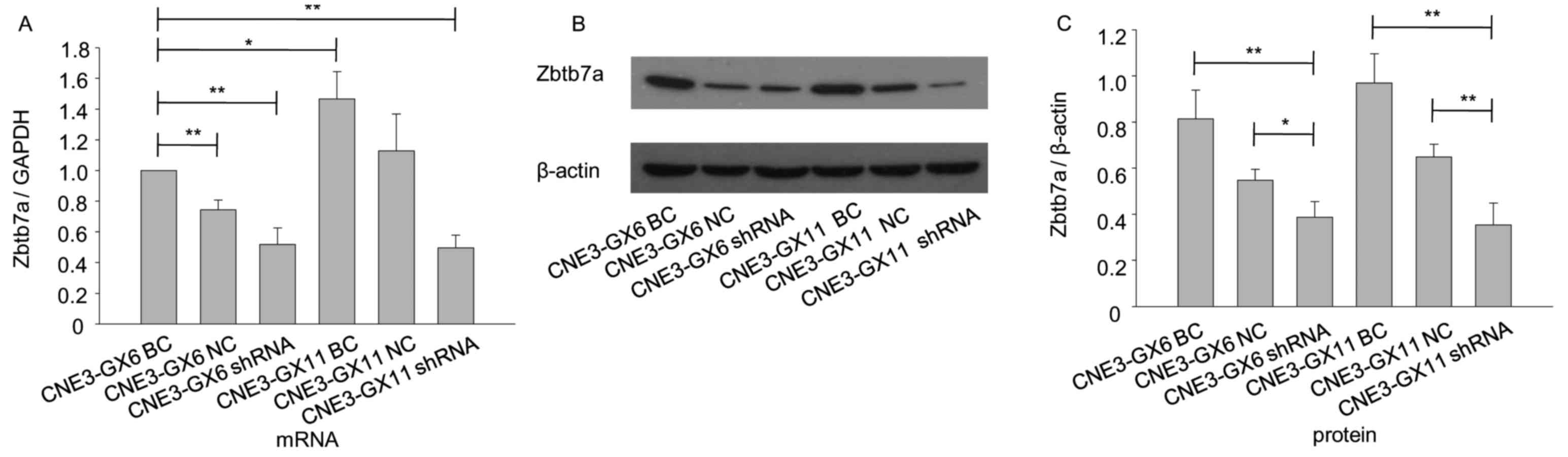
Protein and mRNA levels of Zbtb7a are
decreased following Zbtb7a knockdown in CNE3-GX6 and CNE3-GX11
The shRNA recombinant plasmids of Zbtb7a and blank
plasmid were transiently transfected into CNE3-GX6 and CNE3-GX11
cells. The results indicated that Zbtb7a expression was efficiently
knocked down in transfected CNE3-GX6 and CNE3-GX11 cells (Fig. 4).
Silencing of Zbtb7a reduces growth,
migration and invasion in CNE3-GX6/11
MTT assay showed viability of Zbtb7a
shRNA-transfected cells were reduced compared with cells
transfected with BC and NC (Fig. 5A and
B). Focus forming assay indicated that the number of foci of
shRNA-transfected cells was reduced compared with cells transfected
with NC (Fig. 5C and D). Transwell
migration and invasion assays separately indicated that migratory
and invasion abilities of shRNA cells were reduced compared with
cells transfected with BC and NC-transfected cells (Fig. 5E and F).
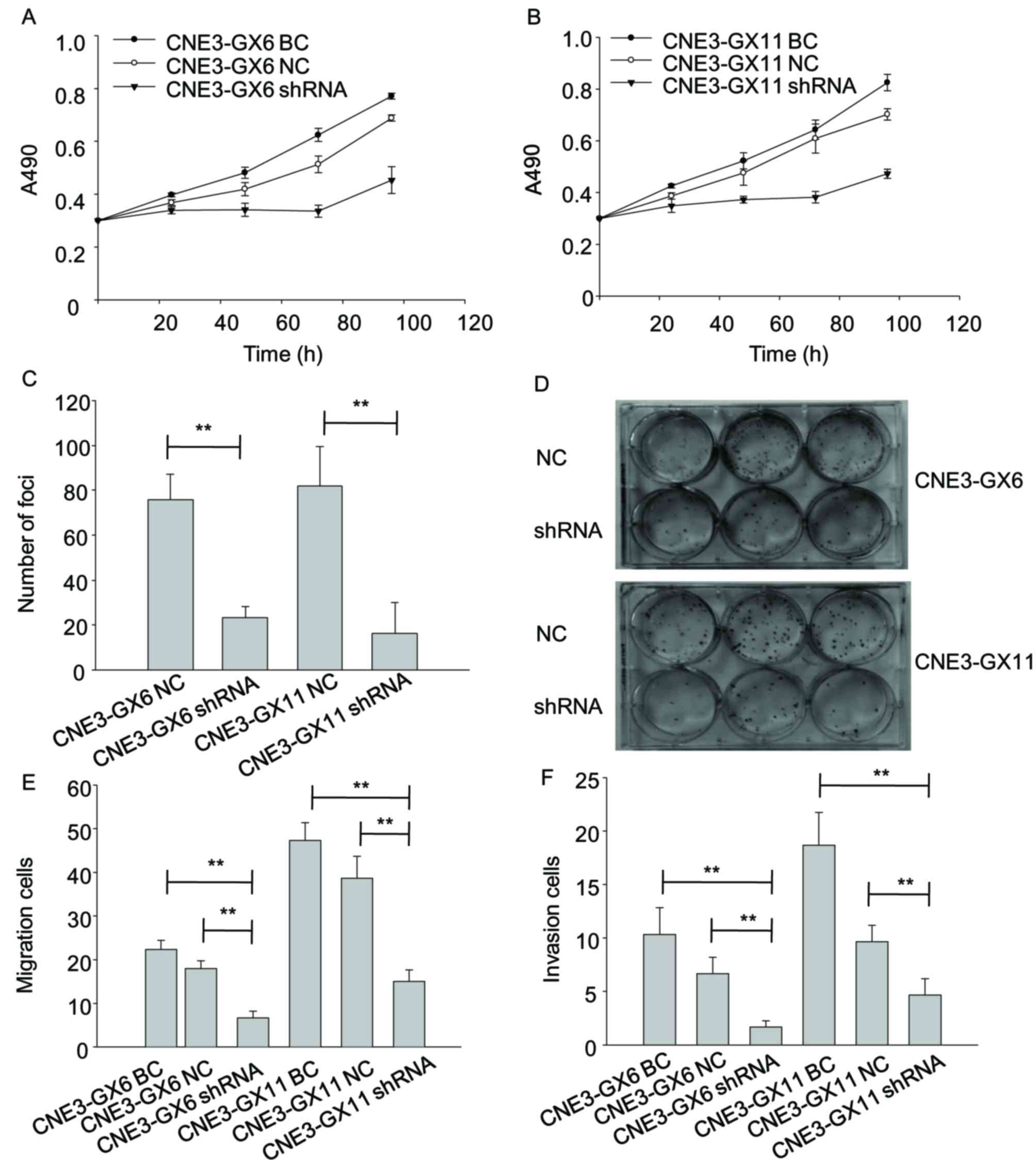 | Figure 5.Knockdown of Zbtb7a reduces growth,
migration and invasion in CNE3-GX6/11 cell lines. (A) The changes
in viability of CNE3-GX6-BC, CNE3-GX6-NC and CNE3-GX6-shRNA
transfected cells were evaluated by MTT assay at 0, 24, 48 and 96
h. (B) The changes in viability of CNE3-GX11-BC, CNE3-GX11-NC and
CNE3-GX11-shRNA transfected cells were evaluated by MTT assay at 0,
24, 48 and 96 h. (C) The proliferative ability of CNE3-GX6/11-NC
and CNE3-GX6/11-shRNA transfected cells was analyzed by
colorimetric focus forming assay after 2 weeks of incubation. (D)
The foci of the cells were stained by 5% crystal violate in 6-well
plates. (E) The migratory abilities of CNE3-GX6/11-BC,
CNE3-GX6/11-NC and CNE3-GX6/11-shRNA-transfected cells were
assessed by Transwell assay. (F) The invasion abilities of
CNE3-GX6/11-BC, CNE3-GX6/11-NC and CNE3-GX6/11-shRNA-transfected
cells were assessed by Transwell assay. **P<0.01. BC, blank
control; NC, negative control; shRNA, short hairpin RNA. |
Discussion
Heterogeneity analysis can be effectively used for
researching invasive and metastatic mechanisms of cancer. For
example, the SUNE-1 subline has been useful tools for studying
tumorigenicity and metastatic potential of NPC, since different
heterogeneity was confirmed (22–24).
In the present study, it was demonstrated that there
are differences in tumorigenicity, viability, migration and
invasion between the CNE3 sublines. Notably, the results indicated
that proliferative capability of CNE3-GX10 cells was the weakest
and the proliferative capability of CNE3-GX11 cells was the
strongest. Similarly, Zbtb7a expression levels and tumorigenicity
of CNE3-GX10 was the lowest, while that of CNE3-GX11was the
highest. The findings suggest that Zbtb7a expression levels may be
associated with heterogeneity of CNE3 sublines.
In order to confirm the hypothesis, RNA interference
was used to effectively silence the expression of specific genes
through microRNA, small interfering RNA (siRNA) and shRNA. When
some genes are transiently knocked down by siRNA or shRNA, some
cellular characteristics are temporarily changed (25–27). The
cell lines knocked down stably by shRNA can provide an ideal model
for studying invasive and metastatic mechanisms of NPC (28). In the present study although high
metastatic potency was not observed in the CNE3 sublines, an
association between Zbtb7a expression and tumorigenicity was
observed. Following transient knockdown of Zbtb7a in CNE3-GX6 and
CNE3-GX11 cells, viability, migration and invasion abilities of the
cells have decreased. The authors of the present study hypothesize
that high Zbtb7a expression may promote tumorigenicity in NPC.
Why do CNE3 cells lose metastatic ability as a liver
metastatic carcinoma cell line? Basing on the ‘seed and soil’
theory (5,6,29,30), the authors of the present study
hypothesize that the tumorigenic characteristics of clonal cells in
distant metastasis are altered in order to adapt to new tumor
microenvironment for metastasis. Primary nasopharyngeal
adenocarcinoma (NAC) is a very rare subtype of NPC (31). CNE3 has typical characteristics of
NAC. Since CNE3 was established, it has been used in various
studies on NPC (32–37). Therefore, CNE3 is a useful tool for
studying characteristics and treatment of NAC.
In summary, the present study suggests that Zbtb7a
may have an important impact on the regulatory mechanism of NPC
sublines, which may be investigated further. Zbtb7a participates in
complex regulation network due to its critical role in cancer.
Notably, Zbtb7a acts as a tumor suppressor in colon carcinoma or
melanoma cell lines by triggering stable shRNA-mediated knockdown
(38,39). It is necessary to investigate whether
Zbtb7a acts as a suppressor or not in NPC cell lines with stable
knockdown of Zbtb7a. For future studies, the authors of the present
study will establish the sublines from high-metastatic potential
NPC cell lines such as 5–8F (40) to
elucidate the role of Zbtb7a in NPC.
Acknowledgements
The present study was supported by grants from the
Natural Science Foundation of Guangxi Province (grant nos.
2011GXNSFA018308, 2015GXNSFAA139166 and 2016GXNSFBA380144) and the
Self Foundation of the Health Department of Guangxi (grant no.
Z2013389).
Glossary
Abbreviations
Abbreviations:
|
NPC
|
nasopharyngeal carcinoma
|
|
NAC
|
nasopharyngeal adenocarcinoma
|
|
IHC
|
immunohistochemistry
|
|
H&E
|
hematoxylin and eosin
|
|
shRNA
|
short hairpin RNA
|
|
BC
|
blank control
|
|
NC
|
negative control
|
|
ln
|
lymph node
|
References
|
1
|
Huang PY, Zeng Q, Cao KJ, Guo X, Guo L, Mo
HY, Wu PH, Qian CN, Mai HQ and Hong MH: Ten-year outcomes of a
randomised trial for locoregionally advanced nasopharyngeal
carcinoma: A single-institution experience from an endemic area.
Eur J Cancer. 51:1760–1770. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jiao W: Establishment of a human
epithelial cell line of nasopharyngeal carcinoma-CNE3 and its
biological characteristics. J Guangxi Med University. 12:187–190.
1995.(In Chinese).
|
|
3
|
Liu F, Jiao W, Mo XL, Lan J, Xiao RP, Zhou
XZ, Huang ZL, Mo XM and Li G: Molecular pathological study of the
human nasopharyngeal carcinoma CNE3 cell line. Oncol Lett.
6:980–984. 2013.PubMed/NCBI
|
|
4
|
Nowell PC: The clonal evolution of tumor
cell populations. Science. 194:23–28. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Greaves M and Maley CC: Clonal evolution
in cancer. Nature. 481:306–313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Paget S: The distribution of secondary
growths in cancer of the breast. 1989. Cancer Metastasis Rev.
8:98–101. 1889.
|
|
7
|
Bonnet D and Dick JE: Human acute myeloid
leukemia is organized as a hierarchy that originates from a
primitive hematopoietic cell. Nat Med. 3:730–737. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li XJ, Ong CK, Cao Y, Xiang YQ, Shao JY,
Ooi A, Peng LX, Lu WH, Zhang Z, Petillo D, et al: Serglycin is a
theranostic target in nasopharyngeal carcinoma that promotes
metastasis. Cancer Res. 71:3162–3172. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ryszawy D, Sarna M, Rak M, SzPak K,
Kedracka-Krok S, Michalik M, Siedlar M, Zuba-Surma E, Burda K,
Korohoda W, et al: Functional links between Snail-1 and Cx43
account for the recruitment of Cx43-positive cells into the
invasive front of prostate cancer. Carcinogenesis. 35:1920–1930.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Albagli O, Dhordain P, Deweindt C, Lecocq
G and Leprince D: The BTB/POZ domain: A new protein-protein
interaction motif common to DNA- and actin-binding Proteins. Cell
Growth Differ. 6:1193–1198. 1995.PubMed/NCBI
|
|
11
|
Collins T, Stone JR and Williams AJ: All
in the family: The BTB/POZ, KRAB, and SCAN domains. Mol Cell Biol.
21:3609–3615. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Maeda T, Hobbs RM, Merghoub T, Guernah I,
Zelent A, Cordon-Cardo C, Teruya-Feldstein J and Pandolfi PP: Role
of the proto-oncogene Pokemon in cellular transformation and ARF
repression. Nature. 433:278–285. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jeon BN, Yoo JY, Choi WI, Lee CE, Yoon HG
and Hur MW: Proto-oncogene FBI-1 (Pokemon/ZBTB7A) represses
transcription of the tumor suppressor Rb gene via binding
competition with Sp1 and recruitment of co-repressors. J Biol Chem.
283:33199–33210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Choi WI, Jeon BN, Park H, Yoo JY, Kim YS,
Koh DI, Kim MH, Kim YR, Lee CE, Kim KS, et al: Proto-oncogene FBI-1
(Pokemon) and SREBP-1 synergistically activate transcription of
fatty-acid synthase gene (FASN). J Biol Chem. 283:29341–29354.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qu H, Qu D, Chen F, Zhang Z, Liu B and Liu
H: ZBTB7 overexpression contributes to malignancy in breast cancer.
Cancer Invest. 28:672–678. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Aggarwal H, Aggarwal A, Hunter WJ III,
Yohannes P, Khan AU and Agrawal DK: Expression of leukemia/lymphoma
related factor (LRF/Pokemon) in human benign prostate hyperplasia
and prostate cancer. Exp Mol Pathol. 90:226–230. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fang F, Yang L, Tao Y and Qin W: FBI-1
promotes cell proliferation and enhances resistance to chemotherapy
of hepatocellular carcinoma in vitro and in vivo. Cancer.
118:134–146. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiao W, Liu F, Tang FZ, Lan J, Xiao RP,
Chen XZ, Ye HL and Cai YL: Expression of proto-oncogene Pokemon in
nasopharyngeal carcinoma cell lines and tissues. Asian Pac J Cancer
Prev. 14:6315–6319. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao GT, Yang LJ, Li XX, Cui HL and Guo R:
Expression of the proto-oncogene Pokemon in colorectal
cancer-inhibitory effects of an siRNA. Asian Pac J Cancer Prev.
14:4999–5005. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu F, Jiao W, Lan J and Xiao RP:
Construction of short hairpin RNA recombinant plasmids targeting
human Pokemon gene and screening in CNE2 cells. J Modern Oncol.
22:994–997. 2014.(In Chinese).
|
|
21
|
Liva KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song LB, Wang HM, Zeng MS, Li MZ and Jian
SW: Study on the tumor heterogeneity of nasopharyngealcarcinoma
cell line (SUNE-1). Ai Zheng. 17:324–326. 1998.(In Chinese).
|
|
23
|
Zhou W, Feng X, Ren C, Jiang X, Liu W,
Huang W, Liu Z, Li Z, Zeng L, Wang L, et al: Over-expression of
BCAT1, a c-Myc target gene, induces cell proliferation, migration
and invasion in nasopharyngeal carcinoma. Mol Cancer. 12:532013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Deng YF, Zhou DN, Pan ZY and Yin P:
Aberrant SATB1 expression is associated with Epstein-Barr virus
infection, metastasis and survival in human nasopharyngeal cells
and endemic nasopharyngeal carcinoma. Int J Clin ExP Pathol.
7:2454–2461. 2014.PubMed/NCBI
|
|
25
|
Niu DL, Li JF, He F, Zou JT, Zhou QF, Wei
X, Li Y and Chen LH: Influence of silencing Pokemon genes using
small-interfering RNA on growth of CNE-2 cells of NPC. Chin J
Cancer Prevention Treatment. 9:1041–1045. 2012.(In Chinese).
|
|
26
|
Ma LS, Yan QI, Huang Y, Zhao W and Zhu YU:
Downregulation of human epidermal growth factor receptor 2 by short
hairpin RNA increases chemosensitivity of human ovarian cancer
cells. Oncol Lett. 9:2211–2217. 2015.PubMed/NCBI
|
|
27
|
Tan X, He X, Jiang Z, Wang X, Ma L, Liu L,
Wang X, Fan Z and Su D: Derlin-1 is overexpressed in human colon
cancer and promotes cancer cell proliferation. Mol Cell Biochem.
408:205–213. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen CY, Lin YS, Chen CL, Chao PZ, Chiou
JF, Kuo CC, Lee FP, Lin YF, Sung YH, Lin YT, et al: Targeting
Annexin A2 reduces tumorigenesis and therapeutic resistance of
nasopharyngeal carcinoma. Oncotarget. 6:26946–26959. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Talmadge JE, Wolman SR and Fidler IJ:
Evidence for the clonal origin of spontaneous metastases. Science.
217:361–363. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Duda DG, Ancukiewicz M, Isakoff SJ, Krop
IE and Jain RK: Seeds and soil: Unraveling the role of local tumor
stroma in distant metastasis. J Natl Cancer Inst. 106:pii: dju 187.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu T, Li ZM, Gu MF, Wei WH, Zhang GY, Wu
QL, Su Y and Hu WH: Primary nasopharyngeal adenocarcinoma: A
review. Asia Pac J Clin Oncol. 8:123–131. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen W, Lee Y, Wang H, Yu GG, Jiao W, Zhou
W and Zeng Y: Suppression of human nasopharyngeal carcinoma cell
growth in nude mice by the wild-type p53 gene. J Cancer Res Clin
Oncol. 119:46–48. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Teng ZP, Ooka T, Huang DP and Zeng Y:
Detection of Epstein-Barr virus DNA in well and poorly
differentiated nasopharyngeal carcinoma cell lines. Virus Genes.
13:53–60. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xia Y, Wong NS, Fona WF and Tideman H:
Upregulation of GADD153 expression in the apoptotic signaling of
N-(4-hydroxyphenyl)retinamide (4HPR). Int J Cancer. 102:7–14. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang XL, Liu XC, Huang L, Lan J, Zhang HY,
Qin MB, Zhong YY and Mo ZN: Effect of TSA on nasopharyngeal
carcinoma CNE3 cells and its mechanism. Chin J Public Health.
26:1029–1030. 2010.(In Chinese).
|
|
36
|
Liu XC, Lan J, Nong CZ, Pan LL and Jiao W:
Effect of mangiferin on induction of apoptosis and intracellular
Ca2+ concentration in nasopharyngeal carcinoma CNE3
cells. Chin J New Clin Med. 3:805–897. 2010.(In Chinese).
|
|
37
|
Peng LX, Chen JX, Cheng JJ, Liu F, Jiao W,
Pang Q, Feng GS, Li SG, Mo XY and Wu XX: Establishment of
radioresistant subline from human nasopharyngeal carcinoma cell
line by repeating irradiation. Chin J Clinical Med. 5:1107–1109.
2012.(In Chinese).
|
|
38
|
Liu XS, Haines JE, Mehanna EK, Genet MD,
Ben-Sahra I, Asara JM, Manning BD and Yuan ZM: ZBTB7A acts as a
tumor suppressor through the transcriptional repression of
glycolysis. Genes Dev. 28:1917–1928. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu XS, Genet MD, Haines JE, Mehanna EK,
Wu S, Chen HI, Chen Y, Qureshi AA, Han J, Chen X, et al: ZBTB7A
suppresses melanoma metastasis by transcriptionally repressing
MCAM. Mol Cancer Res. 13:1206–1217. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zong D, Yin L, Zhong Q, Guo WJ, Xu JH,
Jiang N, Lin ZR, Li MZ, Han P, Xu L, et al: ZNF488 enhances the
invasion and tumorigenesis in nasopharyngeal carcinoma via the Wnt
signaling pathway involving epithelial mesenchymal transition.
Cancer Res Treat. 48:334–344. 2015. View Article : Google Scholar : PubMed/NCBI
|