Introduction
Breast cancer is one of the most common malignant
tumors in women worldwide (1).
Receptor tyrosine-protein kinase erbB2/human epidermal growth
factor receptor 2 (ERBB2/HER2) is a transmembrane tyrosine kinase
receptor that belongs to the family of epidermal growth factor
receptors. HER2 is expressed in 25–30% of invasive breast cancers
and is associated with invasion, metastasis and prognosis (2). Trastuzumab (product name Herceptin) is
the first recombinant humanized monoclonal antibody directed
against the extracellular domain of HER2 that has been approved by
the US Food and Drug Administration for the treatment of
HER2-positive breast cancer. However, 40–60% of patients with
HER2-positive breast cancer do not benefit from it (3). The molecular mechanisms underlying
acquired resistance to trastuzumab remain poorly understood.
Trastuzumab has several possible mechanisms of
action, which include specific binding to HER2 and blocking of
ligand-mediated cell signaling, inhibition of cell growth, inducing
apoptosis, inhibition of tumor angiogenesis, and improving the
ability of immune cells to target tumor cells through
antibody-dependent cellular cytotoxicity (ADCC). ADCC and
complement-dependent cytotoxicity (CDC) are innate immune
mechanisms that destroy tumor cells and serve important roles in
mediating the effects of therapeutic monoclonal antibodies (mAbs)
in the treatment of cancer (4).
Examples of therapeutic mAbs include rituximab, alemtuzumab and
cetuximab. However, it has been reported that CDC does not serve a
role in mediating the effects of trastuzumab (5,6). Since the
contribution of the complement system to the anti-tumor effect of
trastuzumab remains unclear, the deficiency of CDC may explain why
the majority of patients with HER2-positive cancer are not
sensitive to trastuzumab. The efficiency of CDC against tumor
primarily depends on the activation of the complement system, which
is regulated by membrane-bound complement regulatory proteins
(mCRPs), including complement decay-accelerating factor (CD55) and
CD59 glycoprotein precursor (CD59). A number of types of cancer
have been reported to escape from the complement attack due to high
expression of mCRPs, which protects cancer cells from CDC and
anticancer immune responses (7,8).
A previous study from our lab demonstrated that CD55
and CD59 were two important inhibitors of CDC triggered by
heterologous expression of α-gal xenoantigen in colon tumor cell
lines (9). In addition, patients with
breast cancer that overexpressed CD55 or CD59 exhibited a higher
relapse rate following trastuzumab treatment compared with those
with low expression of CD55 or CD59 (10). The mean disease-free survival of
patients with CD55 or CD59 overexpression was significantly shorter
compared with those with low expression of CD55 or CD59, while the
expression of CD46 had no effect on prognosis.
It was hypothesized that CDC induced by trastuzumab
in HER2-positive breast cancer may be limited due to overexpression
of CD55 and CD59. The aim of the present study was to investigate
the association between CD55/CD59 expression and
trastuzumab-induced CDC in a HER2-positive breast cancer cell line.
This was achieved by blocking CD55/CD59 activity using mAbs,
silencing their expression using short hairpin RNA (shRNA), and
modulating their expression via phosphatidylinositol-specific
phospholipase C (PI-PLC).
Materials and methods
Cell lines
The human breast cancer BT474, SK-BR-3, ZR-75-1,
MDA-MB-468, MCF-7, BT549 and MDA-MB-231 cell lines, and a human
colorectal adenocarcinoma (LoVo) and pig iliac artery endothelial
(PIEC) cell line were purchased from the Cell Bank of Type Culture
Collection of the Chinese Academy of Sciences (Shanghai, China).
PIEC, which naturally expresses high level of α-gal xenoantigen,
was used as a positive control to assess complement activity in
cytolysis assays. MCF-7, BT549, LoVo and PIEC cells were cultured
in RPMI-1640 medium containing 10% fetal bovine serum from Gibco
(Thermo Fisher Scientific, Inc., Waltham, MA, USA).SK-BR-3,
MDA-MB-468, MDA-MB-231 and ZR-75-1 cells were cultured in DMEM
medium containing 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.) and 100 IU/ml penicillin/streptomycin (North
China Pharmaceutical Co., Ltd., Shijiazhuang, China). BT474 cells
were cultured in MEM medium containing 10% fetal bovine serum
(Gibco; Thermo Fisher Scientific, Inc.), 100 IU/ml
penicillin/streptomycin (North China Pharmaceutical Co., Ltd.) and
40 U/ml insulin from Tonghua Dongbao Pharmaceutical Co., Ltd.
(Jilin, China). The cells were maintained in an incubator at 37°C
in a 5% CO2 humidified atmosphere.
Sera
Pooled normal human serum (NHS) used as the source
of complement and anti-α-gal-specific antibodies was provided by
the central blood bank at West China Hospital, Sichuan University
(Chengdu, China) and stored at −80°C in aliquots until assayed.
Diluted (50%) NHS was screened using a PIEC killing efficiency
assay, as described previously (9),
and only sera with 100% PIEC killing efficiency was utilized for
subsequent experiments. Pooled heat-inactivated normal human serum
(INHS) was obtained by incubating the sera at 56°C for 30 min,
which was used as negative control in all experiments.
Antibodies and reagents
Fluorescein isothiocyanate (FITC)-conjugated mouse
anti-human CD55 (cat no. FHF055) and CD59 (cat no. FHF0591) mAbs
and a FITC-conjugated immunoglobulin G (IgG)1 isotype Control mAb
(cat no. GMP01103F) for flow cytometry (FCM) were purchased from 4A
Biotech Co., Ltd. (Beijing, China). Mouse anti-HER2/c-erbB-2
monoclonal antibody (cat no. ZM-0065) and Biotin-Streptavidin HRP
detection systems (cat no. SP-9000) for immunohistochemistry was
purchased from Origene Technologies (Beijing, China). The blocking
mAb against CD55 (cat no. BRIC216) was purchased from EMD Millipore
(Billerica, MA, USA). The blocking mAb against CD59 (cat no.
MFM-43) was purchased from Abcam (Cambridge, UK). The mAbs against
CD55 (BRIC216), CD59 (YTH53.1) and β-actin (cat no. SC-47778) for
western blot analysis were purchased from Santa Cruz Biotechnology,
Inc. (Dallas, TX, USA). The horseradish peroxidase (HRP)-labeled
goat anti-mouse IgG (H+L, cat no. 170-6516) was from Bio-Rad
Laboratories, Inc. (Hercules, CA, USA). The HRP-labeled goat
anti-rat IgG (H+L, cat no. sc-2006) was from Santa Cruz
Biotechnology, Inc.
Trastuzumab was obtained from Genentech, Inc. (San
Francisco, CA, USA). PI-PLC 100 U/ml and Lipofectamine®
2000 were purchased from Invitrogen (Thermo Fisher Scientific,
Inc.).
Plasmids and primers
The specified pGPU6/GFP/Neo-shRNA expression
plasmids (shCD55 and shCD59), and the negative control
pGPU6/GFP/Neo-shNC (shNC) were designed and produced by GenePharma
Co., Ltd. (Shanghai, China). The shCD59 targeted sequence
5′-TGAGCTAACGTACTACTACTGC-3′ was designed according to Shi et
al (11). The four specified
shCD55 targeted sequences included shCD55/545,
5′-GCAGTCAATGGTCAGATATTG-3′; shCD55/613,
5′-GCATCCCTCAAACAGCCTTAT-3′; shCD55/829,
5′-GGCATATTATTTGGTGCAACC-3′ and shCD55/1075,
5′-GGAGAGCACTCTATTTATTGT-3′. The shNC targeted sequence was
5′-GTTCTCCGAACGTGTCACGT-3′. The primers for reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis of CD55, CD59 and GAPDH were synthesized by Sangon Biotech
Co., Ltd. (Shanghai, China) as follows: CD55 forward,
5′-TTCCCCCAGATGTACCTAATGC-3′ and reverse,
5′-TTACAGTATCCTCGGGAAAACTTGT-3′; CD59 forward,
5′-TAACCCAACTGCTGACTGCAA-3′ and reverse,
5′-TTTGGTAATGAGACACGCATCAA-3′; GAPDH forward,
5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse,
5′-GAAGATGGTGATGGGATTTC-3′.
Detection of CD55 and CD59 expression
by flow cytometry
Cells were removed from the culture flask using
0.25% trypsin and 0.25% EDTA, washed with 1% bovine serum albumin
(BSA) diluted in PBS and centrifuged at 300 × g for 10 min, then
suspended in 100 µl 1% BSA and incubated with 10 µl FITC-CD55 or
FITC-CD59mAbs for 30 min at 37°C. Flow cytometry was performed
using a FACSAria I and data were analyzed using FACSDiva 6.0 (both
from BD Biosciences, Franklin Lakes, NJ, USA). Cells used FITC-IgG1
isotype control mAb as the negative control. To test the cell
membrane expression of CD55 and CD59 following PI-PLC exposure,
SK-BR-3 and BT474 cells were treated with 0.1 U/ml PI-PLC for 1 h
at 37°C prior to staining and flow cytometry.
Immunocytochemical staining for
HER2
Cells were seeded in 6-well plates at a
concentration of 5×105 cells/well for 24 h, then fixed with cold
methanol for 15 min. Cells were incubated with primary antibodies
against HER2 (dilution, 1:200 in PBS) overnight at 4°C. Cells were
incubated with the appropriate secondary antibodies for 60 min at
37°C. A DAB color developing system, and hematoxylin and eosin
staining were used for the following steps. The negative controls
were created by replacing the primary antibodies with PBS. Stained
cells were observed by light microscope and 5 fields of view were
counted by eye for cell numbers according to the following scoring
system: 0= negative, no dye or <10% cells with cell membrane
staining; 1+= weak positive, >10% cells with thin, fragmented
cell membrane staining; 2+= positive, >10% cells with thin to
moderate intact cell membrane staining; and 3+= strong
positive/high expression, >30% cells with moderate to thick
intact cell membrane staining.
Trypan blue exclusion assay
SK-BR-3, BT474 and PIEC cells were removed from
culture bottle using 0.25% trypsin and 0.25% EDTA, Single cells
were suspended in PBS, counted and divided into 106 cells per
Eppendorf tube. SK-BR-3 and BT474 cells in each Eppendorf tube were
centrifuged at 500 × g and incubated with 50% INHS, 50% NHS, 50
µg/ml trastuzumab or 50 µg/ml trastuzumab +50% NHS in a total
volume of 500 µl for 1 h at 37°C. PIEC cells were incubated with
50% INHS or 50% NHS. A total of 100 µl of cell suspension was added
into an equal volume of 0.4% trypan blue, then the number of
living/dead cells were counted, and the survival and lysis rates
were calculated as follows: Survival rate (%) = Number of living
cells/(number of living cells + number of dead cells) ×100; lysis
rate =100% - survival rate.
In order to block CD55 and CD59, SK-BR-3 and BT474
cells were pre-incubated with 50 µg/ml trastuzumab and 10 µg/ml
anti-CD55 or anti-CD59 mAbs for 10 min at room temperature, then
50% NHS was added to make up a final volume of 500 µl. The samples
were then incubated for 1 h at 37°C.
For the PI-PLC pre-treatment, SK-BR-3 and BT474
cells were incubated with 0.01, 0.05, 0.1 and 0.2 U/ml (diluted in
PBS) for 1 h at 37°C. Cells were then incubated with 50 µg/ml
trastuzumab + 50% NHS (final volume, 500 µl). Each experiment was
repeated three times.
Downregulation of CD55 and CD59
expression with shRNA in SK-BR-3 cells
The shRNA plasmids (4 µg) were mixed with 10 µl
Lipofectamine 2000 in 500 µl serum-free DMEM and transfected into
SK-BR-3 cells to knockdown the expression of CD55 and CD59.
Following transfection with shRNA for 48 h, the interference
efficiencies of shRNAs were determined by RT-qPCR and western blot
analysis. A trypan blue exclusion assay was also tested according
to the aforementioned method.
RNA extraction and RT-qPCR
analysis
Total RNA was isolated from SK-BR-3 cells using
RNeasy Mini kit (cat no. 74104; Qiagen GmbH, Hilden, Germany)
according to the manufacturer's protocol. First-strand cDNAs were
synthesized from total RNA using 5X All-In-One RT MasterMix (G492;
Applied Biological Materials, Inc., Richmond, BC, Canada) according
to the manufacturer's protocol. RT-qPCR was performed using
SsoFast™ Eva Green® Supermix (cat no. 172) in a Chromo4
Real-Time PCR detector (both from Bio-Rad Laboratories, Inc.),
according to the manufacturer's protocol. Cycling conditions for
the RT-qPCR were 95°C for 30 sec, followed by 40 cycles of 95°C,
for 5 sec and 60°C for 10 sec. The primer sequences used were
described in previous sections. The relative quantitation of gene
expression was performed using the 2−ΔΔCq method
(12) with GAPDH as a reference gene.
Experiments were repeated three times.
Western blot analysis
Whole cells were lysed at 4°C in
radioimmunoprecipitation lysis buffer (Beyotime Institute of
Biotechnology, Shanghai, China). Total protein concentration was
determined using the BCA kit (Beyotime Institute of Biotechnology)
according to the manufacturer's protocol. A total of 35 µg protein
from each group was separated by 12% SDS-PAGE and then transferred
to polyvinylidene fluoride membranes. The membranes were blocked
with 5% nonfat milk in PBS-Tween (0.1% Tween in PBS). The membranes
were incubated at 4°C overnight with the primary antibodies against
CD55 (dilution, 1:400), CD59 (dilution, 1:1,000) and β-actin
(dilution, 1:1,000). Subsequent to washing with PBS-Tween three
times for 10 min, the membranes were incubated at room temperature
for 2 h with HRP-labeled goat anti-mouse IgG (dilution, 1:2,000)
and goat anti-rat IgG (dilution, 1:10,000) secondary antibodies.
Subsequent to washing with PBS-Tween 3 times for 10 min, the bands
were visualized using chemiluminescent HRP substrate (cat no.
WBKLS0100) (EMD Millipore), and detected using the ChemiDoc XRS kit
(Bio-Rad, Laboratories, Inc.) according to the manufacturer's
protocol.
Statistical analysis
The data were expressed as the mean ± standard
deviation. Statistical analysis was performed as analysis of
variance followed by one-way analysis of variance for experiments
consisting of more than two groups. A t-test was used for
comparison between two groups. P<0.05 was considered to indicate
a statistically significant difference. SPSS software version 16.0
(SPSS Inc., Chicago, IL, USA) was used.
Results
Expression of CD55 and CD59 in breast
cancer cells
The surface expression of CD55 and CD59 in breast
cancer cells MCF-7, SK-BR-3, MDA-MB-468, BT549, MDA-MB-231, BT474
and ZR-75-1, and the human colorectal adenocarcinoma cell line LoVo
was evaluated by FCM (Table I). All
the breast cancer cell lines evaluated exhibited markedly increased
CD55 and CD59 expression compared with LoVo cells [negative control
(9)].
 | Table I.MFI of CD55 and CD59 in 7 breast
cancer cell lines as detected by flow cytometry. |
Table I.
MFI of CD55 and CD59 in 7 breast
cancer cell lines as detected by flow cytometry.
| Cell line | CD55a, MFI | CD59a, MFI |
|---|
| SK-BR-3 |
741±92 |
5257±283 |
| MCF-7 |
565±55 |
4379±135 |
| ZR-75-1 |
324±99 |
2736±265 |
| MDA-MB-468 |
393±36 |
9172±171 |
| BT549 |
291±57 |
4710±157 |
| MDA-MB-231 |
2242±153 |
4402±153 |
| BT474 |
844±98 |
7965±342 |
| LoVo |
35±21 |
119±17 |
Expression of HER2 in breast cancer
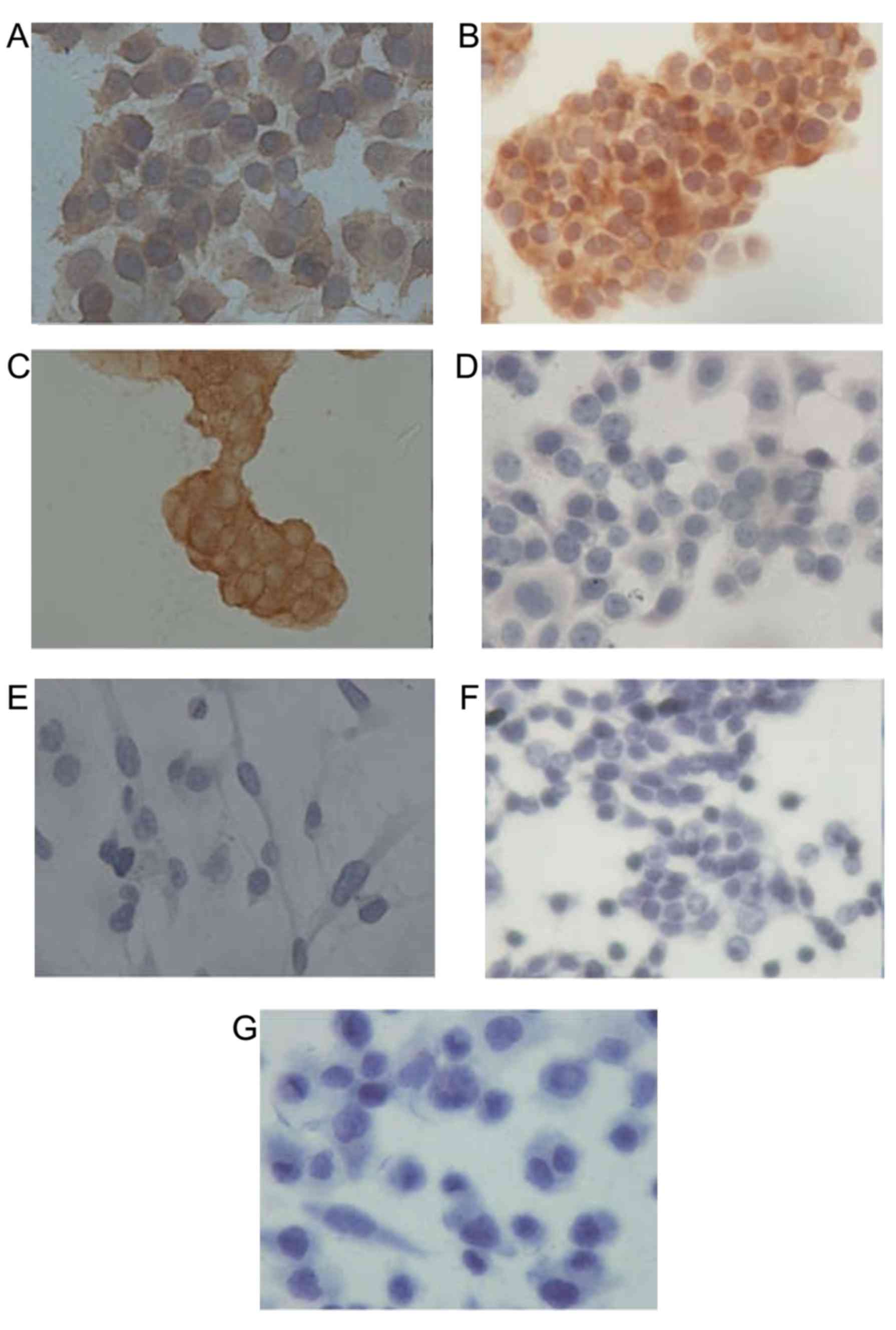
cells
Immunocytochemical staining was performed to
determine the expression level of HER2 protein in the 7 breast
cancer cell lines. The results demonstrated HER2 strong positive
(3+) expression in SK-BR-3, BT474 and ZR-75-1 cells, and negative
(0) expression in other cell lines (Fig.
1). SK-BR-3 and BT474 cells were selected for subsequent
experiments as isolated single ZR-75-1 cells were difficult to
obtain via trypsin digestion, and the resultant cell clusters may
cause inaccuracies in subsequent trypan blue cell counting
experiments.
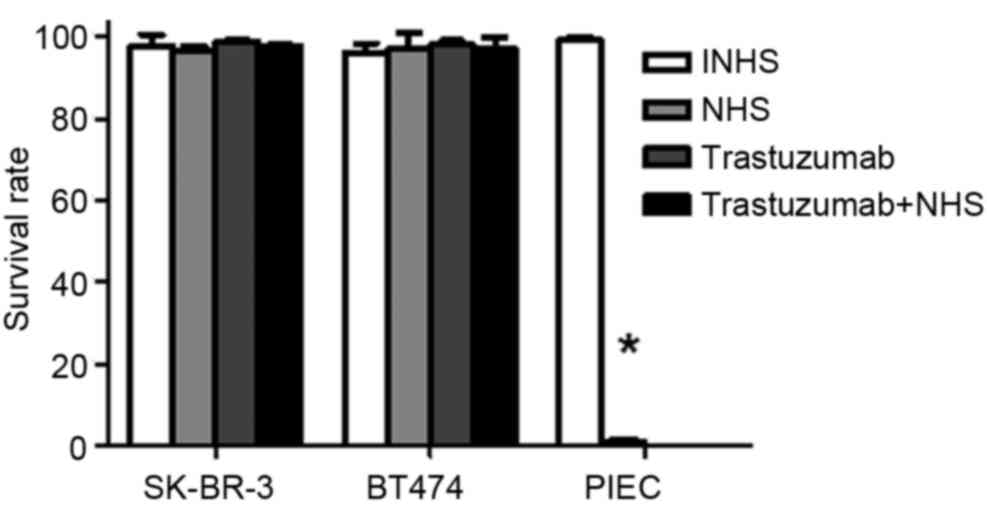
Trastuzumab does not induce CDC in
SK-BR-3 and BT474 cells
Trypan blue exclusion assays demonstrated that NHS
treatment significantly increased PIEC cell death compared with
INHS treatment (P<0.05; Fig. 2).
This confirmed that the NHS was suitable for subsequent
experiments. No significant differences were observed between the
cell lysis rates for SK-BR-3 and BT474 cells treated with INHS,
NHS, trastuzumab and trastuzumab + NHS (P>0.05; Fig. 2). Cell survival rates were >97% in
these treatment groups. The data indicated that trastuzumab does
not induce CDC in SK-BR-3 and BT474 cells.
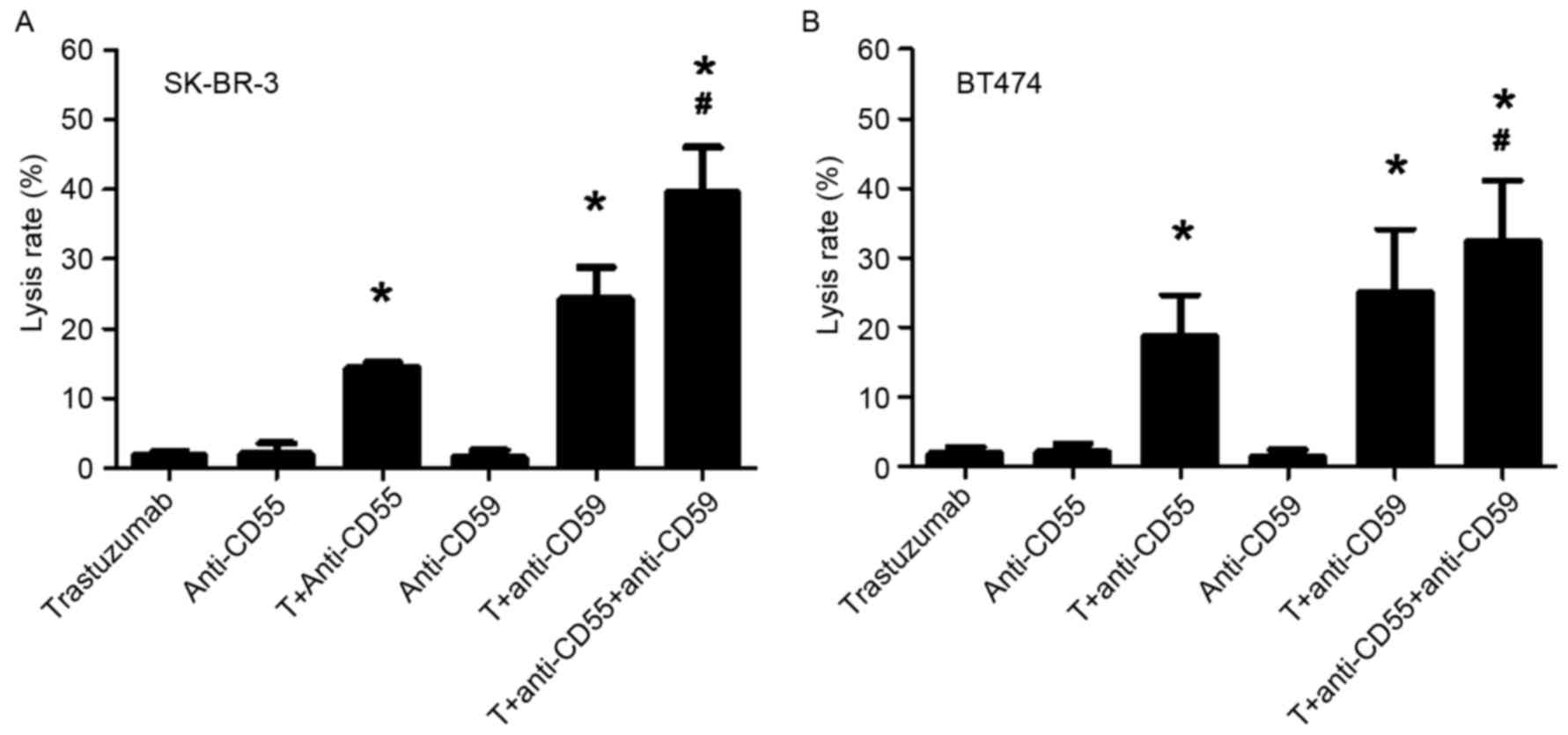
Co-treatment with trastuzumab and
anti-CD55/59mAbs results in CDC-dependent cell lysis
Trypan blue exclusion assays demonstrated that, in
the trastuzumab + anti-CD55, trastuzumab + anti-CD59, trastuzumab +
anti-CD55 + anti-CD59 treatment groups, the cell lysis rates
following NHS treatment for SK-BR-3 were 14.3, 24.2 and 39.5%,
respectively (Fig. 3A), and for BT474
were 18.69, 24.95 and 32.37%, respectively (Fig. 3B). The cell lysis rates in these
groups were significantly increased compared with the trastuzumab
and anti-CD55/59 alone treatment groups (P<0.05). In addition,
the cell lysis rate for the trastuzumab + anti-CD55 + anti-CD59
treatment group was significantly increased compared with the
trastuzumab + anti-CD55 or the trastuzumab + anti-CD59 treatment
groups (P<0.05). These results indicate that co-treatment of
cells with trastuzumab and anti-CD55/59 mAbs results in
CDC-dependent lysis of SK-BR-3 and BT474 cells.
PI-PLC promotes trastuzumab-induced
CDC in SK-BR-3 and BT474 cells in a dose-dependent manner through
cleavage of CD55 and CD59
A trypan blue exclusion assay demonstrated that
following treatment with NHS, trastuzumab-induced CDC was
significantly enhanced in PI-PLC pre-treated SK-BR-3 and BT474
cells in a dose-dependent manner (P<0.05; Fig. 4A). The cell lysis rate reached a
maximum with pre-treatment with 0.1 U/ml PI-PLC (33.5 and 37.7% for
SK-BR-3 and BT474 cells, respectively). Following treatment with
0.1 U/ml PI-PLC for 1 h, FCM analysis revealed a significant
decrease in CD55/59 fluorescence signal compared with before PI-PLC
treatment (P<0.05; Fig. 4B and C),
indicating that CD55 and CD59 had been cleaved.
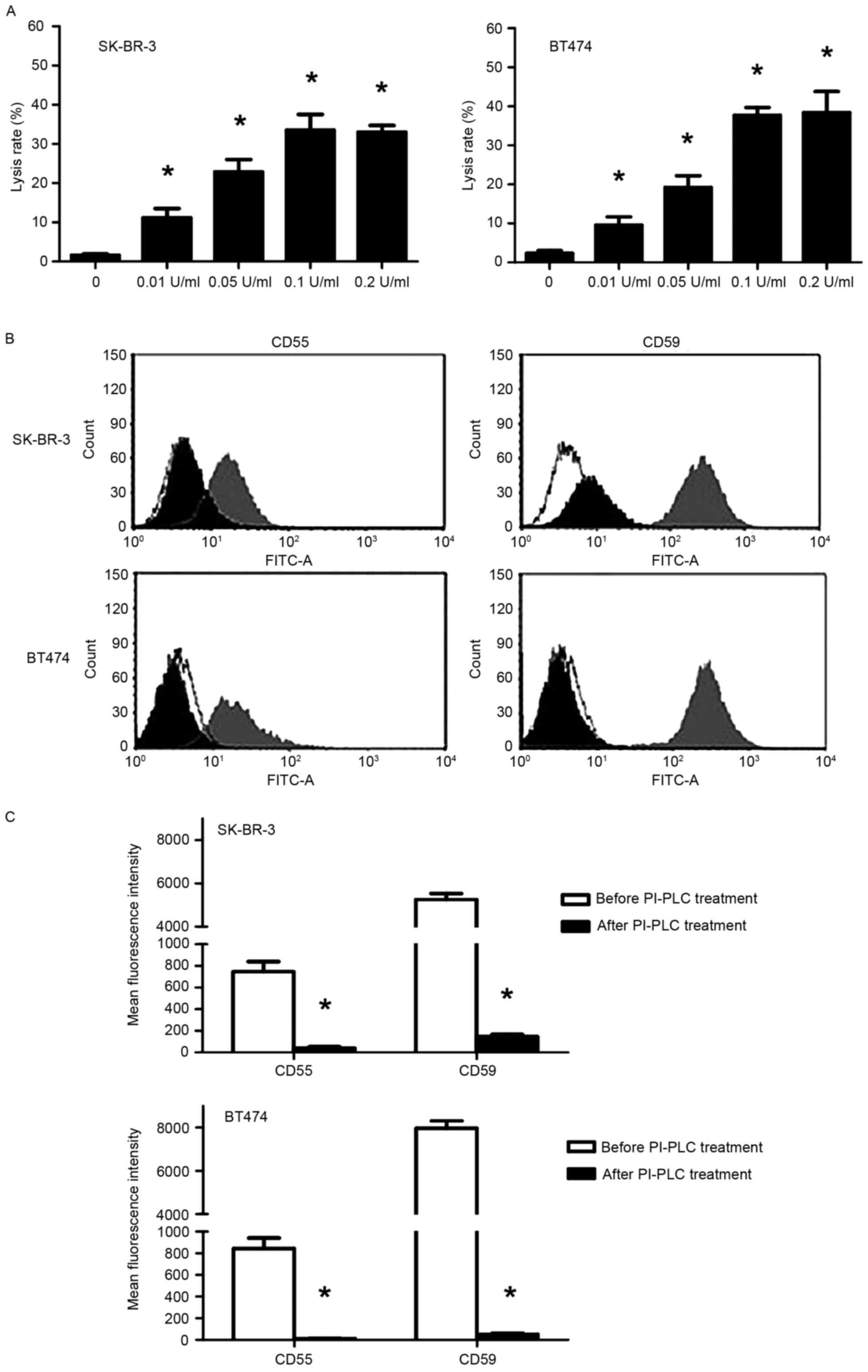 | Figure 4.Effect of PI-PLC pre-treatment on
trastuzumab-induced CDC in SK-BR-3 and BT474 cells. (A) Lysis rates
in SK-BR-3 and BT474 cells following pre-treatment with PI-PLC
(0.01, 0.05, 0.1 and 0.2 U/ml) cells for 1 h at 37°C followed by
incubation with 50 µg/ml trastuzumab + 50% NHS. *P<0.05,
compared with the control (0 U/ml). (B) Representative flow
cytometry histograms (white, control of similar type; gray,
pre-treatment; black, post-treatment) and (C) quantification of
CD55 and CD59 expression in SK-BR-3 and BT474 cells following 0.1
U/ml PI-PLC treatment. *P<0.05, compared with before PI-PLC
treatment. CDC, complement-dependent cytotoxicity; CD55, complement
decay-accelerating factor; CD59, CD59 glycoprotein precursor;
PI-PLC, phosphatidylinositol-specific phospholipase C. |
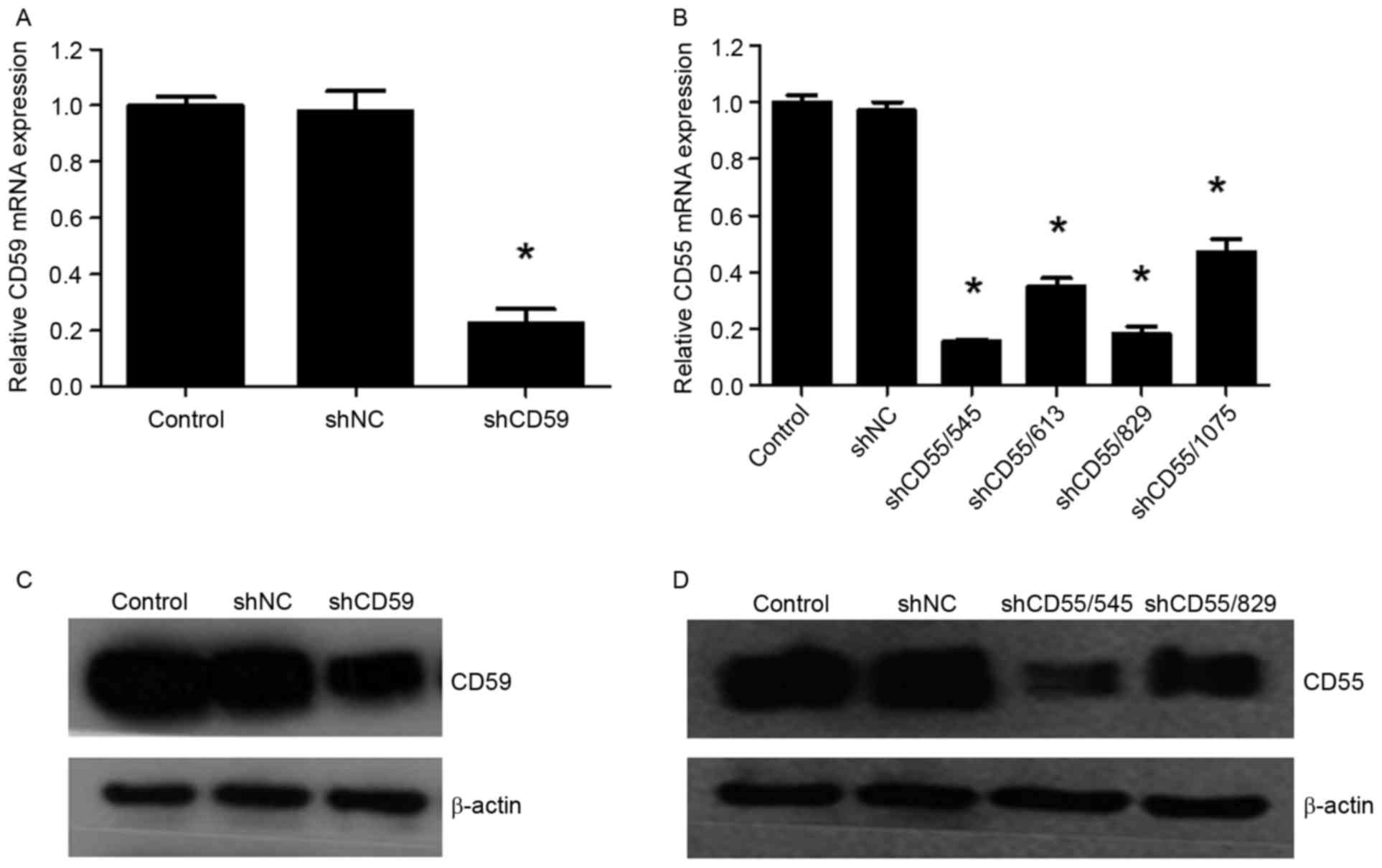
Confirmation of CD55 and CD59
downregulation following shRNA transfection of SK-BR-3 cells
RT-qPCR analysis demonstrated that CD59 mRNA
expression was significantly downregulated following shCD59
transfection compared with the shNC and control groups (P<0.05;
Fig. 5A). The 4 shRNA fragments
targeting CD55 exhibited different efficiencies in downregulating
CD55 mRNA. Treatment with shCD55/545 or shCD55/829 demonstrated the
highest downregulation efficiencies, by 84.5 and 82%, respectively,
compared with the shNC control (P<0.05; Fig. 5B).
Western blot analysis demonstrated that CD59 protein
expression was downregulated following treatment with shCD59
compared with the shNC and control groups (Fig. 5C). In addition, CD55 protein
expression was downregulated following treatment with shCD55/545 or
shCD55/829 comparing with shNC and control groups (Fig. 5D). The shCD55/545 fragment
demonstrated markedly increased downregulation efficiency compared
with shCD55/829; therefore shCD55/545-transfected SK-BR-3 cells
were selected for subsequent experiments.
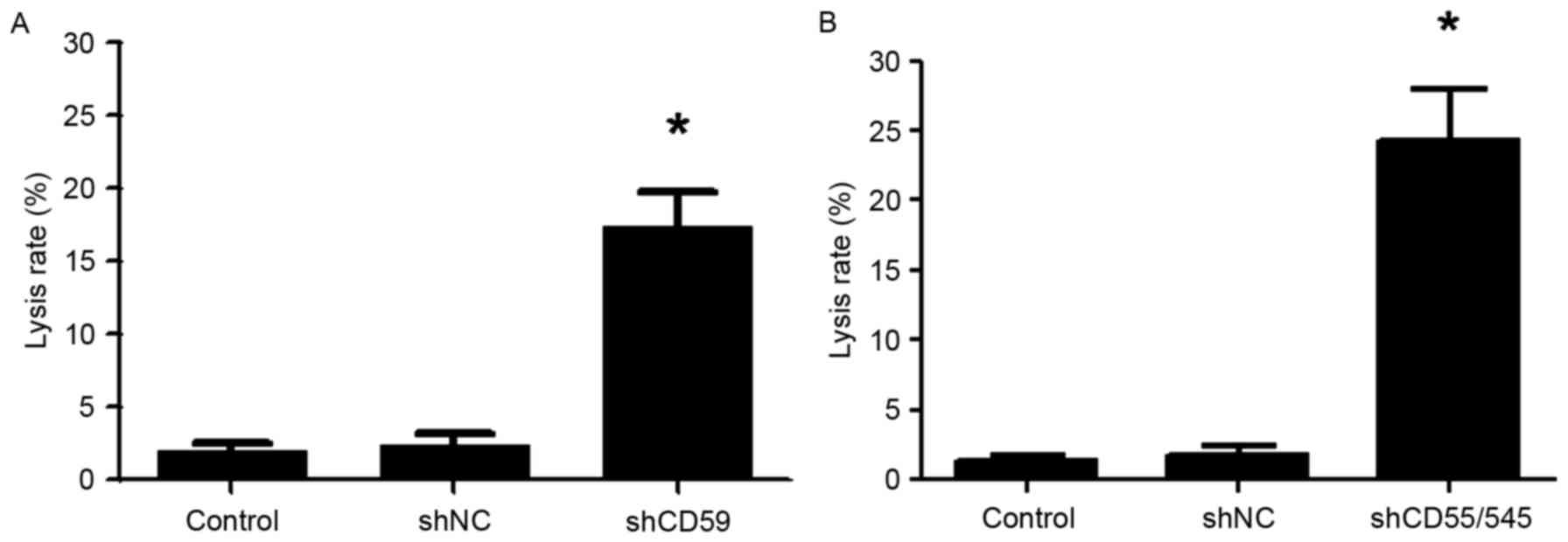
A Trypan blue exclusion assay revealed that
transfection with shCD59 resulted in a significantly increased
lysis rate following trastuzumab treatment compared with control
shNC cells (17.3 vs. 2.3%; P<0.05; Fig. 6A). In addition, transfection with
shCD55/545 resulted in a significantly increased lysis rate
following trastuzumab treatment compared with control shNC cells
(24.3 vs. 1.9%; P<0.05; Fig.
6B).
Discussion
A number of mAbs have been authorized for the
treatment of patients. The molecular mechanisms underlying the
anti-tumor effects of mAbs include the inhibition of signaling
pathways, which lead to downstream effects, including apoptosis,
blocking of growth factor receptor and inhibition of angiogenesis
(4,13). Although a number of studies have
demonstrated that mAbs exhibit low efficacy against their targets
(14), mAbs may exhibit desirable
effects beyond target-related pathways, including enhancement of
CDC.
Trastuzumab comprises a human immunoglobulin G1 Fc
segment, a potent enhancer of CDC (15,16).
However, previous studies have indicated that effects on CDC
effects are not a key molecular mechanism underlying the anti-tumor
effects of trastuzumab (5,6,17–19). Abnormal expression of mCRPs has been
reported in a number of types of tumor (7,20–26), which may inhibit the activation of
complement factors and CDC effects induced by mAbs, therefore
compromising the therapeutic potential of mAbs (23). In accordance with previous studies,
which reported that breast cancer cells overexpress mCRPs (25,27–29), all 7
breast cancer cell lines used in the present study were
demonstrated to overexpress CD55 and CD59. The HER2-positive cell
lines SK-BR-3 and BT474 were selected for subsequent study.
Treatment with trastuzumab was not sufficient to induce CDC in
these cells in the presence of NHS, which is consistent with the
results from Mineo et al (5),
which demonstrated that trastuzumab could not induce CDC in
CD55/59-expressing malignant neuroblastoma cells. Therefore, the
failure of trastuzumab to induce CDC in HER-2-positive breast
cancer cells may be due to the high expression of CD55 and
CD59.
Previous reports have indicated that blocking the
biological function of mCRPs may enhance the anti-tumor effects of
mAbs. For example, rituximab is a human-mouse chimeric antibody
that targets malignant non-Hodgkin's lymphoma B cells that
overexpress CD20. The expression of CD55 and CD59 on the surface of
lymphoma cells was associated with their resistance to CDC.
Lymphoma cells that highly expressed CD55 and CD59 exhibited a
lower cytolysis rate induced by rituximab (30–32). These
previous studies have revealed that blocking, degradation and
downregulation of mCRPs significantly promote CDC effects induced
by rituximab in lymphoma cells.
In the present study, blocking antibodies targeting
CD55 or CD59 significantly enhanced trastuzumab-induced CDC. The
cytolysis rate increased from 1–2% up to 14–25% and the combined
use of the two antibodies further increased the cytolysis rate to
32–40%. The primary biological function of CD55 is to prevent the
assembly of C3 and C5 transferase and accelerate their decay. The
primary biological function of CD59 is preventing the formation of
the membrane attack complex by binding to C8 and C9 in the final
stage of the complement activation phase (33,34).
PI-PLC can specifically separate
glycosyl-phosphatidyl inositol-anchored CD55 and CD59 from the cell
membrane. Previous research demonstrated that the sensitivity of
melanoma (35), lung carcinoma
(36) and ovarian cells (37) to CDC was promoted by PI-PLC. Results
from the present study demonstrated that PI-PLC enhances the
sensitivity of SK-BR-3 and BT474 cells to trastuzumab-induced CDC.
PI-PLC pre-treatment increased the cell lysis rate to 30–40%, which
was comparable with the lysis rate following combined treatment
with CD55 and CD59 blocking antibodies. The effects of PI-PLC on
the cell lysis rate increased in a concentration-dependent manner
up to 0.1 U/ml.
Small interfering RNA (siRNA)-mediated RNA
interference is currently the most effective method for specific
gene silencing (38). Bellone et
al (39) indicated that
downregulation of CD55 and CD59 by siRNA enhances
trastuzumab-induced CDC in uterine serous carcinoma cells, but the
knockdown of CD55 and CD59 were only 11.6 and 10.7%,
respectively.
Mamidi et al (40) reported that knockdown of mCRPs (CD46,
CD55 and CD59) by chemically stabilized siRNAs using cationic
lipoplexes (AtuPLEXes) led to increased CDC in BT474, SK-BR-3
(breast), SKOV3 (ovarian) and Calu-3 (lung) cancer cell lines
following combined treatment with trastuzumab and pertuzumab.
However, knockdown of individual mCRPs did not significantly
increase trastuzumab-induced CDC. Application of in vivo
siRNA technologies is a rapid development field with many
challenges, including the development of transfection systems,
instability and adverse responses in vivo. shRNA plasmids
can integrate into the host genome and achieve long-term gene
silencing. In the present study, shRNA plasmids were designed that
targeted CD55 and CD59 separately. The shRNAs downregulated CD55
and CD59 at mRNA and protein levels, and significantly increased
the cytolytic effect of trastuzumab-induced CDC. Downregulation of
CD55 or CD59 resulted in a cytolysis rate of 24.3 and 17.3%,
respectively. Therefore, downregulation of CD55 and CD59 can
enhance trastuzumab-induced CDC.
The results from the present study indicate that
PI-PLC could almost completely split CD55 and CD59 from cell
membrane specifically, but the cytolysis rate only reached a
maximum of ~40%. Thus, other mCRPs may serve crucial regulatory
roles. Future studies are required to investigate the role of other
complement regulatory proteins, including soluble and
membrane-bound components.
Together with previous research (10), the results from the present study
indicate that mCRP expression may be a predictor of patient
prognosis and response to trastuzumab. The combined application of
mAbs, RNA interference, or other means of downregulating mCRP
expression may improve the clinical efficacy of trastuzumab.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 30972940).
Glossary
Abbreviations
Abbreviations:
|
CDC
|
complement-dependent cytotoxicity
|
|
PI-PLC
|
phosphatidylinositol-specific
phospholipase C
|
|
ADCC
|
antibody-dependent cellular
cytotoxicity
|
|
mAbs
|
monoclonal antibodies
|
|
mCRPs
|
membrane-bound complement regulatory
proteins
|
|
shRNA
|
short hairpin RNA
|
|
NHS
|
normal human serum
|
|
INHS
|
inactivated normal human serum
|
|
FCM
|
flow cytometry
|
|
BSA
|
bovine serum albumin
|
|
MFI
|
mean fluorescence intensity
|
References
|
1
|
Servick K: Breast cancer. Breast cancer: A
world of differences. Science. 343:1452–1453. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thomas E and Berner G: Prognostic and
predictive implications of HER2 status for breast cancer patients.
Eur J Oncol Nurs. 4:10–17. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cardoso F, Piccart MJ, Durbecq V and Di
Leo A: Resistance to trastuzumab: A necessary evil or a temporary
challenge? Clin Breast Cancer. 3:247–259. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Scott AM, Wolchok JD and Old LJ: Antibody
therapy of cancer. Nat Rev Cancer. 12:278–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mineo JF, Bordron A, Quintin-Roué I,
Loisel S, Ster KL, Buhé V, Lagarde N and Berthou C: Recombinant
humanised anti-HER2/neu antibody (Herceptin) induces cellular death
of glioblastomas. Br J Cancer. 91:1195–1199. 2004.PubMed/NCBI
|
|
6
|
Prang N, Preithner S, Brischwein K, Göster
P, Wöppel A, Müller J, Steiger C, Peters M, Baeuerle PA and da
Silva AJ: Cellular and complement-dependent cytotoxicity of
Ep-CAM-specific monoclonal antibody MT201 against breast cancer
cell lines. Br J Cancer. 92:342–349. 2005.PubMed/NCBI
|
|
7
|
Fishelson Z, Donin N, Zell S, Schultz S
and Kirschfink M: Obstacles to cancer immunotherapy: Expression of
membrane complement regulatory proteins (mCRPs) in tumors. Mol
Immunol. 40:109–123. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Macor P and Tedesco F: Complement as
effector system in cancer immunotherapy. Immunol Lett. 111:6–13.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu Y, Wang Y, Qin F, Wang Z, Wang Y, Yang
Y and Zheng H and Zheng H: CD55 limits sensitivity to
complement-dependent cytolysis triggered by heterologous expression
of α-gal xenoantigen in colon tumor cells. Am J Physiol
Gastrointest Liver Physiol. 306:G1056–G1064. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu M, Yang YJ, Zheng H, Zhong XR, Wang Y,
Wang Z, Wang YG and Wang YP: Membrane-bound complement regulatory
proteins are prognostic factors of operable breast cancer treated
with adjuvant trastuzumab: A retrospective study. Oncol Rep.
32:2619–2627. 2014.PubMed/NCBI
|
|
11
|
Shi XX, Zhang B, Zang JL, Wang GY and Gao
MH: CD59 silencing via retrovirus-mediated RNA interference
enhanced complement-mediated cell damage in ovary cancer. Cell Mol
Immunol. 6:61–66. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Reichert JM: Antibody-based therapeutics
to watch in 2011. MAbs. 3:76–99. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Strome SE, Sausville EA and Mann D: A
mechanistic perspective of monoclonal antibodies in cancer therapy
beyond target-related effects. Oncologist. 12:1084–1095. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Clark MR: IgG effector mechanisms. Chem
Immunol. 65:88–110. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dalle S, Thieblemont C, Thomas L and
Dumontet C: Monoclonal antibodies in clinical oncology. Anticancer
Agents Med Chem. 8:523–532. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dean-Colomb W and Esteva FJ: Her2-positive
breast cancer: Herceptin and beyond. Eur J Cancer. 44:2806–2812.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
De Lorenzo C, Tedesco A, Terrazzano G,
Cozzolino R, Laccetti P, Piccoli R and D'Alessio G: A human,
compact, fully functional anti-ErbB2 antibody as a novel antitumour
agent. Br J Cancer. 91:1200–1204. 2004.PubMed/NCBI
|
|
19
|
Yu J, Caragine T, Chen S, Morgan BP, Frey
AB and Tomlinson S: Protection of human breast cancer cells from
complement-mediated lysis by expression of heterologous CD59. Clin
Exp Immunol. 115:13–18. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gelderman KA, Tomlinson S, Ross GD and
Gorter A: Complement function in mAb-mediated cancer immunotherapy.
Trends Immunol. 25:158–164. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yan J, Allendorf DJ, Li B, Yan R, Hansen R
and Donev R: The role of membrane complement regulatory proteins in
cancer immunotherapy. Adv Exp Med Biol. 632:159–174.
2008.PubMed/NCBI
|
|
22
|
Jurianz K, Ziegler S, Garcia-Schüler H,
Kraus S, Bohana-Kashtan O, Fishelson Z and Kirschfink M: Complement
resistance of tumor cells: Basal and induced mechanisms. Mol
Immunol. 36:929–939. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gancz D and Fishelson Z: Cancer resistance
to complement-dependent cytotoxicity (CDC): Problem-oriented
research and development. Mol Immunol. 46:2794–2800. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rushmere NK, Knowlden JM, Gee JM, Harper
ME, Robertson JF, Morgan BP and Nicholson RI: Analysis of the level
of mRNA expression of the membrane regulators of complement, CD59,
CD55, and CD46, in breast cancer. Int J Cancer. 108:930–936. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ravindranath NM and Shuler C: Cell-surface
density of complement restriction factors (CD46, CD55 and CD59):
Oral squamous cell carcinoma versus other solid tumors. Oral Surg
Oral Med Oral Pathol Oral Radiol Endod. 103:231–239. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Loberg RD, Wojno KJ, Day LL and Pienta KJ:
Analysis of membrane-bound complement regulatory proteins in
prostate cancer. Urology. 66:1321–1326. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Madjd Z, Durrant LG, Bradley R, Spendlove
I, Ellis IO and Pinder SE: Loss of CD55 is associated with
aggressive breast tumors. Clin Cancer Res. 10:2797–2803. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zell S, Geis N, Rutz R, Schultz S, Giese T
and Kirschfink M: Down-regulation of CD55 and CD46 expression by
anti-sense phosphorothioate oligonucleotides (S-ODNs) sensitizes
tumour cells to complement attack. Clin Exp Immunol. 150:576–584.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jurianz K, Maslak S, Garcia-Schuler H,
Fishelson Z and Kirschfink M: Neutralization of complement
regulatory proteins augments lysis of breast carcinoma cells
targeted with rhumAb anti-HER2. Immunopharmacology. 42:209–218.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Macor P, Tripodo C, Zorzet S, Piovan E,
Bossi F, Marzari R, Amadori A and Tedesco F: In vivo targeting of
human neutralizing antibodies against CD55 and CD59 to lymphoma
cells increases the antitumor activity of rituximab. Cancer Res.
67:10556–10563. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Golay J, Lazzari M, Facchinetti V,
Bernasconi S, Borleri G, Barbui T, Rambaldi A and Introna M: CD20
levels determine the in vitro susceptibility to rituximab and
complement of B-cell chronic lymphocytic leukemia: Further
regulation by CD55 and CD59. Blood. 98:3383–3389. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou X, Hu W and Qin X: The role of
complement in the mechanism of action of rituximab for B-cell
lymphoma: Implications for therapy. Oncologist. 13:954–966. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nicholson-Weller A and Wang CE: Structure
and function of decay accelerating factor CD55. J Lab Clin Med.
123:485–491. 1994.PubMed/NCBI
|
|
34
|
Fonsatti E, Altomonte M, Coral S, De Nardo
C, Lamaj E, Sigalotti L, Natali PG and Maio M: Emerging role of
protectin (CD59) in humoral immunotherapy of solid malignancies.
Clin Ter. 151:187–193. 2000.PubMed/NCBI
|
|
35
|
Brasoveanu LI, Altomonte M, Fonsatti E,
Colizzi F, Coral S, Nicotra MR, Cattarossi I, Cattelan A, Natali PG
and Maio M: Levels of cell membrane CD59 regulate the extent of
complement-mediated lysis of human melanoma cells. Lab Invest.
74:33–42. 1996.PubMed/NCBI
|
|
36
|
Azuma A, Yamano Y, Yoshimura A, Hibino T,
Nishida T, Yagita H, Okumura K, Seya T, Kannagi R, Shibuya M, et
al: Augmented lung adenocarcinoma cytotoxicity by the combination
of a genetically modified anti-Lewis Y antibody and antibodies to
complement regulatory proteins. Scand J Immunol. 42:202–208. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bjorge L, Hakulinen J, Wahlström T, Matre
R and Meri S: Complement-regulatory proteins in ovarian
malignancies. Int J Cancer. 70:14–25. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Aagaard L and Rossi JJ: RNAi therapeutics:
Principles, prospects and challenges. Adv Drug Deliv Rev. 59:75–86.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bellone S, Roque D, Cocco E, Gasparrini S,
Bortolomai I, Buza N, Abu-Khalaf M, Silasi DA, Ratner E, Azodi M,
et al: Downregulation of membrane complement inhibitors CD55 and
CD59 by siRNA sensitises uterine serous carcinoma overexpressing
Her2/neu to complement and antibody-dependent cell cytotoxicity in
vitro: Implications for trastuzumab-based immunotherapy. Br J
Cancer. 106:1543–1550. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mamidi S, Cinci M, Hasmann M, Fehring V
and Kirschfink M: Lipoplex mediated silencing of membrane
regulators (CD46, CD55 and CD59) enhances complement-dependent
anti-tumor activity of trastuzumab and pertuzumab. Mol Oncol.
7:580–594. 2013. View Article : Google Scholar : PubMed/NCBI
|