Introduction
Colorectal cancer (CRC) has one of the highest cure
rates of all types of malignant tumors (1), however remains ranked as the fourth
leading cause of cancer-associated mortality in the world (2). In previous years, the morbidity and
mortality rates of CRC have significantly increased due to an
ageing population, and with changes in eating habits and lifestyles
(3). The development of distant
metastasis is a major cause of cancer-associated mortalities in CRC
patients (4). Overwhelming evidence
has demonstrated that aberrant expression of microRNA (miRNA/miR)
contributes to CRC development by affecting the expression of the
genes that regulate cancer progression (5).
miRNAs, endogenous small non-coding regulatory RNAs
measuring 18–25 nucleotides long (6),
usually regulate gene expression in a number of tumor-associated
signaling pathways at the post-transcriptional level, including the
Wnt/β-catenin signaling pathway (7).
As miRNAs tend to be localized to fragile chromosomal regions
(8), they have the ability to adjust
the levels of their corresponding mRNAs, and serve critical roles
in the physiological and pathological processes of tumor
development, which are a novel aspect of cancer studies. Previous
evidence has demonstrated that miRNAs are involved in a number of
biological processes, including proliferation, differentiation,
migration, angiogenesis and protein splitting (9–11). miRNAs
also serve as tumor promoter genes or tumor suppressor genes by
negatively regulating their targets. These data suggest a
possibility that miRNA is a novel focus for examining the current
diagnosis and treatment of tumors.
Previously, several studies revealed that miR-124
results in a decrease in the proliferation ability of CRC cells by
targeting ribose-phosphate pyrophosphokinase 1 and ribose
5-phosphate isomerase mRNAs (12),
that miR-146a directs the symmetric division of colorectal cancer
stem cells (13) and that miR-101
serves as an endogenous proteasome inhibitor that suppresses tumor
cell proliferation via targeting of the proteasome assembly factor
proteasome maturation protein (14).
Therefore, attention has been focused on the activities of miRNA in
cancer development.
In the present study, it was demonstrated that the
upregulation of miR-552 is observed in CRC cells and tissues. In
vitro experiments revealed that the high expression of miR-552
was associated with CRC proliferation and migration. Notably, it
was demonstrated that Dachshund family transcription factor 1
(DACH1) is a direct target of miR-552, the levels of which
decreased when miR-552 was overexpressed. In vitro, the
Wnt/β-catenin signaling pathway is a key pathway for the
proliferation of CRC cells (15). The
restoration of normal expression levels of DACH1 affected the
Wnt/β-catenin signaling pathway and the corresponding downstream
targets; therefore it may be possible that miR-552 contributes to
tumor proliferation and migration via inhibiting DACH1 and serves
as a marker for poorer prognoses. Therefore, it may be a potential
target for therapeutic intervention in patients with CRC.
Materials and methods
Total RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues samples and cell
lines using TRIzol reagent (Takara Bio, Inc., Otsu, Japan)
according to the manufacturer's protocol, which efficiently
recovered all RNA species, including miRNAs. RNA quality and
quantity were measured using a spectrophotometer and RNA integrity
was determined by gel electrophoresis. cDNA was reverse-transcribed
and amplified by PCR using Reverse Transcription kit (Takara Bio,
Inc.) and miRNA Real-Time PCR assay kit (GeneCopoeia, Inc.,
Rockville, MD, USA). The expression level of miR-552 was measured
using miR-552 specific primers and SYBR-Green fluorophore (Takara
Bio, Inc.) probes. The primers used are listed in Table I. The reverse transcription product
was 2 µl, and the PCR reaction system was as follows: 40 cycles of
pre-denaturation at 95°C for 10 min, followed by annealing and
stretching at 95°C for 15 sec and 60°C for 1 min, respectively. U6
and GAPDH were used as internal controls for miR-552 and DACH1,
respectively. The relative expression of each gene was quantified
by the 2−∆ΔCq method (16). The experiment was performed in
triplicates.
 | Table I.Reverse transcription-quantitative
polymerase chain reaction primers. |
Table I.
Reverse transcription-quantitative
polymerase chain reaction primers.
| Gene | Sequence (5′→3′) |
|---|
| miR-552 |
AACAGGTGACTGGTTAGACAA |
| U6 |
ATTGGAACGATACAGAGAAGATT |
| Forward |
|
|
Reverse |
CGAACGCTTCACGAATTTG |
| DACH1 |
|
|
Forward |
CCCTCTACAATGACTGCACCA |
|
Reverse |
GCGGCATGATGTGAGAGTTCT |
| c-Myc |
|
|
Forward |
GTCAAGAGGCGAACACACAAC |
|
Reverse |
TTGGACGGACAGGATGTATGC |
| Cyclin D1 |
|
|
Forward |
GTCTGTGCATTTCTGGTTGCA |
|
Reverse |
TTTCTAGACTTTCATGTTTGTCTTTTTGTC |
| GAPDH |
|
|
Forward |
TCATGGGTGTGAACCATGAGAA |
|
Reverse |
GGCATGGACTGTGGTCATGAG |
| GSK3β |
|
|
Forward |
AGACGCTCCCTGTGATTTATGT |
|
Reverse |
CCGATGGCAGATTCCAAAGG |
| β-catenin |
|
|
Forward |
ATGTCCAGCGTTTGGCTGAA |
|
Reverse |
TGGTCCTCGTCATTTAGCAGTT |
Clinical samples, cell lines, in vitro
culture and transfection
The present study was approved by the Ethics Review
Committees of the General Hospital of Ningxia Medical University,
(Yinchuan, Ningxia, China), with written informed consent being
obtained from all patients. A total of 20 pairs of CRC tissues and
matched adjacent normal tissues were collected from patients during
surgical resection. LOVO, SW620, HCT116 and NCM460 cells were all
purchased from the Cell Bank of Type Culture Collection of Chinese
Academy of Sciences (Shanghai, China). Cells were cultured in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) supplemented with 10% fetal bovine serum (FBS) and 1%
antibiotics at 37°C at 5% CO2. The cells were
transfected with 20 µM miR-552 inhibitor (miR-552-in) or
miR-552-inhibitor negative control (NC) at ~50% density using
Effectene transfection reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
miR-552 target gene prediction
Extensive experiments and comprehensive studies have
demonstrated that miRNAs, negative regulators of target genes,
express its biological function directly or indirectly (17). In this study, the putative miR-552
target genes were predicted using MiRanda (Memorial Sloan-Kettering
Cancer Center, New York, NY, USA), PicTar (Computational Medicine
Center, Philadelphia, PA, USA) and TargetScan (Whitehead Institute
for Biomedical Research, Cambridge, MA, USA) software. Followed the
procedure of Betel et al (18), the present study identified DACH1 as a
target gene of miR-552.
Luciferase reporter assay
A total of ~1×104 cells were seeded in a
96-well plate and were co-transfected miR-552-in and wild type (Wt)
or mutant (Mut) DACH1 reporter plasmids (Jikai Gene Chemical Co.,
Ltd., Shanghai, China) into LOVO and SW620 cells using Effectene
transfection reagent. The cells were washed with PBS and harvested
after 48 h. Luciferase activity was measured with dual-luciferase
reporter assay system (Promega Corporation, Madison, WI, USA).
Cell proliferation assay
For the colony formation assay, LOVO and SW620 cells
were grown in 6-well culture plates and transfected with miR-552-in
or NC using Effectene transfection reagent. After 7–10 days, the
cells were fixed with methanol for 15 min and stained with 0.2%
crystal violet for visualization and counting. For the MTT assay,
100 µl cells per well were seeded into 96-well plates, and were
transfected with miR-552-in or the negative control
(miR-552-inhibitor-NC) using Effectene transfection reagent. The
cell proliferation rate was detected at 24, 48, 72 and 96 h. The
cells were disposed with 20 µl MTT solution and incubated at 37°C
for 4 h. Subsequently, 150 µl dimethyl sulfoxide was added and the
absorbance values were detected using Thermo Multiskan Go
Microplate Reader (Thermo Fisher Scientific, Inc.) at 490 nm. The
NC group was the control.
Wound-healing assay
Approximately 70% of the cell density was planted in
12-well plate prior to transfection. The wound-healing assay was
implemented to determine the cell migration ability subsequent to
transfection with miR-552-in or NC, and the cell migration rate was
recorded by microscopy at 0 and 24 h when the monolayer was at 90%
density.
Transwell assay
The cells were grown in 24-well culture plates and
transfected with miR-552-in or NC using Effectene transfection
reagent. Cells were harvested after 48 h. The Transwell assay was
performed using 8-µm pore size Corning chambers (Corning Costar,
Corning, NY, USA). The lower chamber was filled with 600 µl
RPMI-1640 medium containing 20% FBS and Recombinant Human
platelet-derived growth factor-BB (Invitrogen; Thermo Fisher
Scientific, Inc.) was added. The Matrigel-coated insert (BD
Biosciences, Franklin Lakes, NJ, USA) was placed and cells were
counted to 15×104 cells in the upper chamber. The
non-migrated or non-invaded cells were gently removed from the
upper surface of the membrane by cotton swab, and the migrated
cells were attached to the lower surface after 24 h. The cells were
fixed with 100% methanol and stained using 0.2% crystal violet
solution for 15 min. Cells that migrated onto the lower surface
were counted using a CKX-41 inverted fluorescence microscope with
magnification, ×100 (Olympus Corporation, Tokyo, Japan) in five
randomly chosen visual fields.
Western blotting
The cells were lysed in protein extraction
radioimmunoprecipitation buffer [20 mM Tris-HCl (pH 7.4), 100 mM
NaCl, 10% NP-40, 10% sodium deoxycholate and 100 mM EDTA] and each
sample was determined using a bicinchonic acid protein assay
reagent kit (Thermo Fisher Scientific, Inc.) subsequent to
transfection with miR-552-in or NC for 48 h. A total of 20 µg of
lysates were electrophoresed by 10% SDS-PAGE and transferred onto
0.45-µm polyvinylidene fluoride membranes (Merck KGaA, Darmstadt,
Germany). Subsequently, the proteins were blocked with 5% non-fat
milk for 1 h at room temperature. The membranes were incubated with
primary antibodies, including: Anti cyclin D1 (rabbit monoclonal;
dilution, 1:1,000; cat no. 2926), anti c-myc (rabbit monoclonal;
dilution, 1:1,000; cat no. 5605), anti β-catenin (rabbit
monoclonal; dilution, 1:500; cat no. 8480); anti GSK3β (rabbit
monoclonal; dilution 1:1,000; cat no. 12456S) (all from Cell
Signaling Technology, Inc., Danvers, MA, USA); anti DACH1 (rabbit
monoclonal; dilution 1:1,000; cat no. 10914-1-A P; Wuxi AccoBio
BioTech Inc., Wuxi, China), GAPDH (mouse monoclonal; dilution,
1:5,000; cat no. 2D4A7; Novus Biologicals, LLC., Littleton, CO,
USA) overnight at 4°C, followed by incubation with the horseradish
peroxidase conjugated secondary antibodies (anti-rabbit: dilution,
1:10,000; cat no. #7074; or anti-mouse: dilution, 1:10,000; cat no.
#7076) (both from Cell Signaling Technology, Inc.) for 1.5 h at
room temperature. The protein expression was normalized against
GAPDH and grayscale analysis was performed using Image-Pro plus
(version 6.0) software (Media Cybernetics, Rockville, MD, USA). The
experiment was performed in triplicate.
Statistical analysis
All statistical analyses were performed using the
SPSS 17.0 statistical software package (SPPS, Inc. Chicago, IL,
USA) or GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA,
USA). The difference between two groups was determined as mean ±
standard deviation and statistical significance was analyzed using
a t-test or one-way analysis of variance followed by Bonferroni's
multiple comparison test. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-552 is upregulated in CRC tissues
and cell lines
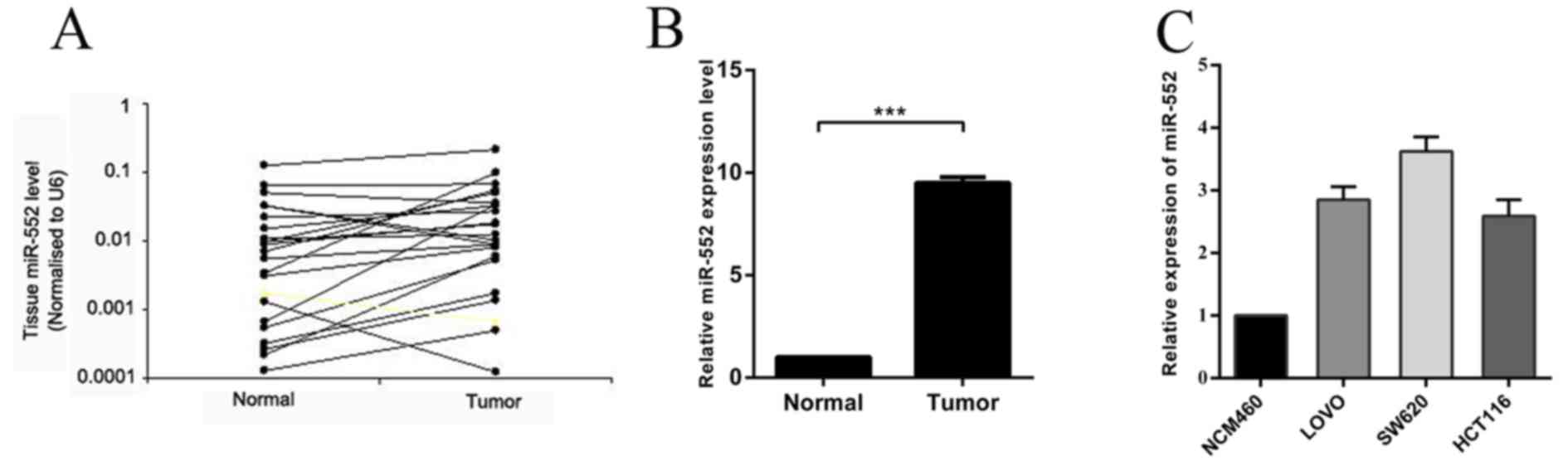
In order to confirm the involvement of miR-552 in
CRC carcinogenesis, the expression of miR-552 was measured by
RT-qPCR in 20 pairs of CRC tissues and their adjacent normal
tissues. The results indicated that the expression level of miR-552
in CRC tissues was significantly higher compared with that in
matched normal tissues (Fig. 1A and
B). It was also demonstrated that miR-552 was upregulated in
the CRC LOVO, SW620 and HCT116 cell lines, when compared with
normal colorectal NCM460 cell line (Fig.
1C), which is consistent with the results identified in CRC
tissues. In conclusion, these observations suggest that the
overexpression of miR-552 may serve important roles in CRC
carcinogenesis and progression.
Ectopic expression of miR-552 promoted
CRC cell proliferation in vitro
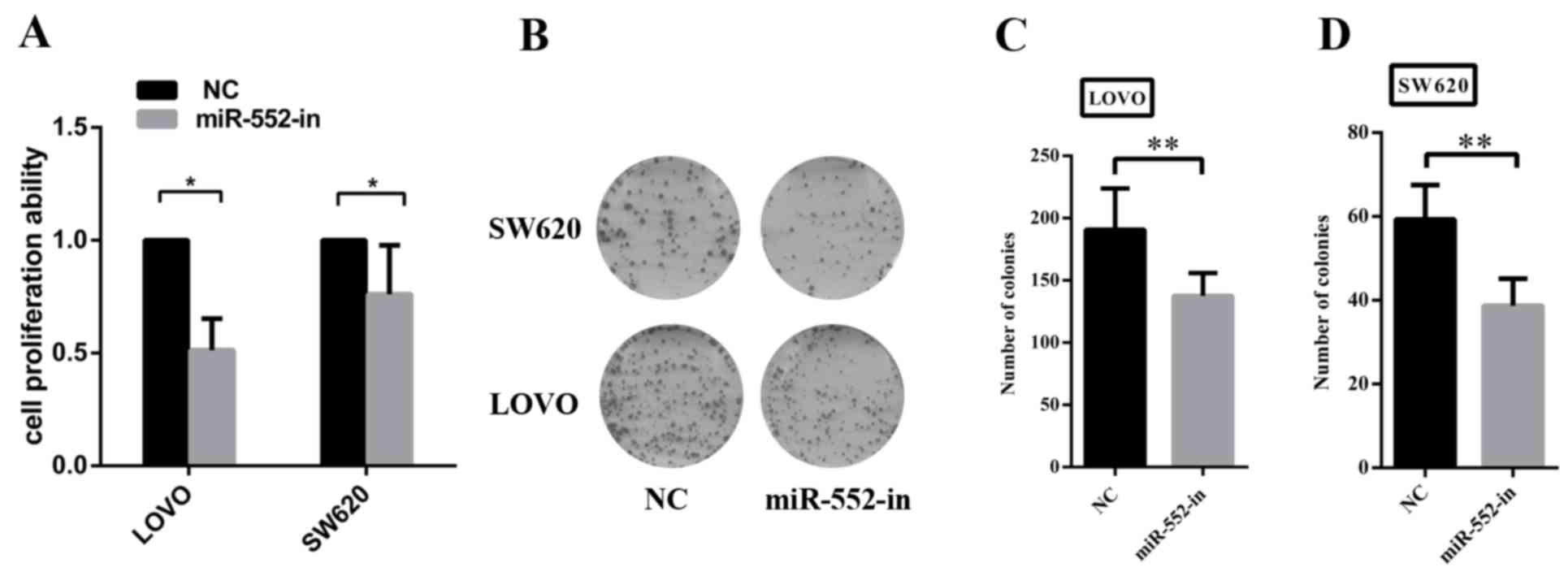
The increased expression of miR-552 in CRC cell
lines suggested that miR-552 may be an oncogene. To additionally
investigate the biological functions of miR-552 in CRC development,
MTT and colony formation assays were performed to assess the role
of miR-552 in CRC cell viability and proliferation. The results of
the MTT assay illustrate that miR-552-in significantly reduced the
viability of CRC cells compared with the control group (Fig. 2A). The colony formation assay also
revealed that the overexpression of miR-552 increased the
proliferation ability in the two CRC cell lines (Fig. 2B). The inhibition efficiency was
additionally measured by calculating the entire field of vision on
the number of cells (Fig. 2C and
D).
Overexpression of miR-552 promotes
migration of CRC cell in vitro
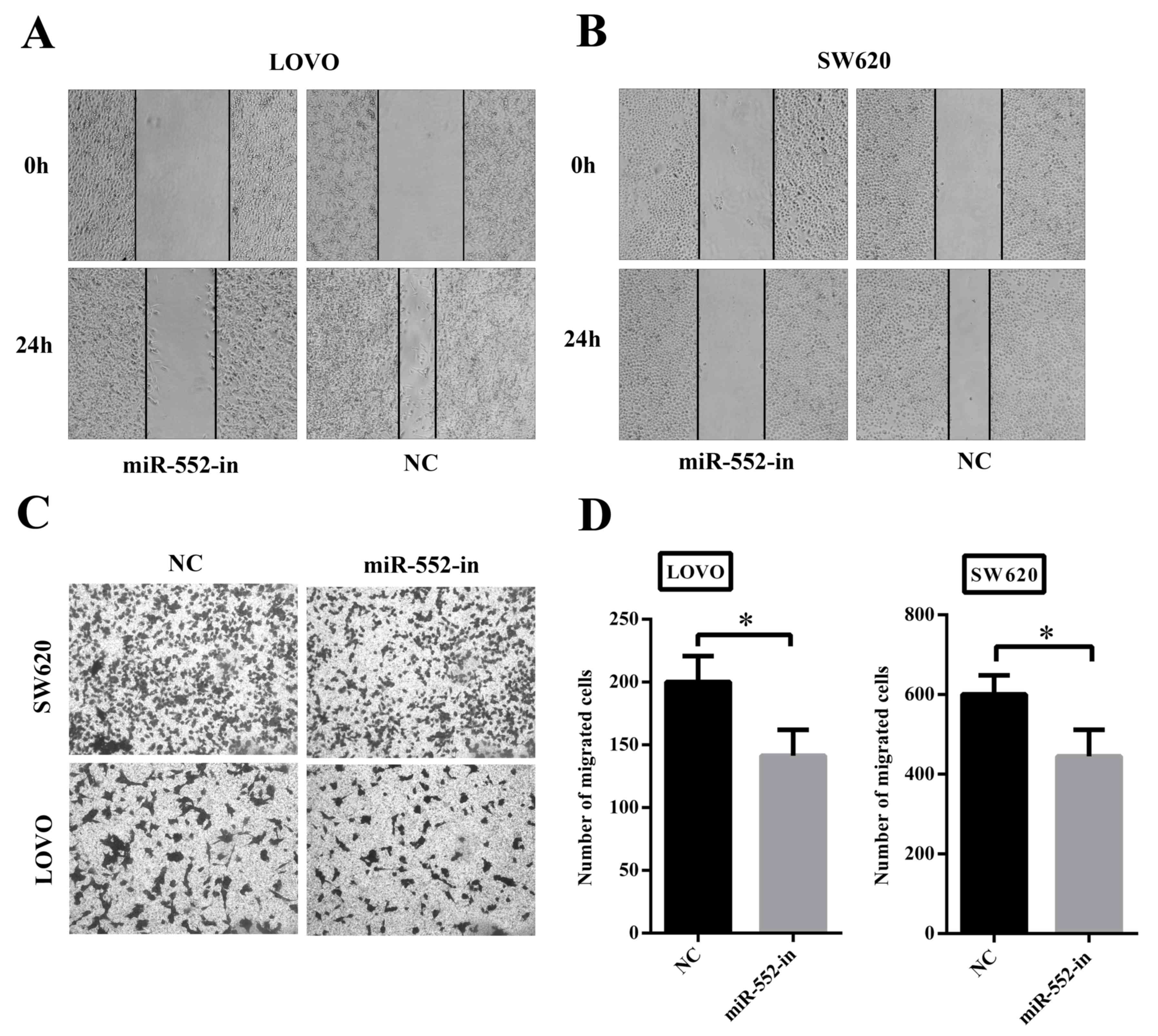
To determine whether overexpression of miR-552
exhibited a crucial role in migration and invasion, wound-healing
and Transwell assays were performed. Fig.
3A and B illustrates that decreased expression of miR-552
(miR-552-in) was associated with significantly slower wound closure
in LOVO and SW620 cells compared with their corresponding controls.
Secondly, the Transwell assay also demonstrated that transfection
with miR-552-in exhibited lower migratory and invasive activities
compared with the control groups (Fig.
3C). In addition, the number of miR-552-in transfected
migrating cells was calculated using statistical software (Fig. 3D). Thus, it is suggested that miR-552
clearly facilitates CRC cell migration in vitro.
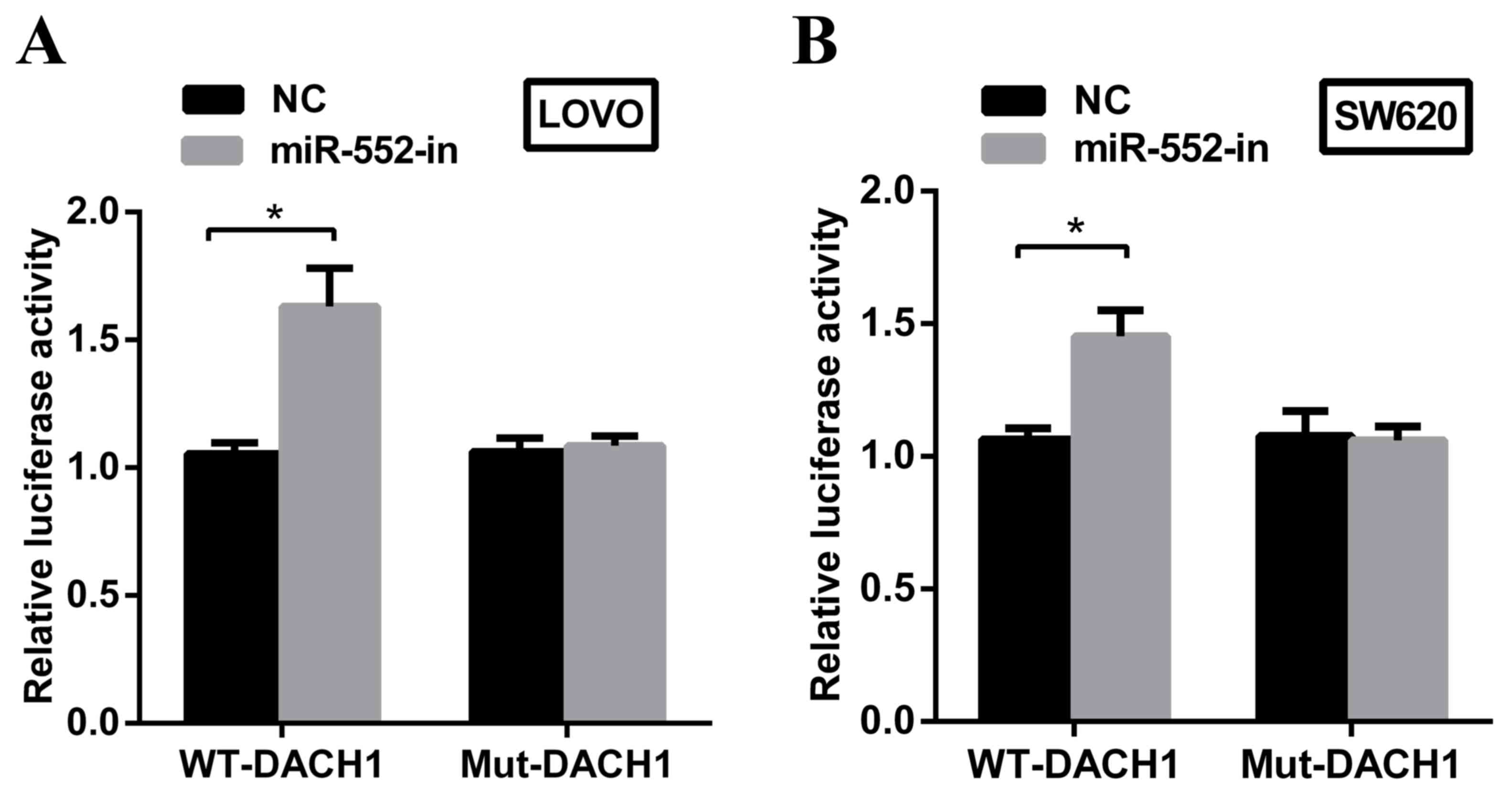
DACH1 is a direct target of miR-552 in
CRC cells
DACH1 is frequently downregulated in CRC and is
closely associated with poorer prognoses (19). Based on MiRanda software, it was
predicted that DACH1 may be a potential downstream target of
miR-552. To explore whether DACH1 is a direct target of miR-552 in
the CRC cellular environment, a dual-luciferase reporter assay was
performed. miR-552-in or NC with plasmids containing 3′untranslated
regions (UTR) of wt-DACH1 or mut-DACH1 were transfected into LOVO
and SW620 cell lines. The results demonstrated that the
downregulation of miR-552 significantly increased the relative
luciferase activity of wt-DACH1-3′UTR in the two cell lines.
However, no difference in luciferase activity was observed in
mut-DACH1-3′UTR (Fig. 4A and B).
miR-552 negatively regulates DACH1
expression at the post-transcriptional level
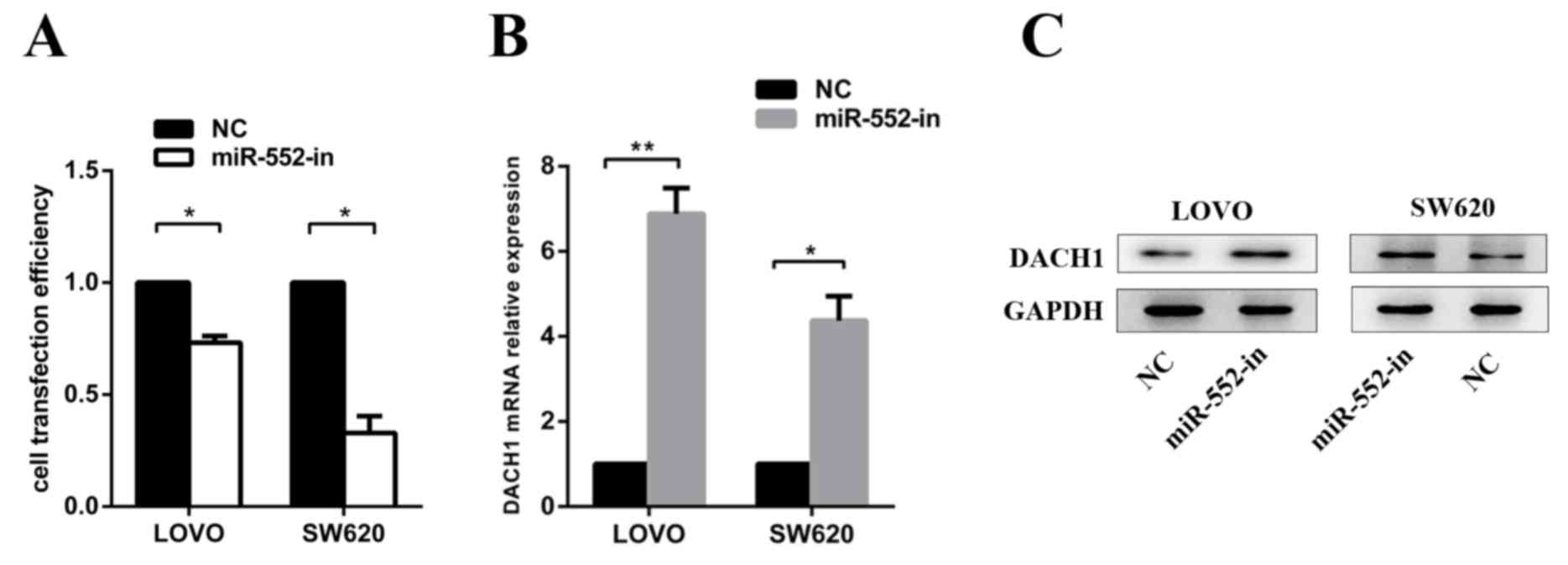
The LOVO and SW620 cells were transfected with
miR-552-in or NC to measure transfection efficiency (Fig. 5A) and to verify the expression of
DACH1 mRNA and protein levels. It was revealed with RT-qPCR
(Fig. 5B) and western blot analysis
(Fig. 5C) that miR-552-in markedly
increased the DACH1 mRNA and protein expression levels.
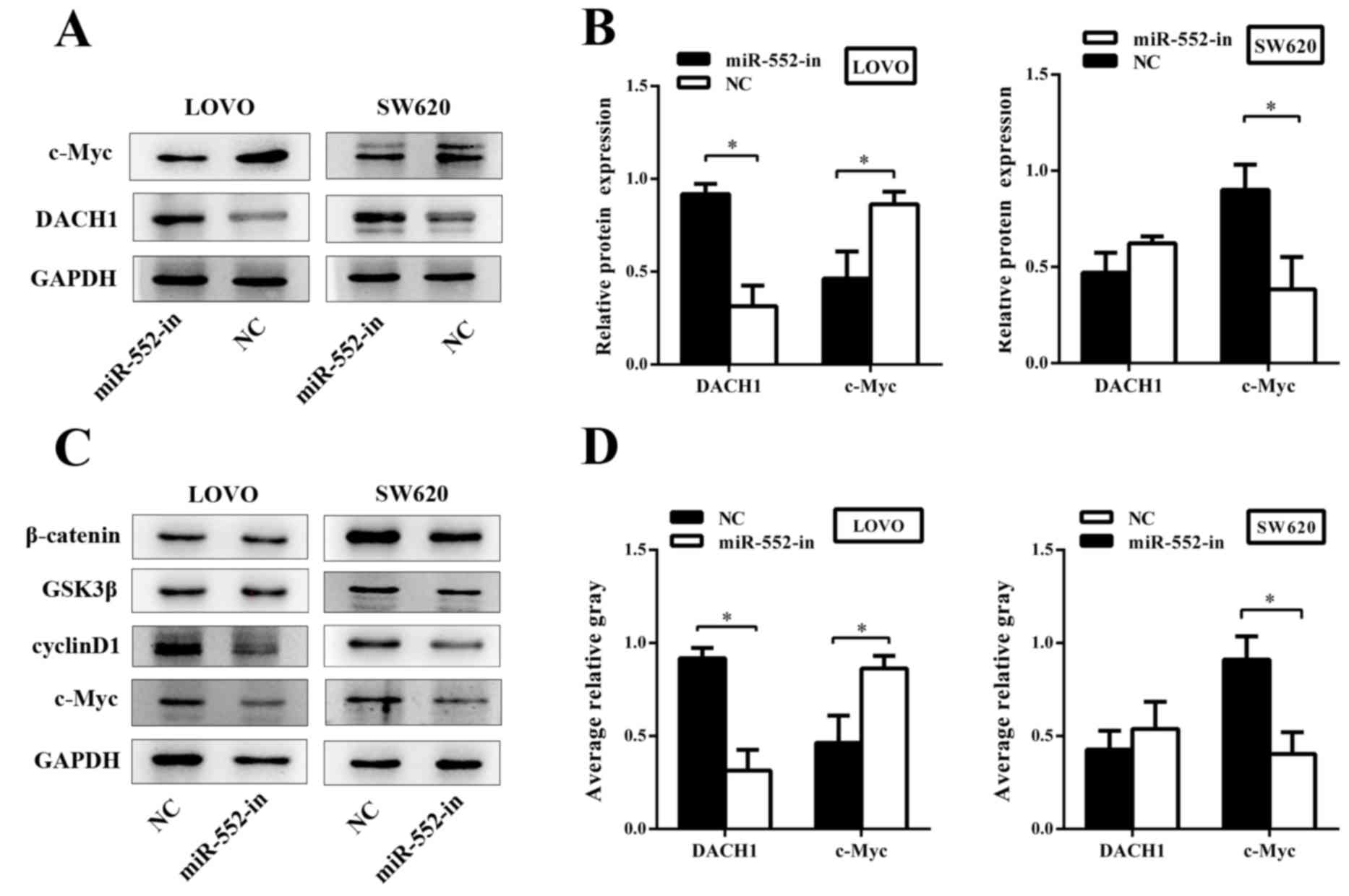
miR-552 affects the Wnt/β-catenin
signaling pathway
To investigate the underlying mechanism of miR-552
in CRC, the biological functions of the genes associated with the
Wnt/β-catenin signaling pathway were analyzed. Western blotting was
used to explore gene expression of the Wnt/β-catenin signaling
pathway at the protein level. The results demonstrated that an
abnormal expression of DACH1 significantly inhibited the abundance
of c-Myc, and that the higher expression of DACH1 was usually
accompanied by a lowered expression of c-Myc (Fig. 6A). DACH1 was dramatically upregulated
in response to miR-552-in. By contrast, the expression of c-Myc was
decreased (Fig. 6B). In addition,
other members of the Wnt/β-catenin signaling pathway including
glycogen synthase kinase 3β (GSK3β), cyclin D1 and β-catenin were
examined via western blotting subsequent to transfection. The
results demonstrated that miR-552-in inhibited the expression of
cyclin D1 and c-Myc without altering the expression of GSK3β and
β-catenin when compared with the corresponding negative controls
(Fig. 6C). The average relative gray
value of DACH1 and c-Myc are also presented (Fig. 6D). In conclusion, miR-552 promotes
proliferation and migration of CRC and activates the Wnt/β-catenin
signaling pathway by directly regulating corresponding target
proteins.
Discussion
The molecular mechanisms of proliferation and
migration in CRC requires additional study in vivo and in
vitro (20). Data from several
previous studies have indicated that the interactions between
miRNAs and their corresponding mRNA regulate the expression of
proto-oncogenes or tumor suppressor genes (21) and determine the malignant
characteristics of tumor cells. Therefore, a comprehensive
understanding of the association between miRNAs and tumor
development is crucial for the diagnosis and treatment of cancer
(22). In the present study, the
expression level of miR-552 was initially measured in CRC cells and
tissues, and the results demonstrated that the expression of
miR-552 was significantly increased in the CRC cells and tissues
compared with that in normal cells and tissues. Notably, these data
indicated that a high expression of miR-552 was clearly associated
with the proliferation and migration of CRC. The present study may
explain the function of miR-552 in CRC.
It has been demonstrated that miR-552 is
overexpressed in CRC cells and tissues (23). Additionally, the expression of miR-552
is markedly upregulated in the side population (SP) of CRC cells,
which is closely associated to the resistance to multiple
chemotherapeutics demonstrated in CRC (24). Although the importance of miR-552 in
CRC has received attention in previous years, the hypothesis that
miR-552 promotes the proliferation and migration of CRC has not
been demonstrated. A notable aspect of the present study was that
miR-552 facilitated CRC cell proliferation and migration in
vitro using the colony formation and MTT assays to detect CRC
cell proliferation ability. The wound-healing and Transwell assays
were performed to test migration ability of CRC. The results
demonstrated that an overexpression of miR-552 markedly promoted
the proliferation and migration capabilities of CRC cells. Taken
together, these data highlight the function of miR-552 as a tumor
promoter in the progression of CRC.
As a cancer suppress gene, DACH1 is a crucial member
of the Retinal Determination Gene Network (25). Generally, it has been demonstrated to
be downregulated in a wide number of tumors, such as breast,
endometrial and prostate cancer (19). Additionally, DACH1 has been identified
to inhibit transforming growth factor-β signaling pathway in breast
and ovarian cancer (26) and to
restrain the Wnt/β-catenin signaling pathway in CRC (27). However, the epigenetic regulation and
mechanisms of DACH1 remain unclear. In the present study, it was
revealed that DACH1 was a direct functional target of miR-552 in
CRC cells. Firstly, the miR-552-dependent promotion of CRC cell
proliferation and migration may be completely restored by DACH1
overexpression. Secondly, the upregulation of miR-552 leads to the
downregulation of its target, DACH1, which in turn leads to
proliferation and migration in CRC. Thirdly, miR-552 overexpression
markedly reduced DACH1 mRNA and protein levels. Conversely,
inhibition of the expression of miR-552 may significantly increase
the activity of luciferase reporter containing the 3′UTR sequence
of DACH1. All these data demonstrate that miR-552 exhibits a
negative correlation with DACH1.
The Wnt/β-catenin pathway is an evolutionarily
conserved pathway, which is important in initiating and regulating
a diverse range of cellular activities, including cell
proliferation, calcium homeostasis and cell polarity (28). There are a number of target proteins
in the Wnt/β-catenin signaling pathway, including c-Myc, cyclin D1,
MMP3 and LEF. Previous studies have revealed that DACH1 exhibited
an inverse correlation with c-Myc and cyclin D1 (19). In combination with the results from
the present study, it is hypothesized that miR-552 may have
specific effects on the Wnt/β-catenin signal pathway. To
investigate these effects of miR-552 on the Wnt/β-catenin pathway,
DACH1, c-Myc and cyclin D1 protein expression levels were measured
using western blot analysis. The results demonstrated that the
restoration of expression levels of DACH1 may reduce the expression
of c-Myc and cyclin D1. As DACH1 is associated with the
Wnt/β-catenin pathway, and c-Myc and cyclin D1 are major downstream
targets of the Wnt/β-catenin signaling pathway, the results of this
experiment suggested that miR-552 regulates the expression of DACH1
and c-Myc, therefore affecting the Wnt/β-catenin signaling pathway
in CRC.
In conclusion, the data of the present study
demonstrates that miR-552 is significantly upregulated in CRC. This
miRNA can potently promote CRC cell proliferation and migration
in vitro by mediating DACH1, and activating the
Wnt/β-catenin signaling pathway by directly regulating the
corresponding target proteins. The identification of tumor-specific
miRNAs and their targets is crucial for the comprehensive
understanding of CRC progression and development. These results
suggest that miR-552 may be a predictive biomarker and a novel
therapeutic target for patients with advanced CRC, and may improve
the prognosis of all patients.
Acknowledgements
The present study was supported in part by grants
from the Special Personnel Project of Ningxia Medical University
(grant no. XT201414), and from the National Natural Science
Foundations of China (grant no. 81560474). The authors would like
to thank the Affiliated Hospital of Qingdao University for
providing technical support.
References
|
1
|
Chen MB, Wei MX, Han JY, Wu XY, Li C, Wang
J, Shen W and Lu PH: MicroRNA-451 regulates AMPK/mTORC1 signaling
and fascin1 expression in HT-29 colorectal cancer. Cell Signal.
26:102–109. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dong Y, Yu J and Ng SS: MicroRNA
dysregulation as a prognostic biomarker in colorectal cancer.
Cancer Manag Res. 6:405–422. 2014.PubMed/NCBI
|
|
3
|
Gellad ZF and Provenzale D: Colorectal
cancer: National and international perspective on the burden of
disease and public health impact. Gastroenterology. 138:2177–2190.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cristóbal I, Caramés C, Madoz-Gúrpide J,
Rojo F, Aguilera O and García-Foncillas J: Downregulation of
miR-214 is specific of liver metastasis in colorectal cancer and
could play a role determining the metastatic niche. Int J
Colorectal Dis. 29:8852014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bonfrate L, Altomare DF, Di Lena M,
Travaglio E, Rotelli MT, De Luca A and Portincasa P: MicroRNA in
colorectal cancer: New perspectives for diagnosis, prognosis and
treatment. J Gastrointestin Liver Dis. 22:311–320. 2013.PubMed/NCBI
|
|
6
|
Giordano S and Columbano A: MicroRNAs: New
tools for diagnosis, prognosis, and therapy in hepatocellular
carcinoma? Hepatology. 57:840–847. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka
H, Boland CR and Goel A: Circulating microRNA-203 predicts
prognosis and metastasis in human colorectal cancer. Gut.
66:654–665. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bai R, Weng C, Dong H, Li S, Chen G and Xu
Z: MicroRNA-409-3p suppresses colorectal cancer invasion and
metastasis partly by targeting GAB1 expression. Int J Cancer.
137:2310–2322. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jinushi T, Shibayama Y, Kinoshita I,
Oizumi S, Jinushi M, Aota T, Takahashi T, Horita S, Dosaka-Akita H
and Iseki K: Low expression levels of microRNA-124-5p correlated
with poor prognosis in colorectal cancer via targeting of SMC4.
Cancer Med. 3:1544–1552. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rokavec M, Öner MG, Li H, Jackstadt R,
Jiang L, Lodygin D, Kaller M, Horst D, Ziegler PK, Schwitalla S, et
al: IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated
colorectal cancer invasion and metastasis. J Clin Invest.
124:1853–1867. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qiu Z, Guo W, Wang Q, Chen Z, Huang S,
Zhao F, Yao M, Zhao Y and He X: MicroRNA-124 reduces the pentose
phosphate pathway and proliferation by targeting PRPS1 and RPIA
mRNAs in human colorectal cancer cells. Gastroenterology.
149:1587–1598, e11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hwang WL, Jiang JK, Yang SH, Huang TS, Lan
HY, Teng HW, Yang CY, Tsai YP, Lin CH, Wang HW and Yang MH:
MicroRNA-146a directs the symmetric division of Snail-dominant
colorectal cancer stem cells. Nat Cell Biol. 16:268–280. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang X, Schulz R, Edmunds S, Krüger E,
Markert E, Gaedcke J, Cormet-Boyaka E, Ghadimi M, Beissbarth T,
Levine AJ, et al: MicroRNA-101 suppresses tumor cell proliferation
by acting as an endogenous proteasome inhibitor via targeting the
proteasome assembly factor POMP. Mol Cell. 59:243–257. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Slaby O, Svoboda M, Michalek J and Vyzula
R: MicroRNAs in colorectal cancer: Translation of molecular biology
into clinical application. Mol Cancer. 8:1022009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Della Vittoria Scarpati G, Calura E, Di
Marino M, Romualdi C, Beltrame L, Malapelle U, Troncone G, De
Stefano A, Pepe S, De Placido S, et al: Analysis of differential
miRNA expression in primary tumor and stroma of colorectal cancer
patients. Biomed Res Int. 2014:8409212014.PubMed/NCBI
|
|
18
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36:D149–D153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yan W, Wu K, Herman JG, Brock MV, Fuks F,
Yang L, Zhu H, Li Y, Yang Y and Guo M: Epigenetic regulation of
DACH1, a novel Wnt signaling component in colorectal cancer.
Epigenetics. 8:1373–1383. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Y, Zhou R, Yuan X, Han N, Zhou S, Xu
H, Guo M, Yu S, Zhang C, Yin T and Wu K: DACH1 is a novel
predictive and prognostic biomarker in hepatocellular carcinoma as
a negative regulator of Wnt/β-catenin signaling. Oncotarget.
6:8621–8634. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Amirkhah R, Schmitz U, Linnebacher M,
Wolkenhauer O and Farazmand A: MicroRNA-mRNA interactions in
colorectal cancer and their role in tumor progression. Genes
Chromosomes Cancer. 54:129–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shen G, Rong X, Zhao J, Yang X, Li H,
Jiang H, Zhou Q, Ji T, Huang S, Zhang J and Jia H: MicroRNA-105
suppresses cell proliferation and inhibits PI3K/AKT signaling in
human hepatocellular carcinoma. Carcinogenesis. 35:2748–2755. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim J, Lim NJ, Jang SG, Kim HK and Lee GK:
miR-592 and miR-552 can distinguish between primary lung
adenocarcinoma and colorectal cancer metastases in the lung.
Anticancer Res. 34:2297–2302. 2014.PubMed/NCBI
|
|
24
|
Xia ZS, Wang L, Yu T, Zhong W, Lian GD, Wu
D, Zhou HM and Chen GC: MiR-5000-3p, miR-5009-3P and miR-552:
Potential microRNA biomarkers of side population cells in colon
cancer. Oncol Rep. 32:589–596. 2014.PubMed/NCBI
|
|
25
|
Atkins M, Jiang Y, Sansores-Garcia L,
Jusiak B, Halder G and Mardon G: Dynamic rewiring of the Drosophila
retinal determination network switches its function from selector
to differentiation. PLoS Genet. 9:e10037312013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang P: Suppression of DACH1 promotes
migration and invasion of colorectal cancer via activating
TGF-β-mediated epithelial-mesenchymal transition. Biochem Biophys
Res Commun. 460:314–319. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chu Q, Han N, Yuan X, Nie X, Wu H, Chen Y,
Guo M, Yu S and Wu K: DACH1 inhibits cyclin D1 expression, cellular
proliferation and tumor growth of renal cancer cells. J Hematol
Oncol. 7:732014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mohammed MK, Shao C, Wang J, Wei Q, Wang
X, Collier Z, Tang S, Liu H, Zhang F, Huang J, et al: Wnt/β-catenin
signaling plays an ever-expanding role in stem cell self-renewal,
tumorigenesis and cancer chemoresistance. Genes Dis. 3:11–40. 2016.
View Article : Google Scholar : PubMed/NCBI
|