Introduction
The high risk of venous thromboembolism (VTE)
occurring in patients with cancer is a worldwide public health
problem, particularly in patients with lymphoma, and leads to high
mortality (1,2). Patients with cancer exhibit an 8-fold
higher risk of mortality compared with patients without cancer in
incidences of acute VTE (3), and
patients with carcinogenic malignancies were more prone to
developing VTE (4). Additionally,
compared to patients with other types of cancer, patients with
lymphoma have been reported exhibit a higher frequency of VTE
(7.7%) (5). The clinical risk factors
for VTE comprise exogenous and endogenous factors: Exogenous
factors include surgery and chemotherapy, immobility, trauma and
hormone use, and endogenous factors include cancer, inherited
elevated risk (family history of VTE) and hypercoagulation
(6).
Lymphoma are a heterogeneous group of lymphoid
disorders that originate in the lymphatic tissues and share a
common characteristic of clonal expansion of malignant lymphocytes
(7). Although numerous types of
cancer eventually spread to parts of the lymphatic system, lymphoma
is different as it actually originates in this area (8,9). A
previous study demonstrated that the greatest risk of VTE occurred
in patients with lymphoma, followed by patients with lung and
gastrointestinal cancer, which suggests that the incidence of VTE
in patients with lymphoma was higher compared with patients with
solid tumors (10). The majority of
patients with lymphoma receive anti-neoplastic therapy and thus
increase their risk of VTE (2). A
previous study suggested that the rate of VTE was 59.5% in patients
with lymphoma, and that it occurred earlier due to the period of
intensive therapy compared with patients with VTE without lymphoma
(11). However, there are few studies
examining procoagulants and the molecular mechanism of VTE in
patients with malignant lymphoma. The risk factors of VTE in
lymphoma patients have not been fully characterized, and the
majority of thromboses occur early in the course of the disease
(12).
The use of anticoagulation may lead to an increased
incidence of bleeding, which may result in thromboprophylaxis.
Analysis of the risk factors of thrombosis may allow appropriate
prophylaxis of VTE occurrence, to protect against the fatal or
detrimental consequences, such as bleeding or mortality (12). Although some risk factors such as the
site of cancer, platelet count, level of hemoglobin, leukocyte
count and body mass index have been proposed, genetic risk factors
should also be investigated (13). To
date, a small number of studies have evaluated congenital
thrombophilia in patients with lymphoma (14,15). A
previous study found that there was an increased risk of venous
thrombosis in patients with cancer and demonstrated that patients
possessing the Factor V Leiden mutation exhibited a 12-fold
increase of thrombosis, whilst patients without this mutation only
had 5-fold increase (10).
However, the molecular pathogenesis of thrombosis in
patients with lymphoma remains unknown. In the present study, the
gene associated with VTE in patients with lymphoma was identified
by bioinformatic approaches, which may provide novel insights for
additional application of thromboprophylaxis.
Materials and methods
Microarray data preprocessing
Microarray data (GSE17078) was downloaded from the
Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) and contained 27
normal blood outgrowth endothelial cell (BOECs) samples, the
control group and 3 BOECs samples of venous thrombosis with Protein
C deficiency, the case group. The affy package of R was used to
normalize the raw data. Probes were switched into gene symbols with
microarray platform of GPL96 [HG-U133A] Affymetrix Human Genome
U133A Array. Probes without gene symbols and expression values were
filtered.
Identification of differentially
expressed genes and functional enrichment analysis
With the cut-offs of adjusted to P<0.05 and an
absolute value of log (fold change) of >1, differentially
expressed genes (DEGs) were identified by the limma package of R.
The following gene ontology (GO) and Kyoto encyclopedia of genes
and genomes (KEGG) pathway analyses were performed by the Database
for Annotation, Visualization and Integrated Discovery (DAVID;
david.abcc.ncifcrf.gov) for annotation,
visualization and integrated discovery. GO described DEGs in terms
of their associated biological processes, cellular components and
molecular functions in a species-independent manner. The KEGG
database is used to display the biochemical pathways of the DEGs of
interest.
Identification of differential
coexpression pairs
DCGL is a package that may serve as an effective
tool for differential coexpression analysis (DCEA). DCEA determines
the change of expression correlation of gene pairs and aids the
investigation of the global transcriptional mechanisms underlying
phenotypic changes (16). In the
present study, differential coexpression pairs were identified by
the DCGL package (version 2.1.2) (16) of R and P<0.05 was used as the
cut-off criterion.
Construction of protein-protein
interaction (PPI) network and gene coexpression network
Therefore, PPI and gene coexpression networks were
constructed by the Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING) database and visualized by Cytoscape
(version 3.4.0).
Results
DEGs
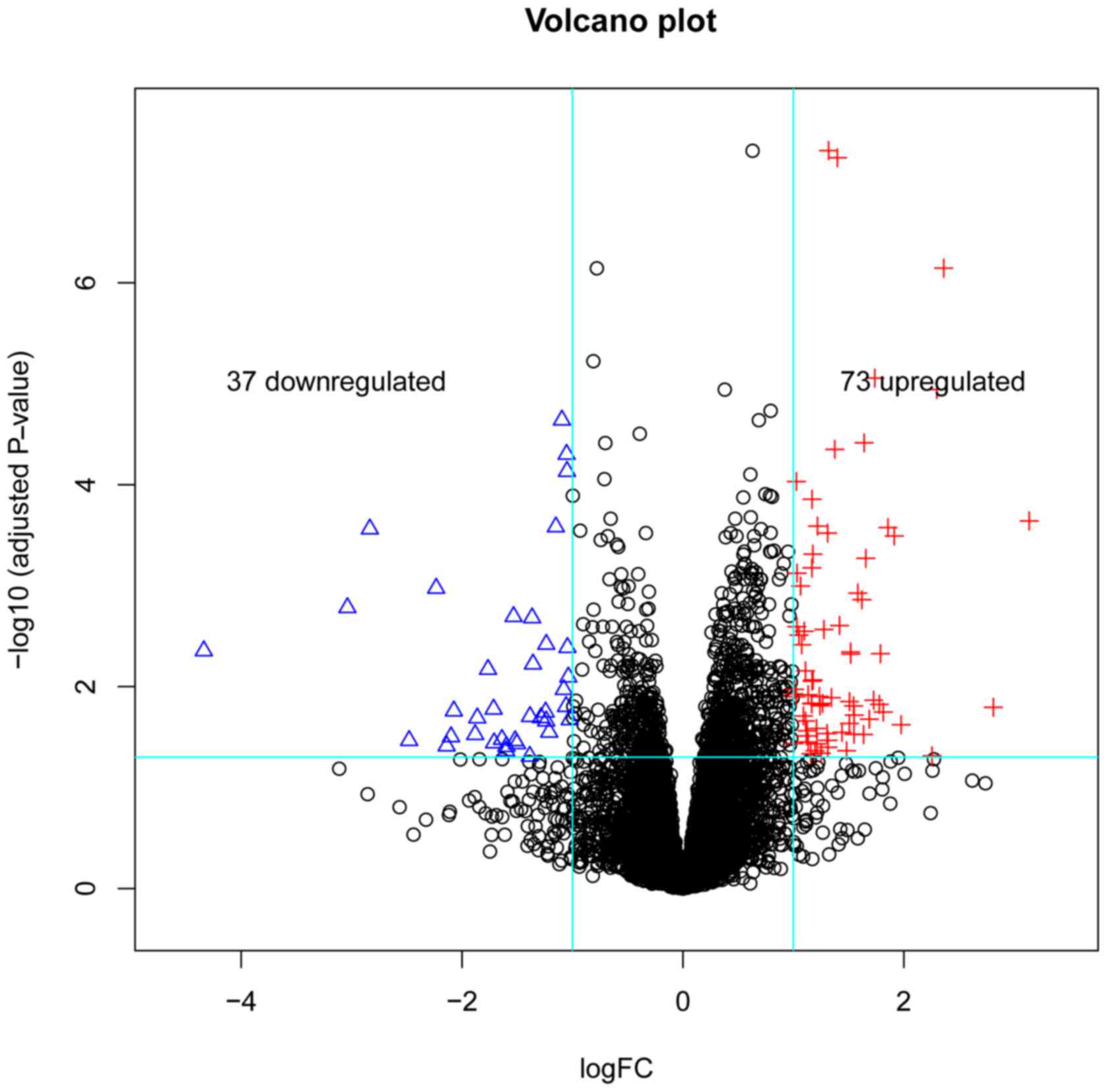
A total of 110 DEGs were obtained, including 73
upregulated and 37 downregulated genes. A volcano plot of DEGs is
demonstrated in Fig. 1. Genes were
arranged along axes of biological and statistical significance in
the volcano plot.
Functional enrichment analysis
GO enrichment analyses illustrated that the DEGs
were enriched in 132 GO terms. A total of 98 GO terms were in
biological processes, 18 terms in molecular function and 16 terms
in cellular component. Additionally, 9 significant KEGG pathways
were identified, including the two most significant types of
pathways: The intestinal immune network for IgA production and the
cytokine-cytokine receptor interaction pathway. The nine KEGG
pathways are listed in Table I.
 | Table I.Significant KEGG pathways. |
Table I.
Significant KEGG pathways.
| Category | Pathway Name | Gene Number | P-value | Genes |
|---|
| KEGG_PATHWAY | Intestinal immune
network for Immunoglobulin A production | 5 | 0.001001303 | HLA-DPA1, HLA-DPB1,
ITGA4, HLA-DMA, CXCL12 |
| KEGG_PATHWAY | Cytokine-cytokine
receptor interaction | 8 | 0.009523061 | IL1R1, CXCL5,
CCL20, IL10RB, CXCL3, KITLG, CXCL6, CXCL12 |
| KEGG_PATHWAY | Viral
myocarditis | 4 | 0.027292882 | RAC2, HLA-DPA1,
HLA-DPB1, HLA-DMA |
| KEGG_PATHWAY | Chemokine signaling
pathway | 6 | 0.028009403 | RAC2, CXCL5, CCL20,
CXCL3, CXCL6, CXCL12 |
| KEGG_PATHWAY | Asthma | 3 | 0.028974901 | HLA-DPA1, HLA-DPB1,
HLA-DMA |
| KEGG_PATHWAY | Cell adhesion
molecules | 5 | 0.032728891 | HLA-DPA1, HLA-DPB1,
ITGA4, SELE, HLA-DMA |
| KEGG_PATHWAY | Antigen processing
and presentation | 4 | 0.040649303 | PDIA3, HLA-DPA1,
HLA-DPB1, HLA-DMA |
| KEGG_PATHWAY | Allograft
rejection | 3 | 0.043169157 | HLA-DPA1, HLA-DPB1,
HLA-DMA |
| KEGG_PATHWAY | Graft-versus-host
disease | 3 | 0.049900633 | HLA-DPA1, HLA-DPB1,
HLA-DMA |
Differential coexpression pairs
A total of 309 differential coexpression pairs were
identified by the DCGL package, which included 166 positive
correlative regulation of coexpression pairs, 80 upregulated and 86
downregulated simultaneously, and 143 negative correlative
regulation of coexpression pairs.
PPI network and gene coexpression
network
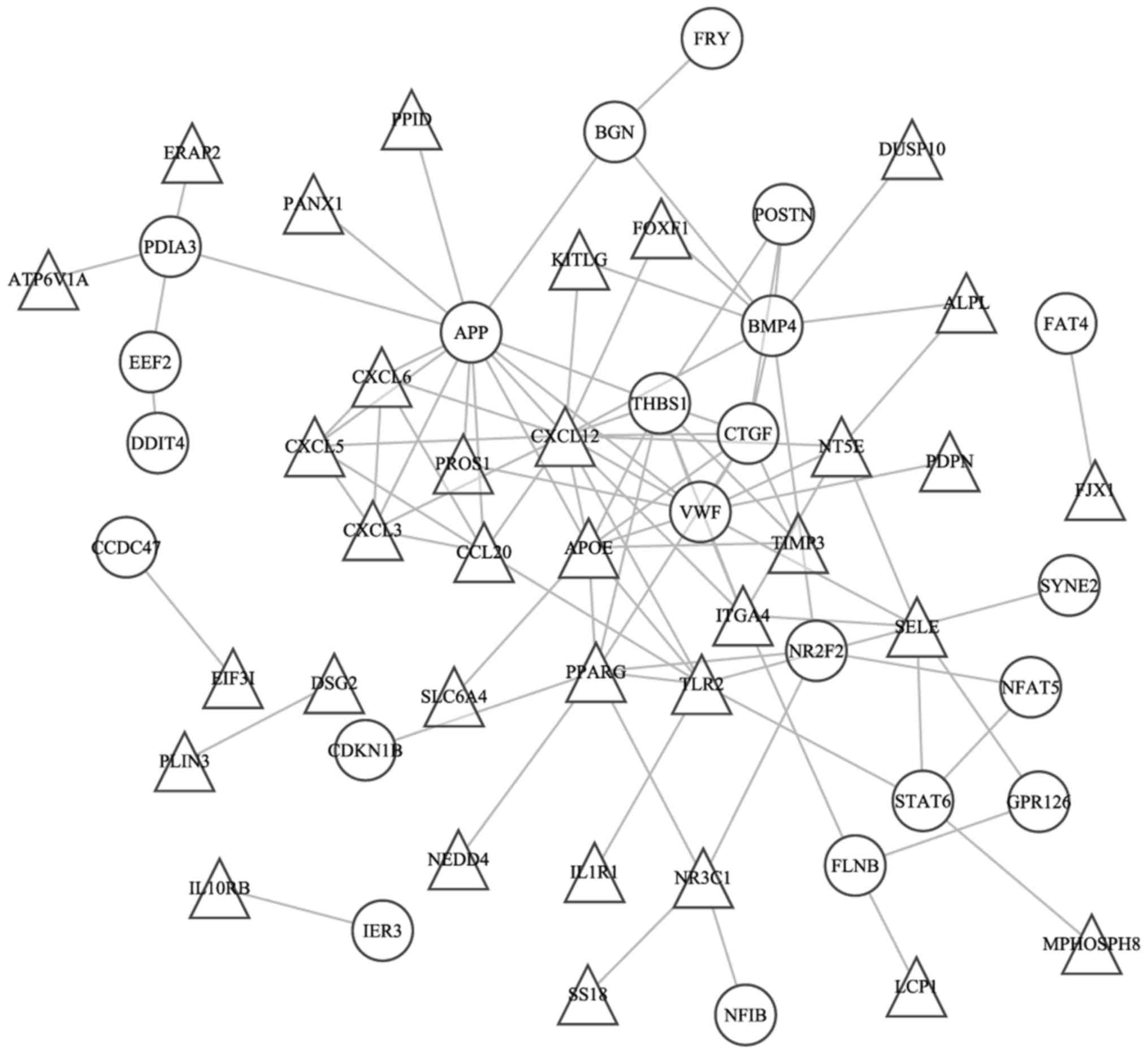
The PPI and coexpression networks were constructed
by STRING, as demonstrated in Fig. 2.
The degree of the node was calculated based on the number of links
of 1 node, which directly contacted with other nodes. The C-x-C
motif chemokine ligand 12 (CXCL12), amyloid β precursor
protein (APP), Vvon Willebrand factor (VWF),
apolipoprotein E (APOE), bone morphogenic protein 4
(BMP4), thrombospondin 1 (THBS1), peroxisome
proliferator activated receptor γ (PPARG) and connective
tissue growth factor genes (CTGF) exhibited high degrees in
PPI network. The coexpression network contained 309 coexpression
pairs and 105 genes (Fig. 3). The
lymphocyte cytosolic protein 1 (LCP1), popeye domain
containing 3 (POPDC3), nuclear receptor subfamily 2 group F
member 2 (NR2F2), Yip1 domain family member 5
(YIPF5), epiregulin (EREC), glutaminyl-peptide
cyclotransferase (QPCT), nBAF chromatin remodeling complex
subunit (SS18), neuron navigator 2 (NAV2), podoplanin
(PDPN), CTGF, nuclear factor I B (NFIB) and perilipin
3 genes (PLIN3) possessed high degrees >10 in this
coexpression network.
Discussion
The occurrence of VTE associated with cancer is a
common complication of patients with malignancies (17). The clinical relevance of the VTE
phenomenon is associated with the presence of cancer itself and
solid tumors are typically considered to present higher risks
(18–20). However, a number of previous studies
have suggested that the high risk of VTE associated with patients
with hematological malignancies such as lymphoma and leukemia may
be comparable to patients with solid tumors (10,21). In
fact, the incidence of VTE in hematological-associated malignancies
exceeded the incidence rate of solid tumors (22,23). A
previous study revealed that the global incidence rate of
thrombosis in patients with lymphoma was >6%, and the majority
of thromboses occurred during the treatment stages of this disease
(12). Therefore, microarray data was
analyzed by bioinformatics to investigate the molecular mechanism
of VTE in patients with lymphoma, which may provide novel insights
for antithrombotic prophylaxis.
In total, two significant KEGG pathways, the
intestinal immune network for IgA production and the
cytokine-cytokine receptor interaction pathway, were identified in
the present study. The intestines are the largest lymphoid tissue
in the body, and serve a critical role in intestinal immunity: A
number of non-inflammatory immunoglobulin A (IgA) antibodies are
produced by the immunological factors in the intestines as the
first line of defense against foreign microorganisms (24). Secreted IgA molecules may promote
immune exclusion via entrapping dietary antigens and microorganisms
in the mucus, and function to neutralize pathogenic microbes and
toxins (25). Cytokines, which are
soluble extracellular proteins or glycoproteins, are crucial
intercellular regulators and mobilizers of cells that are involved
in the innate and adaptive immune responses, cell death, cell
growth, differentiation, angiogenesis and the repair processes
involved in the restoration of homeostasis (26). However, the underlying mechanisms of
complex diseases such as lymphoma and VTE are affected by a number
of environmental and genetic factors (27). Therefore, the PPI and gene
coexpression networks were utilized to illustrate the molecular
basis of lymphoma. It was notable that the PPI network was used to
identify the differences between normal and disease patient groups
and helped to recognize potentially disease-associated gene
candidates (28). The current
computational approach to predicting PPI networks was fast and
useful for selecting potential targets for additional experimental
screening (29). The gene
coexpression network may provide potential specific molecular
mechanisms in disease. It is common to use gene coexpression
networks to recognize highly connected genes, which are usually
associated with diseases (30).
PPI networks usually serve an important role in
identifying novel protein functions (31) and examining the associations between
protein network structure and function (32). Gene expression profiles may describe
the pairwise associations of proteins with a biological network.
Similarly, gene coexpression networks may be used to describe genes
that exhibit similar expression patterns in regulatory processes,
pathways and certain complexes (33).
Gene coexpression network is based on a gene coexpression measure
and used for gene filtering and outcome prediction. The main
feature of these two networks is the degree distribution- the
number of connections a node possesses. Key nodes, such as hubs,
which possess high degrees are highly connected with other genes
and may be potential disease targets (34). The results of the present study
suggest that CTGF possessed the highest degree in the PPI
and coexpression networks. Therefore, it was logical to hypothesize
that CTGF was the target gene of VTE. CTGF, also
termed CCN2, is a member of the CCN family (35). CTGF has been demonstrated to
serve important roles in a number of biological processes such as
cell proliferation, differentiation, adhesion, angiogenesis, tissue
wound repair and fibrotic disease (36). It is a secreted multifunctional
matricellular protein comprising four distinct conserved structural
modules that include a thrombospondin type 1 repeat (TSR), a von
Willebrand type C repeats domain, an amino-terminal insulin-like
growth factor binding domain and a carboxyl-terminal (CT)
cysteine-knot motif (37). These
structural modules of CTGF confer a variety of functions.
For instance, the TSR domain interacts with vascular endothelial
growth factor (38) and the CT domain
binds to transforming growth factor (TGF)-β superfamily (39). In addition, CTGF has been
identified in almost all fibrotic pathologies (40): It has been revealed that CTGF
is upregulated during the pathogenesis of several fibrotic diseases
such as kidney and liver fibrosis, scleroderma and atherosclerosis
(41). It was also indicated that
CTGF may induce sustained fibrosis during interaction with
TGF-β (42) and intensify the
production of the extracellular matrix, which is associated with
other fibrosis-inducing conditions (40). Additionally, fibrosis, cardiovascular
diseases (41) and numerous types of
malignancy (43) were associated with
aberrant CTGF expression. In pancreatic cancer,
CTGF-specific antibodies diminished tumor growth and
metastases (44). However, in
hematological malignancies, overexpressed CTGF was
associated with poor outcome in patients with precursor-B acute
lymphoblastic leukemia (45).
Evidence suggests that the tumor type, the stage of
the cancer and treatment with antineoplastic agents may contribute
to the absolute risk of VTE development (46). Furthermore, age, immobilization,
surgery and other comorbidities will also affect the overall
likelihood of thrombotic complications, and even affect patients
without cancer. At present, the mechanisms of hereditary
thrombophilia in patients with cancer and thrombosis remain
unclear. Additional studies will assist to improve the prophylactic
and treatment strategies for VTE in these complex patients.
In conclusion, the present study screened important
genes, which may reveal the molecular mechanisms of VTE in patients
with lymphoma. These results may help to define the lymphoma
populations at high thrombotic risk, who require the development of
effective prophylactic treatments. However, confirmation of these
findings and additional studies investigating lymphoma are
required.
Acknowledgements
The present study was supported by the Health Bureau
Science and Technology Foundation of Tianjin (grant no., 2014KZ102)
and the Municipal Science and Technology Commission of Tianjin
(grant no. 15ZLZLZF00440 and 16ZLZXZF00120).
References
|
1
|
Khorana AA, Francis CW, Culakova E,
Kuderer NM and Lyman GH: Frequency, risk factors, and trends for
venous thromboembolism among hospitalized cancer patients. Cancer.
110:2339–2346. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wun T and White RH: Venous thromboembolism
in patients with acute leukemia, lymphoma and multiple myeloma.
Thromb Res. 125:(Suppl 2). S96–S102. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou X, Teegala S, Huen A, Ji Y, Fayad L,
Hagemeister FB, Gladish G and Vadhan-Raj S: Incidence and risk
factors of venous thromboembolic events in lymphoma. Am J Med.
123:935–941. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Khalil J, Bensaid B, Elkacemi H, Afif M,
Bensaid Y, Kebdani T and Benjaafar N: Venous thromboembolism in
cancer patients: An underestimated major health problem. World J
Surg Oncol. 13:2042015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mohren M, Markmann I, Jentsch-Ullrich K,
Koenigsmann M, Lutze G and Franke A: Increased risk of
thromboembolism in patients with malignant lymphoma: A
single-centre analysis. Br J Cancer. 92:1349–1351. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cushman M: Epidemiology and risk factors
for venous thrombosis. Semin Hematol. 44:62–69. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zheng RL, Jiang YJ and Wang X: Role of
microRNAs on therapy resistance in Non-Hodgkin's lymphoma. Int J
Clin Exp Med. 7:3818–3832. 2014.PubMed/NCBI
|
|
8
|
Benson N, Whipple M and Kalet IJ: A Markov
model approach to predicting regional tumor spread in the lymphatic
system of the head and neck. AMIA Annu Symp Proc. 2006:31–35.
2006.
|
|
9
|
Kewitz S, Kurch L, Volkmer I and Staege
MS: Stimulation of the hypoxia pathway modulates chemotherapy
resistance in Hodgkin's lymphoma cells. Tumor Biol. 37:8229–8237.
2016. View Article : Google Scholar
|
|
10
|
Blom JW, Doggen CJ, Osanto S and Rosendaal
FR: Malignancies, prothrombotic mutations, and the risk of venous
thrombosis. JAMA. 293:715–722. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Goldschmidt N, Linetsky E, Shalom E, Varon
D and Siegal T: High incidence of thromboembolism in patients with
central nervous system lymphoma. Cancer. 98:1239–1242. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Caruso V, Di Castelnuovo A, Meschengieser
S, Lazzari MA, de Gaetano G, Storti S, Iacoviello L and Donati MB:
Thrombotic complications in adult patients with lymphoma: A
meta-analysis of 29 independent cohorts including 18 018 patients
and 1149 events. Blood. 115:5322–5328. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ikeda M, Kan-No H, Hayashi M, Tsukada H,
Shida M, Hirasawa T, Muramatsu T, Ogushi Y and Mikami M: Predicting
perioperative venous thromboembolism in Japanese gynecological
patients. PLoS One. 9:e892062014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Boersma RS, Hamulyak K, Cate HT and
Schouten HC: Congenital thrombophilia and central venous
catheter-related thrombosis in patients with cancer. Clin Appl
Thromb/Hemost. 16:643–649. 2010. View Article : Google Scholar
|
|
15
|
Achkar A, Horellou MH, Leclercq X, Lambert
Y, Conard J, Laaban JP and Samama MM: PO73 prevalence of cancer and
congenital thrombophilia in 435 patients with acute venous
thromboembolism (VTE). Thromb Res. 120:(Suppl 2). S1682007.
View Article : Google Scholar
|
|
16
|
Liu BH, Yu H, Tu K, Li C, Li YX and Li YY:
DCGL: An R package for identifying differentially coexpressed genes
and links from gene expression microarray data. Bioinformatics.
26:2637–2638. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Khorana AA, Kuderer NM, Culakova E, Lyman
GH and Francis CW: Development and validation of a predictive model
for chemotherapy-associated thrombosis. Blood. 111:4902–4907. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Korte W: Cancer and thrombosis: An
increasingly important association. Support Care Cancer.
16:223–228. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rodriguez AO, Wun T, Chew H, Zhou H,
Harvey D and White RH: Venous thromboembolism in ovarian cancer.
Gynecol Oncol. 105:784–790. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Semrad TJ, O'Donnell R, Wun T, Chew H,
Harvey D, Zhou H and White RH: Epidemiology of venous
thromboembolism in 9489 patients with malignant glioma. J Neurosur.
106:601–608. 2007. View Article : Google Scholar
|
|
21
|
Franchini M: Thromboembolic risk in
hematological malignancies. Clin Chem Lab Med. 53:1139–1147. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Falanga A, Marchetti M and Russo L: Venous
thromboembolism in the hematologic malignancies. Curr Opin Oncol.
24:702–710. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Elice F and Rodeghiero F: Hematologic
malignancies and thrombosis. Thromb Res. 129:360–366. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kunisawa J and Kiyono H: Alcaligenes is
commensal bacteria habituating in the gut-associated lymphoid
tissue for the regulation of intestinal IgA responses. Front
Immunol. 3:652012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Avula LR, Knapen D, Buckinx R, Vergauwen
L, Adriaensen D, Van Nassauw L and Timmermans JP: Whole-genome
microarray analysis and functional characterization reveal distinct
gene expression profiles and patterns in two mouse models of ileal
inflammation. BMC Genomics. 13:3772012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang L and Wu X: Cytokines of defender
against cell death 1 and allograft inflammatory factor-1 with
regard to innate immunity of fish. J Fisher Sci Chin. 18:237–242.
2013. View Article : Google Scholar
|
|
27
|
Ghasemi S, Tavakoli A, Moghadam M, Zargar
MA, Abbaspour M, Hatamnejadian N and Ebrahimi A: Risk of prostate
cancer and thrombosis-related factor polymorphisms. Biomed Rep.
2:53–56. 2014.PubMed/NCBI
|
|
28
|
Sarajlić A, Janjić V, Stojković N, Radak D
and Pržulj N: Network topology reveals key cardiovascular disease
genes. PLoS One. 8:e715372013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
von Mering C, Krause R, Snel B, Cornell M,
Oliver SG, Fields S and Bork P: Comparative assessment of
large-scale data sets of protein-protein interactions. Nature.
417:399–403. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gaiteri C, Ding Y, French B, Tseng GC and
Sibille E: Beyond modules and hubs: The potential of gene
coexpression networks for investigating molecular mechanisms of
complex brain disorders. Genes Brain Behav. 13:13–24. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sharan R, Ulitsky I and Shamir R:
Network-based prediction of protein function. Mol Syst Biol.
3:882007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yook SH, Oltvai ZN and Barabási AL:
Functional and topological characterization of protein interaction
networks. Proteomics. 4:928–942. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Horvath S and Dong J: Geometric
interpretation of gene coexpression network analysis. PLoS Comput
Biol. 4:e10001172008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Safari-Alighiarloo N, Taghizadeh M,
Rezaei-Tavirani M, Goliaei B and Peyvandi AA: Protein-protein
interaction networks (PPI) and complex diseases. Gastroenterol
Hepatol Bed Bench. 7:17–31. 2014.PubMed/NCBI
|
|
35
|
Jun JI and Lau LF: Taking aim at the
extracellular matrix: CCN proteins as emerging therapeutic targets.
Nat Rev Drug Discov. 10:945–963. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hall-Glenn F and Lyons KM: Roles for CCN2
in normal physiological processes. Cell Mol Life Sci. 68:3209–3217.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
de Winter P, Leoni P and Abraham D:
Connective tissue growth factor: Structure-function relationships
of a mosaic, multifunctional protein. Growth Factors. 26:80–91.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yin D, Chen W, O'Kelly J, Lu D, Ham M,
Doan NB, Xie D, Wang C, Vadgama J, Said JW, et al: Connective
tissue growth factor associated with oncogenic activities and drug
resistance in glioblastoma multiforme. Int J Cancer. 127:2257–2267.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Leask A and Abraham DJ: All in the CCN
family: Essential matricellular signaling modulators emerge from
the bunker. J Cell Sci. 119:4803–4810. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Brigstock DR: Connective tissue growth
factor (CCN2, CTGF) and organ fibrosis: Lessons from transgenic
animals. J Cell Commun Signal. 4:1–4. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Koshman YE, Patel N, Chu M, Iyengar R, Kim
T, Ersahin C, Lewis W, Heroux A and Samarel AM: Regulation of
connective tissue growth factor gene expression and fibrosis in
human heart failure. J Card Fail. 19:283–294. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mori T, Kawara S, Shinozaki M, Hayashi N,
Kakinuma T, Igarashi A, Takigawa M, Nakanishi T and Takehara K:
Role and interaction of connective tissue growth factor with
transforming growth factor-beta in persistent fibrosis: A mouse
fibrosis model. J Cell Physiol. 181:153–159. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Charrier A and Brigstock DR: Regulation of
pancreatic function by connective tissue growth factor (CTGF,
CCN2). Cytokine Growth Factor Rev. 24:59–68. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Aikawa T, Gunn J, Spong SM, Klaus SJ and
Korc M: Connective tissue growth factor-specific antibody
attenuates tumor growth, metastasis, and angiogenesis in an
orthotopic mouse model of pancreatic cancer. Mol Cancer Ther.
5:1108–1116. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sala-Torra O, Gundacker HM, Stirewalt DL,
Ladne PA, Pogosova-Agadjanyan EL, Slovak ML, Willman CL, Heimfeld
S, Boldt DH and Radich JP: Connective tissue growth factor (CTGF)
expression and outcome in adult patients with acute lymphoblastic
leukemia. Blood. 109:3080–3083. 2007.PubMed/NCBI
|
|
46
|
Lee AY and Levine MN: Venous
thromboembolism and cancer: Risks and outcomes. Circulation.
107:(23 Suppl 1). I17–I21. 2003. View Article : Google Scholar : PubMed/NCBI
|