Introduction
Histone deacetylases (HDACs) was the name originally
given to a family of proteins responsible for deacetylation of
histone proteins, which were later shown to be also involved in the
deacetylation of non-histone proteins (1,2). HDACs are
divided into four classes based on structure: Class I including
HDAC1, HDAC2, HDAC3 and HDAC8; class II, which is further divided
into class IIa (HDAC4, HDAC5, HDAC7 and HDAC9) and class IIb (HDAC6
and HDAC10); class III comprising SIRT1 to SIRT7; and class IV,
consisting of only HDAC11. While HDAC6 is a well-investigated class
II HDAC, little is known about HDAC10.
HDAC10 has been reported to be involved in
homologous recombination (3),
melanogenesis (4), cells autophagy
(5–7),
cell cycle regulation (8), DNA
mismatch repair (9) and cancer
progression (10–16). While HDAC10 was reported to suppresses
the proliferation and invasion of clear cell renal cell carcinoma
(13), it was also demonstrated to
promote cell proliferation via AKT phosphorylation in lung cancer
(15). Based on these contradictory
observations, we hypothesized that the function of HDAC10 in cancer
is more complex and may be dependent on the type of tissue.
Colorectal carcinoma is among the five most commonly
diagnosed cancers accounting for over 50% of the top five cancers
all cases in China (17). Owing to
the high risk of relapse and metastasis, the treatment for advanced
colon cancer poses a significant challenge. Thus, it is crucial to
discover new and competent therapeutic targets for colon cancer,
thereby enabling the discovery of new diagnostic and therapeutic
drugs.
To date, the expression of HDAC10, especially the
prognostic role and its association with clinicopathological
features in colon cancer has not been investigated. In this study
we analyzed 100 colon cancer specimens in a tissue microarray (TMA)
to assess HDAC10 expression and to evaluate the clinical
significance of HDAC10 in colon cancer.
Patients and methods
Patients
A total of 100 colon cancer patients (54 male, 45
female and one missed gender information) aged between 24 and 90
were recruited in this study. All patients underwent surgery during
July 2006 to May 2007 and received no prior extra therapy. Of
these, 6 were cTNM stage I, 54 were cTNM stage II, 35 were cTNM
stage III, and 3 were cTNM stage IV according to AJCC. Following
surgery, a long-term follow-up was implemented for all patients up
to July 2015. During the follow-up time, 61 patients died of colon
carcinoma with a median overall survival time of 26 months.
Detailed patient information is listed in Table I.
 | Table I.Clinical parameters of colon carcinoma
patients. |
Table I.
Clinical parameters of colon carcinoma
patients.
| Clinical factors | No. of patients |
|---|
| Gender |
|
| Male | 54 |
|
Female | 45 |
| Age |
|
| ≤60 | 21 |
|
>60 | 73 |
| Tumor size |
|
| ≤5
cm | 55 |
| >5
cm | 43 |
| Pathological
grade |
|
| 1 | 2 |
| 2 | 48 |
| 3 | 50 |
| T stage |
|
|
T1+T2 | 7 |
|
T3+T4 | 89 |
| N stage |
|
| N0 | 60 |
| N1 | 27 |
| N2 | 11 |
| M stage |
|
| M0 | 97 |
| M1 | 3 |
| cTNM stage |
|
| 1 | 6 |
| 2 | 54 |
| 3 | 35 |
| 4 | 3 |
| Tumor location |
|
|
Right | 61 |
| Left | 38 |
Immunohistochemistry
Colon adenocarcinoma TMA (HColA180Su09) containing
100 tumor tissues and 80 paired adjacent tissues was obtained from
Shanghai Outdo Biotech Co., Ltd. (Shanghai, China) for
standardization. Deparaffinization of the TMA was performed by
xylene and graded alcohol following incubation at high temperature
for an hour. After antigen retrieval by EDTA and blocking with goat
serum, the TMA was incubated with the primary anti-HDAC10 antibody
(24913-1-AP) obtained from ProteinTech Group, Inc. (Chicago, IL,
USA) at a dilution of 1:2,500 at 4°C overnight, and subsequently
incubated with horse radish peroxidase (HRP) labeled
secondary-antibody (K8000; Dako; Agilent Technologies, Inc., Santa
Clara, CA, USA) for 30 min. Diaminobenzidine (DAB) and hematoxylin
redyeing were performed for visualization. Three random fields
having more than 100 cells were visually analyzed and scored by
pathologists. The colon cancer patients were divided into three
subgroups based on differences in HDAC10 expression as follows:
0–60%, low expression; 61–90%, median expression; 91–100%, high
expression. The expression of mutL homolog 1 (MLH1) (1:5,000,
sc-56160; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), mutS
homolog (MSH)2 (1:100, sc-56163; Santa Cruz Biotechnology, Inc.),
MSH6 (1:5,000, 66172-1-Ig; ProteinTech Group, Inc.) and PMS2
(1:1,500, sc-618; Santa Cruz Biotechnology, Inc.) was also detected
in these patients with the same protocol. The specificity of the
anti-HDAC10 antibody used in the present study was validated by
Wang et al in a previous study (18).
Statistical analysis
The difference in HDAC10 expression between colon
adenocarcinoma and adjacent tissues was evaluated by paired t-test.
Spearman's rank correlation coefficient and two-tailed test were
performed to evaluate the correlation between HDAC10 expression and
clinical parameters. Pearson analysis was performed to assess the
association between HDAC10 and MLH1/MSH2/MSH6/PMS2 expression.
Based on HDAC10 and clinical parameters, overall survival curves
were drawn according to the Kaplan-Meier method and log-rank test.
Subsequently, COX multivariate regression survival analysis was
performed to determine the independent prognostic marker. All
statistical analyses were conducted using SPSS 17.0 software (SPSS,
Inc., Chicago, IL, USA), with P<0.05 being considered
significant.
Results
Evaluation of HDAC10 mRNA expression
in colon carcinoma
The HDAC10 mRNA expression level in colon
adenocarcinoma was investigated in the Oncomine database. As
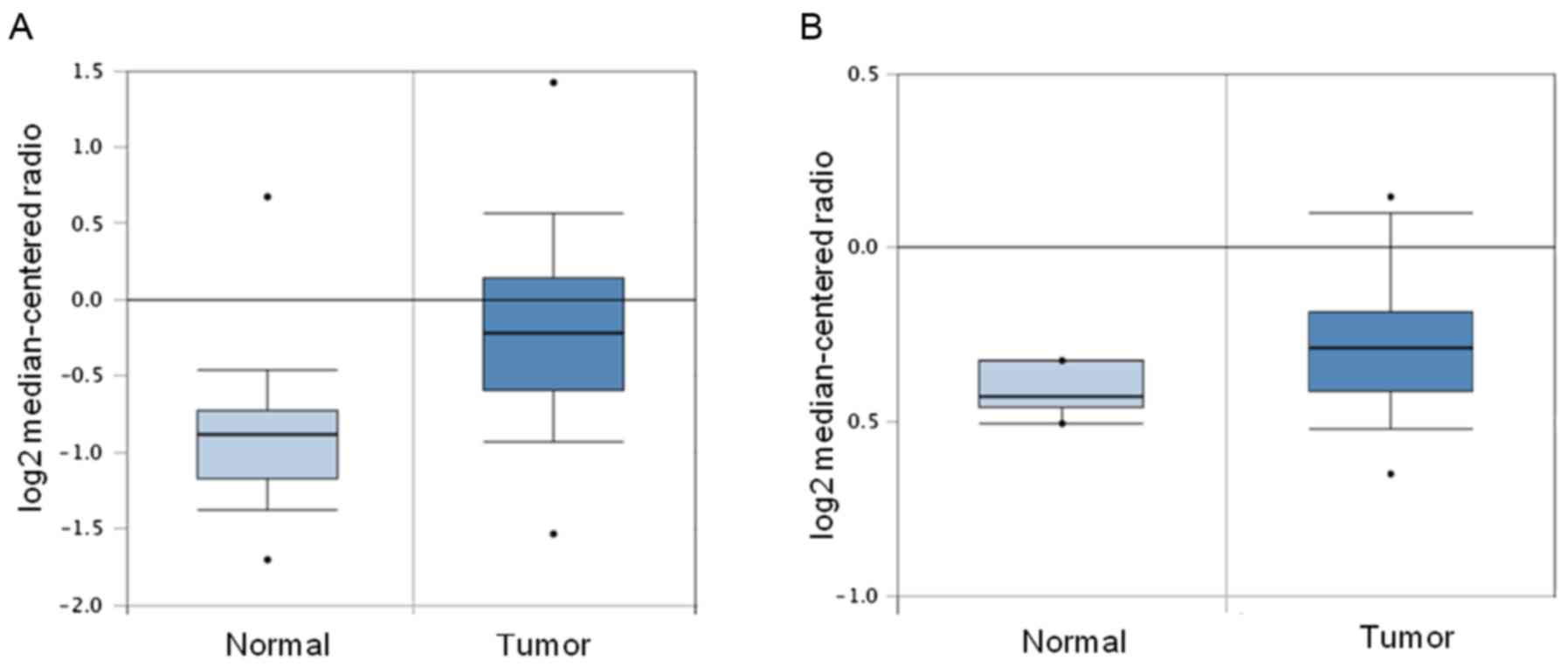
depicted in Fig. 1, HDAC10 mRNA
expression in colon adenocarcinoma tissues was found to be
significantly upregulated both in the Kaiser colon statistics
involving 41 colon carcinoma tissues and 5 normal tissues (fold
change=1.103, P<0.05; Fig. 1A) as
well as in the TCGA colorectal database containing 101 colon
adenocarcinoma tissues and 19 normal colon tissues (fold
change=1.656, P=1.07e-7; Fig.
1B).
HDAC10 expression is high in colon
carcinoma
To investigate HDAC10 expression in colon
adenocarcinoma, two-step immunohistochemistry was performed on the
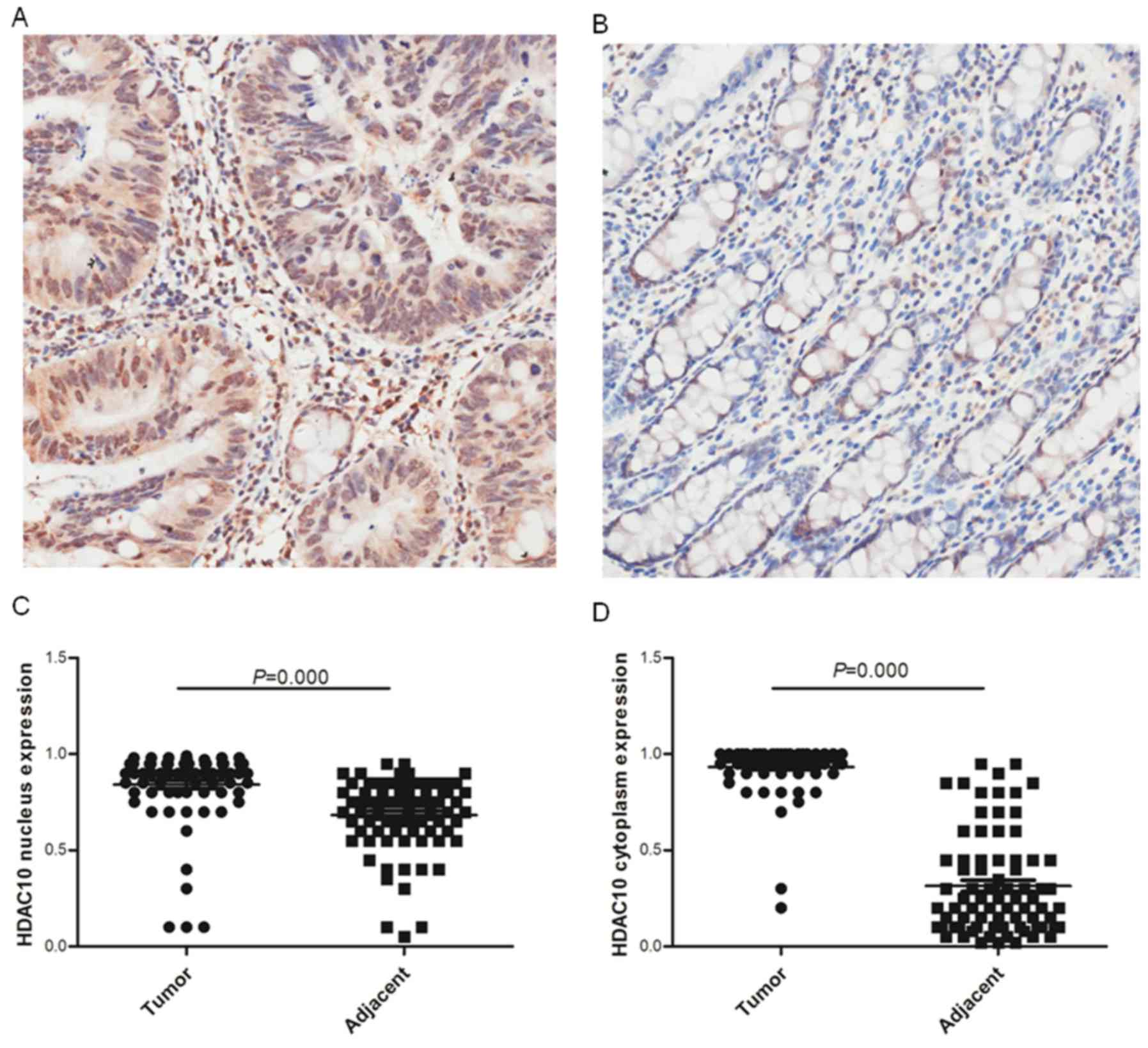
TMA. As shown in Fig. 2,
constitutively high HDAC10 expression was visualized both in the
cytoplasm and nucleus in most colon cancer tissues when compared
with the adjacent normal tissues. Subsequently, HDAC10 expression
in colon cancer was systemically investigated by statistical
analysis with the adjacent normal tissue as a control (cytoplasm,
93.12±12.98 vs. 31.65±26.50%; Fig.
2C; nucleus, 84.16±19.23 vs. 68.64±19.00%; Fig. 2D). The results indicated significantly
elevated expression of HDAC10 protein at the tissue level.
Regardless of the location, HDAC10 expression did not show a
significant correlation with the tumor tissue when compared with
the adjacent normal tissue (Table I).
Interestingly, HDAC10 expression in the nucleus was associated with
its cytoplasmic expression, both in the tumor tissue as well as in
the adjacent normal tissue (Table
II).
 | Table II.Correlation between HDAC10 expression
in tumor and para-carcinoma tissues. |
Table II.
Correlation between HDAC10 expression
in tumor and para-carcinoma tissues.
|
|
| Cytoplasm | Nucleus |
|---|
|
|
|
|
|
|---|
| Location | Tissue | Tumor | Para-carcinoma | Tumor | Para-carcinoma |
|---|
| Cytoplasm | Tumor |
|
|
|
|
|
| Pearson
correlation | 1 | 0.100 | 0.227a | −0.107 |
|
| Sig.
(2-tailed) |
| 0.387 | 0.024 | 0.356 |
|
|
Number | 98 | 77 | 98 | 77 |
|
| Para-carcinoma |
|
|
|
|
|
| Pearson
correlation | 0.100 | 1 | −0.094 | 0.333b |
|
| Sig.
(2-tailed) | 0.387 |
| 0.416 | 0.003 |
|
|
Number | 77 | 79 | 77 | 79 |
| Nucleus | Tumor |
|
|
|
|
|
| Pearson
correlation | 0.227a | −0.094 | 1 | 0.037 |
|
| Sig.
(2-tailed) | 0.024 | 0.416 |
| 0.747 |
|
|
Number | 98 | 77 | 98 | 77 |
|
| Para-carcinoma |
|
|
|
|
|
| Pearson
correlation | −0.107 | 0.333b | 0.037 | 1 |
|
| Sig.
(2-tailed) | 0.356 | 0.003 | 0.747 |
|
|
|
Number | 77 | 79 | 77 | 79 |
Correlation between HDAC10 expression
and clinical parameters
HDAC10 expression in colon adenocarcinoma tissues
showed no significant correlation with any clinicopathological
factors, apart from a link between cytoplasmic HDAC expression and
gender (r=0.265, P<0.05). Intriguingly however, HDAC10
expression in para-carcinoma tissues was highly associated with
clinicopathological factors. Cytoplasmic HDAC10 expression was
found to be positively correlated with lymph node metastasis (N
stage, r=0.256, P<0.05) and distant metastasis (M stage,
r=0.331, P<0.05). Detailed results of the correlation analysis
are listed in Table III.
 | Table III.Association between HDAC10 and
clinical parameters in colon adenocarcinoma. |
Table III.
Association between HDAC10 and
clinical parameters in colon adenocarcinoma.
| HDAC10
expression |
|
|
|
|
|
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| Location | Tissue | Gender | Age | Tumor size | Pathological
grade | T | N | M | cTNM | Tumor location |
|---|
| Cytoplasm | Tumor |
|
|
|
|
|
|
|
|
|
|
|
Correlation | 0.194 | 0.108 | 0.025 | −0.181 | −0.030 | 0.146 | 0.087 | 0.101 | 0.001 |
|
|
Coefficient |
|
|
|
|
|
|
|
|
|
|
| Sig.
(2-tailed) | 0.057 | 0.305 | 0.811 | 0.074 | 0.773 | 0.157 | 0.395 | 0.326 | 0.996 |
|
|
Number | 97 | 92 | 96 | 98 | 94 | 96 | 98 | 96 | 97 |
|
| Para-carcinoma |
|
|
|
|
|
|
|
|
|
|
|
Correlation | 0.179 | 0.133 | −0.015 | −0.095 | −0.107 | 0.256a | 0.331b | 0.180 | −0.085 |
|
|
coefficient |
|
|
|
|
|
|
|
|
|
|
| Sig.
(2-tailed) | 0.118 | 0.259 | 0.899 | 0.403 | 0.356 | 0.024 | 0.003 | 0.115 | 0.461 |
|
|
Number | 78 | 74 | 78 | 79 | 77 | 78 | 79 | 78 | 78 |
| Nucleus | Tumor |
|
|
|
|
|
|
|
|
|
|
|
Correlation | 0.265b | 0.100 | 0.199 | −0.007 | 0.059 | 0.073 | −0.082 | 0.065 | −0.046 |
|
|
Coefficient |
|
|
|
|
|
|
|
|
|
|
| Sig.
(2-tailed) | 0.009 | 0.344 | 0.052 | 0.946 | 0.571 | 0.477 | 0.421 | 0.529 | 0.655 |
|
|
Number | 97 | 92 | 96 | 98 | 94 | 96 | 98 | 96 | 97 |
|
| Para-carcinoma |
|
|
|
|
|
|
|
|
|
|
|
Correlation | 0.119 | −0.067 | 0.045 | 0.119 | −0.090 | 0.207 | 0.075 | 0.122 | 0.037 |
|
|
Coefficient |
|
|
|
|
|
|
|
|
|
|
| Sig.
(2-tailed) | 0.301 | 0.569 | 0.694 | 0.298 | 0.438 | 0.069 | 0.510 | 0.287 | 0.746 |
|
|
Number | 78 | 74 | 78 | 79 | 77 | 78 | 79 | 78 | 78 |
Different prognostic role of HDAC10 in
colon carcinoma and para-carcinoma
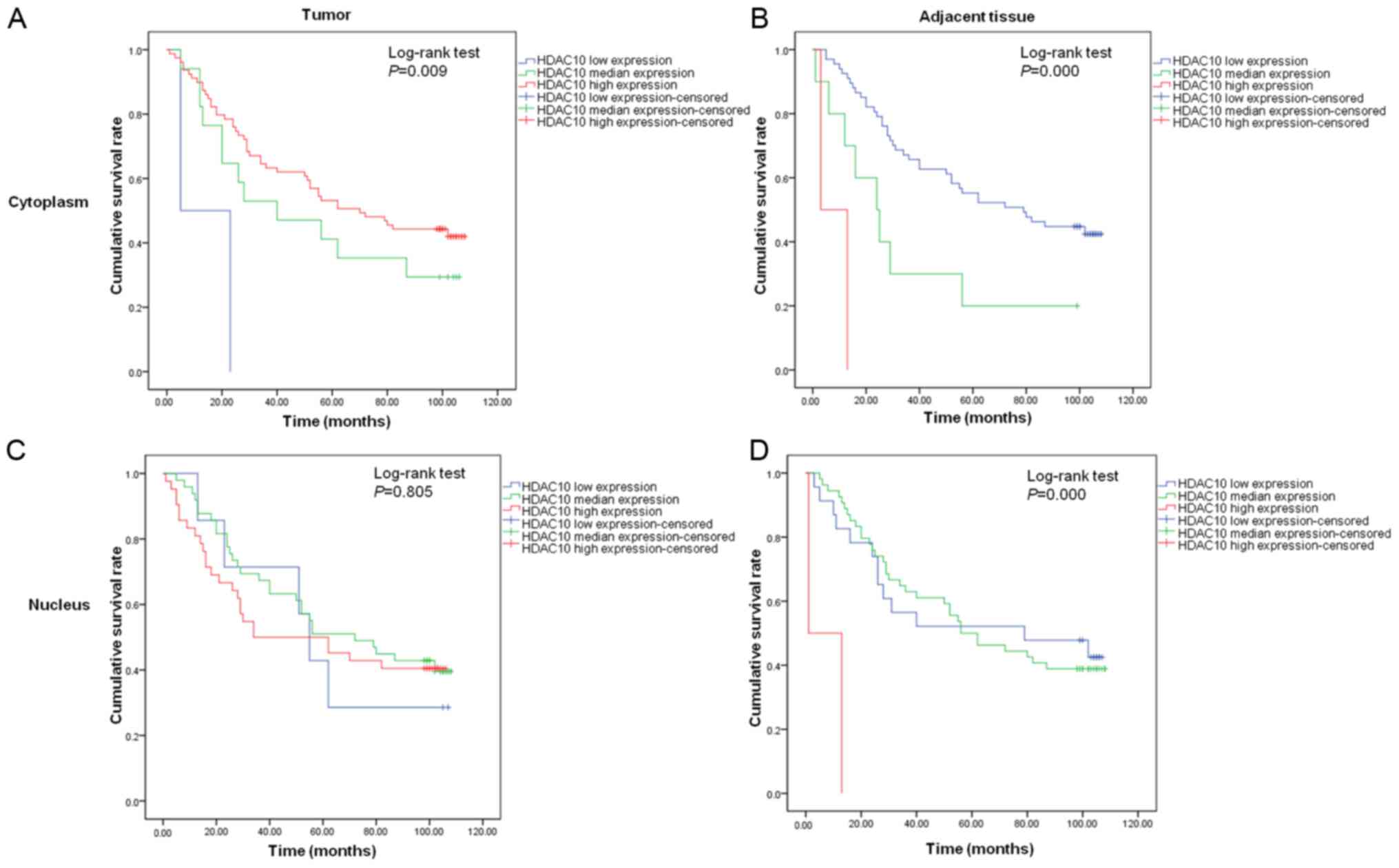
High cytoplasmic expression of HDAC10 in tumor
tissues predicted good prognosis in colon cancer patients, with 0%
survival in the population with low HDAC10 expression after 8 years
of follow-up, in contrast with 29.4% for population with median
expression and 43.0% for population with high expression (Fig. 3A). Conversely, cytoplasmic HDAC10
expression in para-carcinoma tissues was associated with poor
outcome of patients (43.3 vs. 20.0 vs. 0%, P<0.001; Fig. 3B). Nuclear HDAC10 expression in the
tumor tissues did not correlate with overall increased survival of
colon cancer patients (28.6 vs. 40.8 vs. 40.5%, P>0.05; Fig. 3C), while high nuclear HDAC10
expression in the para-carcinoma tissues was correlated with
increased survival rate (43.5 vs. 38.9 vs. 0%, P<0.001; Fig. 3D). Furthermore, regional lymph node
metastasis (N stage, 51.7 vs. 25.9 vs. 9.1%, P=0.000), distant
metastasis (M stage, 40.2 vs. 0.0%, P<0.001), tumor location
(right vs. left, 31.1 vs. 52.6%, P<0.05) and clinical stage
(cTNM, 66.7 vs. 50.0 vs. 22.9 vs. 0.0%, P<0.001) were all
correlated with overall survival time. Subsequent multivariate
analysis indicated that only cytoplasmic HDAC10 expression was an
independent prognostic marker for colon cancer (Table IV).
 | Table IV.Multivariate analysis of factors
associated with survival in colon carcinoma. |
Table IV.
Multivariate analysis of factors
associated with survival in colon carcinoma.
|
|
|
| 95.0% CI for Exp
(B) |
|---|
|
|
|
|
|
|---|
| Factors | Sig. | Exp (B) | Lower | Upper |
|---|
| N stage | 0.928 | 0.954 | 0.343 | 2.657 |
| M stage | 0.986 | 0.981 | 0.109 | 8.845 |
| cTNM stage | 0.120 | 2.378 | 0.798 | 7.086 |
| Tumor location
(right vs. left) | 0.469 | 0.788 | 0.413 | 1.503 |
| Cytoplasmic HDAC10
in tumor | 0.093 | 0.539 | 0.262 | 1.109 |
| Cytoplasmic HDAC10
in para-carcinoma | 0.020 | 2.896 | 1.181 | 7.105 |
| Nuclear HD10 in
para-carcinoma | 0.974 | 0.988 | 0.488 | 2.000 |
HDAC10 may be associated with DNA
mismatch repair
The implication of HDAC10 in DNA repair pathway via
interaction with DNA mismatch repair gene MSH2 in HeLa cells
prompted us to explore the possibility that HDAC10 is involved in
the progression of colon cancer via its interaction with the DNA
mismatch repair genes (9). Four major
DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 were
investigated by immunohistochemistry. Pearson analysis was
performed to evaluate the their association with HDAC10. The
results, listed in Table V, predicted
that cytoplasmic HDAC10 expression in the tumor tissue was not
associated with any of the four DNA mismatch repair genes. Instead
it showed negative association with MLH1 (r=−0.244, P<0.05),
MSH2 (r=−0.410, P<0.001) and MSH6 (r=−0.240, P<0.05) in the
para-carcinoma tissues. Similarly, nuclear HDAC10 expression was
negatively correlated with MLH1 expression (r=−0.288,
P<0.05).
 | Table V.Association between HDAC10 expression
and expression of DNA mismatch repair genes in colon cancer. |
Table V.
Association between HDAC10 expression
and expression of DNA mismatch repair genes in colon cancer.
| HDAC10
expression |
|
|
|
|
|---|
|
|
|
|
|
|---|
| Location | Tissue | MLH1 | MSH2 | MSH6 | PMS2 |
|---|
| Cytoplasm | Tumor |
|
|
|
|
|
| Pearson
correlation | −0.072 | −0.208 | −0.071 | 0.158 |
|
| Sig.
(2-tailed) | 0.530 | 0.068 | 0.526 | 0.165 |
|
|
Number | 79 | 78 | 81 | 79 |
|
| Para-carcinoma |
|
|
|
|
|
| Pearson
correlation | −0.244a | −0.410b | −0.240a | −0.066 |
|
| Sig.
(2-tailed) | 0.038 | 0.000 | 0.040 | 0.580 |
|
|
Number | 72 | 71 | 73 | 72 |
| Nucleus | Tumor |
|
|
|
|
|
| Pearson
correlation | 0.164 | 0.118 | 0.230a | 0.297b |
|
| Sig.
(2-tailed) | 0.148 | 0.302 | 0.039 | 0.008 |
|
|
Number | 79 | 78 | 81 | 79 |
|
| Para-carcinoma |
|
|
|
|
|
| Pearson
correlation | −0.288a | −0.126 | −0.097 | −0.031 |
|
| Sig.
(2-tailed) | 0.014 | 0.295 | 0.413 | 0.798 |
|
|
Number | 72 | 71 | 73 | 72 |
Discussion
To the best of our knowledge, our findings highlight
for the first time, the clinical significance of HDAC10 in colon
cancer. HDAC10 was demonstrated to be a tumor suppresser in some
types of cancer, including clear cell renal cell carcinoma
(13), cervical cancer (19), gastric cancer (12,20), and
ovarian cancer (16). The function of
HDAC10 in lung cancer is a matter of debate (10,15). High
HDAC10 expression was associated with good prognosis in non-small
cell lung cancer (10) but has been
lately demonstrated to promote lung cancer proliferation (15).
Class II HDACs have been reported to be able to
shuttle between the nucleus and cytoplasm. In this study, we found
similar behavior of HDAC10 expression in colon cancer tissues. Yang
et al reported that HDAC10 is mainly expressed in the
cytoplasm of lung cancer cells but is mainly located in the nucleus
of normal lung cells, and suggested different functions of
cytoplasmic and nuclear HDAC10 in lung cancer progression (15). The association of cytoplasmic and
nuclear HDAC10 expression and clinicophathological and prognostic
effect is inconsistent even in colon cancer. Moreover, in tumor
tissues HDAC10 acted as a tumor suppressor, but showed quite
different effect in adjacent tissues, suggesting that HDAC10 might
act as a tumor suppressor in colon cancer tissues but may also
function as a tumor promoter by promoting tumor metastasis to
regional lymph node or to distant tissues.
A growing number of studies point to quite different
prognosis of patients between right-sided and left-sided colon
cancers (21). The probability of
relapse of colon carcinoma in different sides is dependent on
different molecular pathways (22).
Our observation is consistent with the previous studies that
survival of patients with right-sided colon cancer is less than
those with left-sided colon cancer.
In summary, our findings suggest that HDAC10
expression in tumor tissues is associated with good prognosis of
colon cancers but predicted poor prognostic outcomes in
para-carcinoma tissues, probably owing to regulation of the DNA
mismatch repair pathway. Further studies by altering the HDAC10
expression in colon cancer cells and normal colon cells to
investigate the potential functionin invasion and metastasis is
needed to test our notion that HDAC10 has different roles in tumor
and para-carcinoma tissues.
Acknowledgements
The present study was supported by Foundation for
Key Project of Natural Science Research Education Department of
Anhui Province (KJ2016A726 and KJ2017A249).
References
|
1
|
Yuan Z, Peng L, Radhakrishnan R and Seto
E: Histone deacetylase 9 (HDAC9) regulates the functions of the
ATDC (TRIM29) protein. J Biol Chem. 285:39329–39338. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Clocchiatti A, Florean C and Brancolini C:
Class IIa HDACs: From important roles in differentiation to
possible implications in tumourigenesis. J Cell Mol Med.
15:1833–1846. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kotian S, Liyanarachchi S, Zelent A and
Parvin JD: Histone deacetylases 9 and 10 are required for
homologous recombination. J Biol Chem. 286:7722–7726. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lai IL, Lin TP, Yao YL, Lin CY, Hsieh MJ
and Yang WM: Histone deacetylase 10 relieves repression on the
melanogenic program by maintaining the deacetylation status of
repressors. J Biol Chem. 285:7187–7196. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oehme I, Lodrini M, Brady NR and Witt O:
Histone deacetylase 10-promoted autophagy as a druggable point of
interference to improve the treatment response of advanced
neuroblastomas. Autophagy. 9:2163–2165. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Oehme I, Linke JP, Böck BC, Milde T,
Lodrini M, Hartenstein B, Wiegand I, Eckert C, Roth W, Kool M, et
al: Histone deacetylase 10 promotes autophagy-mediated cell
survival. Proc Natl Acad Sci USA. 110:E2592–E2601. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pinto G, Shtaif B, Phillip M and
Gat-Yablonski G: Growth attenuation is associated with histone
deacetylase 10-induced autophagy in the liver. J Nutr Biochem.
27:171–180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Y, Peng L and Seto E: Histone
deacetylase 10 regulates the cell cycle G2/M phase transition via a
novel let-7-HMGA2-cyclin A2 pathway. Mol Cell Biol. 35:3547–3565.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Radhakrishnan R, Li Y, Xiang S, Yuan F,
Yuan Z, Telles E, Fang J, Coppola D, Shibata D, Lane WS, et al:
Histone deacetylase 10 regulates DNA mismatch repair and may
involve the deacetylation of MutS homolog 2. J Biol Chem.
290:22795–22804. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Osada H, Tatematsu Y, Saito H, Yatabe Y,
Mitsudomi T and Takahashi T: Reduced expression of class II histone
deacetylase genes is associated with poor prognosis in lung cancer
patients. Int J Cancer. 112:26–32. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Park BL, Kim YJ, Cheong HS, Lee SO, Han
CS, Yoon JH, Park JH, Chang HS, Park CS, Lee HS and Shin HD: HDAC10
promoter polymorphism associated with development of HCC among
chronic HBV patients. Biochem Biophys Res Commun. 363:776–781.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee JH, Jeong EG, Choi MC, Kim SH, Park
JH, Song SH, Park J, Bang YJ and Kim TY: Inhibition of histone
deacetylase 10 induces thioredoxin-interacting protein and causes
accumulation of reactive oxygen species in SNU-620 human gastric
cancer cells. Mol Cells. 30:107–112. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fan W, Huang J and Xiao H: Histone
deacetylase 10 suppresses proliferation and invasion by inhibiting
the phosphorylation of β-catenin and serves as an independent
prognostic factor for human clear cell renal cell carcinoma. Int J
Clin Exp Med. 8:3734–3742. 2015.PubMed/NCBI
|
|
14
|
Powers J, Lienlaf M, Perez-Villarroel P,
Deng S, Knox T, Villagra A and Sahakian E: Expression and function
of histone deacetylase 10 (HDAC10) in B cell malignancies. Methods
Mol Biol. 1436:129–145. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang Y, Huang Y, Wang Z, Wang HT, Duan B,
Ye D, Wang C, Jing R, Leng Y, Xi J, et al: HDAC10 promotes lung
cancer proliferation via AKT phosphorylation. Oncotarget.
7:59388–59401. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Islam MM, Banerjee T, Packard CZ, Kotian
S, Selvendiran K, Cohn DE and Parvin JD: HDAC10 as a potential
therapeutic target in ovarian cancer. Gynecol Oncol. 144:613–620.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang X, Liu J, Zhen J, Zhang C, Wan Q, Liu
G, Wei X, Zhang Y, Wang Z, Han H, et al: Histone deacetylase 4
selectively contributes to podocyte injury in diabetic nephropathy.
Kidney Int. 86:712–725. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Song C, Zhu S, Wu C and Kang J: Histone
deacetylase (HDAC) 10 suppresses cervical cancer metastasis through
inhibition of matrix metalloproteinase (MMP) 2 and 9 expression. J
Biol Chem. 288:28021–28033. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jin Z, Jiang W, Jiao F, Guo Z, Hu H and
Wang L and Wang L: Decreased expression of histone deacetylase 10
predicts poor prognosis of gastric cancer patients. Int J Clin Exp
Pathol. 7:5872–5879. 2014.PubMed/NCBI
|
|
21
|
Petrelli F, Tomasello G, Borgonovo K,
Ghidini M, Turati L, Dallera P, Passalacqua R, Sgroi G and Barni S:
Prognostic survival associated with left-sided vs right-sided colon
cancer: A systematic review and meta-analysis. JAMA Oncol. Oct
27–2016.(Epub ahead of print).
|
|
22
|
Bauer KM, Hummon AB and Buechler S:
Right-side and left-side colon cancer follow different pathways to
relapse. Mol Carcinog. 51:411–421. 2012. View Article : Google Scholar : PubMed/NCBI
|