Introduction
Chordoma is a rare locally invasive low-grade
malignant bone tumor that is considered to originate from remnants
of the embryonic notochord (1).
Chordoma accounts for between 1 and 4% of malignant bone tumors
with an annual incidence of 0.08/100,000 individuals (2). Complete removal of chordoma is difficult
owing to the requirement to preserve adjacent vital structures, the
large tumor size and extensive intraoperative blood loss (3). Additionally, chordoma is largely
resistant to chemotherapy and radiotherapy (4). Accordingly, local recurrence rates of
sacral chordoma were >40%, and the 5- and 10-year overall
survival rates were 70 and 40% respectively, which leads to a poor
quality of life and poor prognosis of patients with chordoma
(5). Therefore, there is a clinical
requirement for improving therapeutic options for this
life-threatening tumor. Currently, there is inadequate
understanding of the genetics and molecular biology of chordoma,
and information about the potential molecular therapeutic targets
is lacking.
MicroRNAs (miRNAs/miRs) are small non-coding
regulatory RNA molecules that exert a range of effects on the
regulation of gene expression. Dysregulation of miRNAs that target
the expression of oncogenes or tumor suppressor genes may therefore
influence a number of essential biological functions, including
cancer initiation and progression (6,7). The role
of miRNAs in chordoma has been studied when compared with muscle
tissue or adult nucleus pulposus tissues (8–10). Certain
miRNAs are differentially expressed in chordoma and, in particular,
miR-1 and miR-31 may have a functional impact on chordoma tumor
pathogenesis (8,9). Nevertheless, the unique expression
profiles of miRNAs and their downstream signaling pathways in
chordoma remain incompletely characterized. Additionally, to the
best of our knowledge, it has been demonstrated that chordoma
originate from the remnant notochord (11), and notochordal cells disappear by
early childhood and are replaced by the nucleus pulposus in the
intervertebral discs (12).
Therefore, fetal nucleus pulposus tissues were selected as a
control in the present study. The aim of the present study was to
delineate the global miRNA expression profile and associated
signaling networks in chordoma.
Materials and methods
Tissue samples
A total of 15 chordoma tissues from patients who had
undergone surgical resection of chordoma were obtained from the
Department of Orthopedic Surgery, The First Affiliated Hospital of
Soochow University (Suzhou, China) between January 2005 and June
2015. The clinical and pathological features of the patients are
listed in Table I. A total of 10
fetal nucleus pulposus tissues from the intervertebral discs of
aborted fetuses between the 20 and 28th week of gestation were
included as a control group. Fresh specimens were collected
immediately following resection, snap-frozen in liquid nitrogen and
stored at-80°C until total RNA extraction. Samples were taken from
areas exhibiting neither hemorrhage nor necrosis. These samples
were confirmed by two pathologists. The present study was approved
by the Ethics Committee of The First Affiliated Hospital of Soochow
University and written informed consent was provided by all
patients.
 | Table I.Clinical and pathological features of
patients. |
Table I.
Clinical and pathological features of
patients.
| Patient no. | Age, years | Sex | Tumor site | Tumor size, mm | Surrounding muscle
invasion |
|---|
| 1 | 40 | Male | S2 | 110 | Yes |
| 2 | 66 | Male | S1 | 135 | Yes |
| 3 | 48 | Female | S3 | 90 | No |
| 4 | 44 | Male | S2 | 105 | Yes |
| 5 | 51 | Male | S3 | 95 | Yes |
| 6 | 55 | Female | S2 | 115 | Yes |
| 7 | 44 | Female | S2 | 120 | No |
| 8 | 46 | Female | S3 | 100 | Yes |
| 9 | 45 | Male | S4 | 60 | No |
| 10 | 50 | Female | S2 | 110 | Yes |
| 11 | 37 | Female | S1 | 155 | Yes |
| 12 | 60 | Male | S3 | 85 | No |
| 13 | 38 | Female | S2 | 110 | Yes |
| 14 | 22 | Male | S1 | 130 | Yes |
| 15 | 56 | Male | S2 | 115 | No |
RNA isolation and miRNA microarray
profiling
Total RNA was extracted from three chordoma and
three fetal nucleus pulposus fresh-frozen tissue samples using
mirVana™ RNA Isolation Kit (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), according to the
manufacturer's protocol. Total RNA was quantified using a NanoDrop
ND-2000 instrument (Thermo Fisher Scientific, Inc.) and RNA
integrity was assessed using an Agilent Bioanalyzer 2100 instrument
(Agilent Technologies, Inc., Santa Clara, CA, USA). RNAs with a
2100RIN (RNA integrity number) 6.0 and 28S/18S ratio 0.7 were used
for the miRNA array analysis and reverse transcription (RT).
miRNA expression profiling was performed using
Agilent Human miRNA Microarray kit (Agilent Technologies, Inc.)
containing 2,006 human mature miRNAs. Total RNAs were
dephosphorylated, denaturized and then labeled with
cyanine-3-cytosine triphosphate. Following purification, the
labeled RNAs were hybridized onto the microarray. Following
washing, the arrays were scanned using an Agilent Scanner G2505C
(Agilent Technologies, Inc.). Genespring software (version 12.5,
Agilent Technologies, Inc.) was used for microarray analysis, with
the raw data normalized using the quantile algorithm.
Differentially expressed miRNAs were identified through fold
changes as well as P-values calculated using a Student's t-test.
The thresholds set for up- and downregulated genes were a fold
change ≥2.0 and P≤0.05.
Bioinformatics analysis
Hierarchical clustering was performed to identify
the distinguishable miRNA expression pattern among samples.
Potential target genes of differentially expressed miRNAs were
predicted by the intersection of three databases (Targetscan,
http://www.targetscan.org; microRNA.org, http://www.microrna.org; and PITA, http://genie.weizmann.ac.il/pubs/mir07/mir07_dyn_data.html).
For functional analyses of potential miRNA targets, Gene Ontology
(GO) term analysis was applied to organize genes into categories on
the basis of biological processes, cellular components and
molecular functions. Biological pathways analysis, which was
defined by the Kyoto Encyclopedia of Genes and Genomes (KEGG;
www.genome.ad.jp/kegg), provided an
improved understanding of gene expression information as a complete
network. Fisher's exact test, χ2 test, and the threshold
of significance were defined by the P-value and false discovery
rate (FDR). The screening criterion was P<0.05.
RT-quantitative polymerase chain
reaction (qPCR)
RT-qPCR was used to measure miRNA expression using
TaqMan MicroRNA assays (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Primer sequences used were as follows: miR-21
forward, 5′-TAGCTTATCAGACTGATGTTGA-3′ and reverse,
5′-TCAACATCAGTCTGATAAGCTA-3′; miR-150 forward
5′-CAGTATTCTCTCCCAACCCTTGTA-3′ and reverse,
5′-AATGGATGATCTCGTCAGTCTGTT-3′; miR-1290 forward,
5′-CAGTGCTGGATTTTTGGAT-3′ and reverse,
5′-TATGGTTGTTCACGACTCCTTCAC-3′; miR-623 forward,
5′-CAGAAGCGTAATGGACCTTTCG-3′ and reverse,
5′-TATCGCTGATCACGACTCGTTGAT-3′; and U6 forward,
5′-ATTGGAACGATACAGAGAAGATT-3′ and reverse,
5′-GGAACGCTTCACGAATTTG-3′. miRNA-specific reverse transcription was
performed with 5 ng total RNA using the TaqMan MicroRNA reverse
transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. qPCR was performed
using the TaqMan MicroRNA assay kit on the Step-One Plus Real-Time
PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Thermal cycling conditions were set according to the manufacturer's
fast protocol and all reactions were performed in triplicate.
Relative expression was determined using the 2−ΔΔCq
method (13), and expression values
were normalized to U6, which has been demonstrated to be a suitable
reference gene.
Results
miRNA array analysis
Three primary chordoma and three fetal nucleus
pulposus fresh-frozen tissue samples were selected for the
identification of differentially expressed miRNAs. In total, 42 of
the 2006 miRNAs analyzed were significantly dysregulated in the
chordoma group compared with normal controls at a level of
P<0.05 and a fold change >2. In total, 23 downregulated and
19 upregulated known miRNAs were identified in chordoma tissues
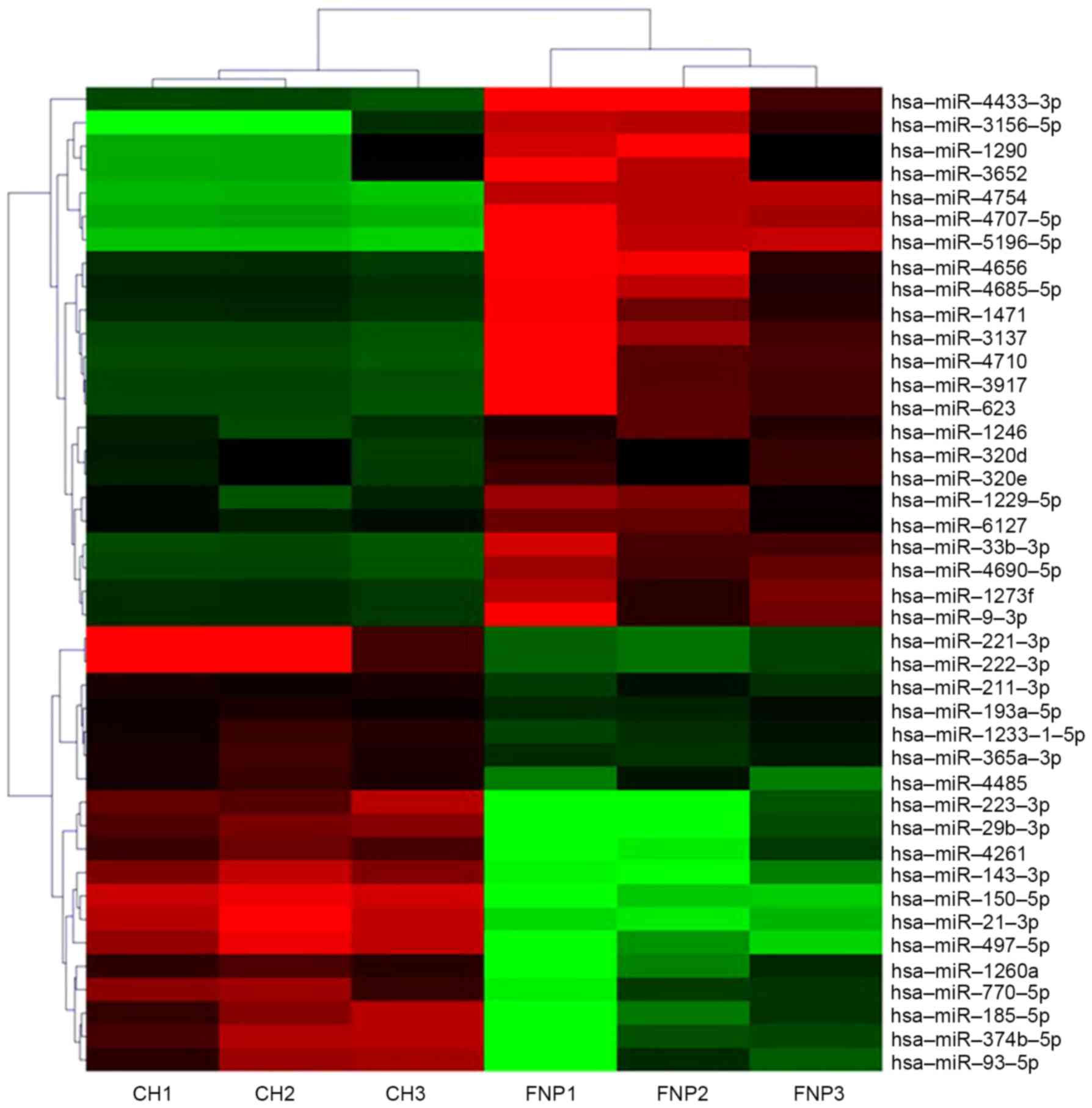
compared with fetal nucleus pulposus tissues (Table II). A non-supervised 2D-cluster
analysis was applied for all tumors and the 42 significantly
dysregulated miRNAs (Fig. 1).
 | Table II.Significantly dysregulated miRNAs in
the chordoma tissues vs. fetal nucleus pulposus tissues. |
Table II.
Significantly dysregulated miRNAs in
the chordoma tissues vs. fetal nucleus pulposus tissues.
|
| Fold change | P-value | Regulation |
|---|
| hsa-miR-150-5p | 341.1334 | 0.0001 | Up |
| hsa-miR-497-5p | 321.3715 | 0.0042 | Up |
| hsa-miR-21-3p | 240.6141 | <0.0001 | Up |
| hsa-miR-29b-3p | 226.4459 | 0.0205 | Up |
| hsa-miR-223-3p | 188.2892 | 0.0202 | Up |
| hsa-miR-143-3p | 117.0034 | 0.0011 | Up |
| hsa-miR-222-3p | 78.3672 | 0.0155 | Up |
| hsa-miR-221-3p | 76.1249 | 0.0164 | Up |
| hsa-miR-4261 | 66.7500 | 0.0320 | Up |
|
hsa-miR-374b-5p | 46.1905 | 0.0136 | Up |
| hsa-miR-185-5p | 45.5516 | 0.0222 | Up |
| hsa-miR-93-5p | 44.2137 | 0.0399 | Up |
| hsa-miR-770-5p | 24.4578 | 0.0183 | Up |
| hsa-miR-1260a | 19.4710 | 0.0406 | Up |
| hsa-miR-4485 | 6.2146 | 0.0192 | Up |
|
hsa-miR-1233-1-5p | 3.5351 | 0.0064 | Up |
|
hsa-miR-365a-3p | 3.4641 | 0.0033 | Up |
| hsa-miR-211-3p | 2.7623 | 0.0052 | Up |
|
hsa-miR-193a-5p | 2.2088 | 0.0049 | Up |
|
hsa-miR-5196-5p | 244.3847 | 0.0001 | Down |
|
hsa-miR-4433-3p | 146.8665 | 0.0452 | Down |
| hsa-miR-4754 | 130.6493 | <0.0001 | Down |
|
hsa-miR-4707-5p | 130.4238 | 0.0001 | Down |
|
hsa-miR-3156-5p | 108.7725 | 0.0173 | Down |
| hsa-miR-3652 | 33.9637 | 0.0467 | Down |
| hsa-miR-1290 | 33.7951 | 0.0455 | Down |
| hsa-miR-3137 | 29.5932 | 0.0158 | Down |
| hsa-miR-623 | 26.7450 | 0.0368 | Down |
| hsa-miR-3917 | 25.9233 | 0.0422 | Down |
| hsa-miR-4656 | 25.3885 | 0.0245 | Down |
| hsa-miR-4710 | 25.1405 | 0.0253 | Down |
| hsa-miR-33b-3p | 15.6259 | 0.0085 | Down |
|
hsa-miR-4685-5p | 14.0308 | 0.0302 | Down |
|
hsa-miR-4690-5p | 13.1652 | 0.0015 | Down |
| hsa-miR-9-3p | 12.2306 | 0.0286 | Down |
| hsa-miR-1471 | 11.6128 | 0.0325 | Down |
| hsa-miR-1273f | 9.7157 | 0.0105 | Down |
|
hsa-miR-1229-5p | 7.1305 | 0.0399 | Down |
| hsa-miR-1246 | 4.7649 | 0.0071 | Down |
| hsa-miR-6127 | 3.7462 | 0.0391 | Down |
| hsa-miR-320e | 2.9118 | 0.0438 | Down |
| hsa-miR-320d | 2.6649 | 0.0459 | Down |
GO analysis
To further determine the biological functions of
these miRNAs, intersection predicted with three databases
(Targetscan, microRNA.org and PITA) was used to
predict the target genes of the 42 miRNAs, which resulted in the
identification of 10,292 putative target genes. The functions of
these target genes were then determined using GO analysis.
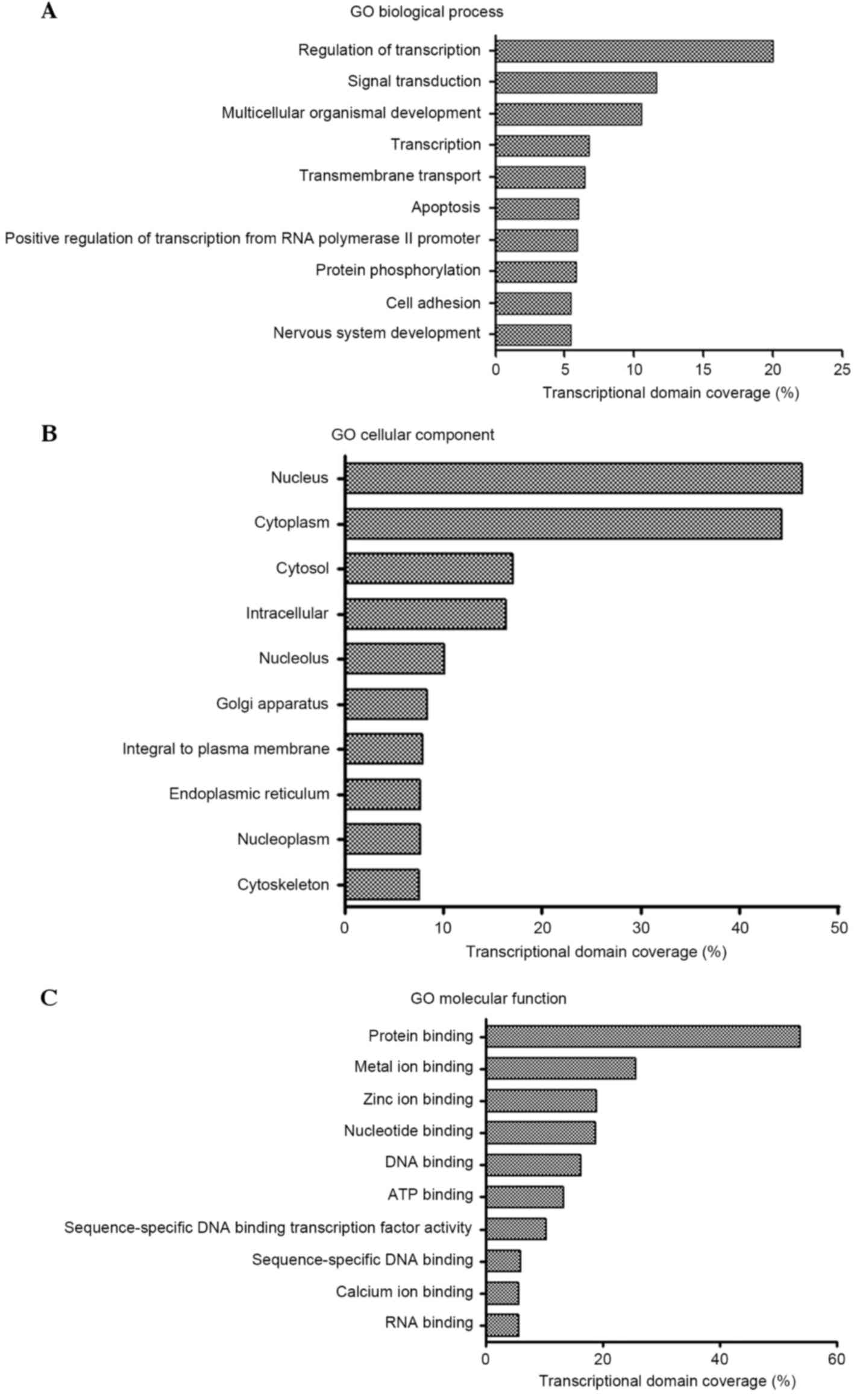
According to the results, 496, 151 and 152 GO terms were identified
from three ontologies including biological processes, cellular
components and molecular functions, respectively. Significant GO
terms corresponding to biological processes included regulation of
transcription (DNA-dependent), signal transduction and
multicellular organismal development. The main GO terms for
cellular components included nucleus, cytoplasm and cytosol. The
significant GO categories corresponding to molecular functions
included protein binding, metal ion binding and zinc ion binding
(Fig. 2A-C). These functions are
known to be markedly associated with tumor generation and
progress.
KEGG pathway analysis
To achieve an improved understanding of the
functions and regulatory networks of predicted target genes, target
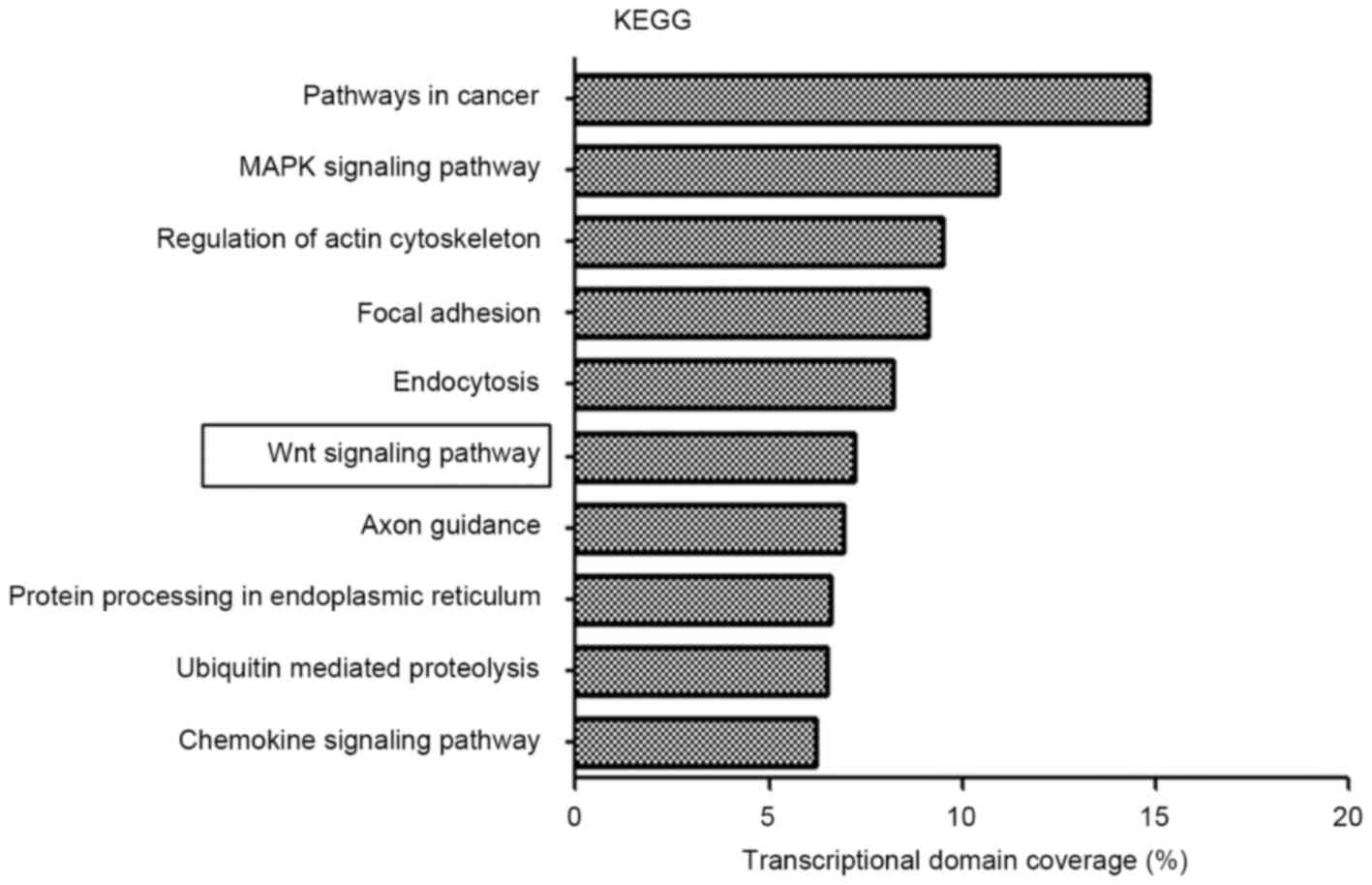
enrichment was searched for using KEGG. There were 1,805
differentially expressed genes identified in 65 pathways with a
threshold of P<0.01 and FDR<0.05 using KEGG pathway analysis.
Overall, a genetic cluster summarizing the functions of signaling
pathways in cancer, mitogen-activated protein kinase (MAPK)
signaling pathway, regulation of actin cytoskeleton, focal adhesion
and endocytosis was identified to exhibit the most marked
association with the chordoma group (Fig.
3). Notably, the Wnt signaling pathway was dysregulated in
chordoma; aberrant Wnt signaling is associated with tumorigenesis
in a number of types of tumor.
Validation of miRNA array data
To validate miRNA microarray data, RT-qPCR was
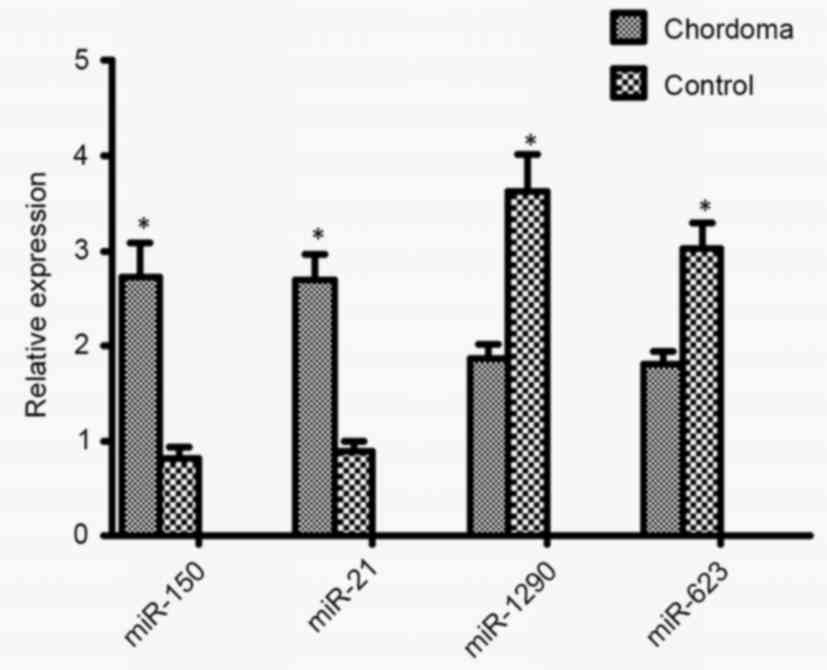
performed using TaqMan MicroRNA assays. In total, four miRNAs
(hsa-miR-21-3p, hsa-miR-150-5p, hsa-miR-1290 and hsa-miR-623) were
randomly selected for validation from the 42 significantly
dysregulated miRNAs. As presented in Fig.
4, the expression levels of hsa-miR-21-3p and hsa-miR-150-5p
were significantly increased in chordoma tissues compared with in
the control group, whereas expression levels of hsa-miR-1290 and
hsa-miR-623 were significantly decreased in chordoma tissues
compared with the control group. These four miRNAs may therefore
serve a role in the malignant progression of chordoma.
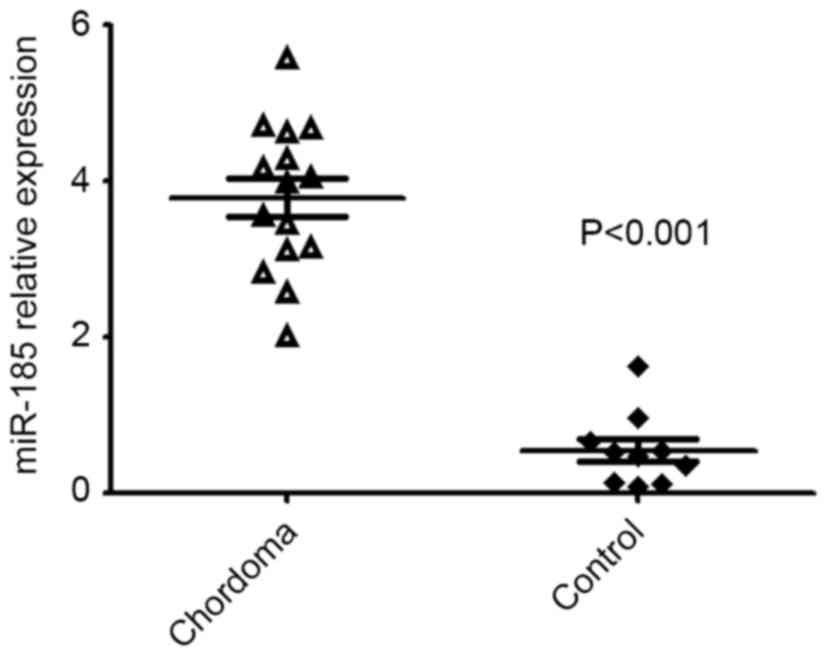
Additionally, due to the Wnt signaling pathway
potentially serving a pivotal role in chordoma, hsa-miR-185-5p was
included for validation which had the potential to target the Wnt
signaling pathway. It was confirmed that the expression of
hsa-miR-185-5p was dysregulated in chordoma tissues (Fig. 5).
Discussion
Chordoma is a malignant bone tumor known to arise
from the embryonic remnants of the notochord (1). However, the biological features of
chordoma remain largely unknown. miRNAs have emerged as key
regulators in numerous oncogenic processes. A number of miRNAs have
been identified to be involved in the initiation and progression of
several types of human cancer (6,7).
Therefore, it is relevant to delineate the expression profiles,
functions and potential regulatory mechanisms of miRNAs in
chordoma. In the present study, the expression signature of miRNAs
in chordoma was investigated, and their functions, downstream
targets and signaling pathways were analyzed.
First, miRNA expression profiles were described in
chordoma. Compared with fetal nucleus pulposus tissues, 42 miRNAs
were significantly dysregulated in the chordoma group. The
differential expression of hsa-miR-21-3p, hsa-miR-150-5p,
hsa-miR-1290 and hsa-miR-623 was validated using RT-qPCR.
Previously, hsa-miR-21-3p was demonstrated to exhibit oncogenic
functions in osteosarcoma, breast cancer and colorectal cancer
(14–16). The results of the present study
demonstrated that hsa-miR-21-3p expression was increased in
chordoma and may regulate invasion and metastasis, and
hsa-miR-150-5p is overexpressed in chordoma tissues which acts as
an oncogene. Furthermore, hsa-miR-1290 and hsa-miR-623 were
downregulated in chordoma tissues. The results suggest that the
tumorigenesis of chordoma is characterized by marked changes in
miRNA expression profiles. A number of dysregulated miRNAs that
were identified in the present study are reported for the first
time in chordoma. Therefore, the results of the present study
provide a rationale for further study of these novel dysregulated
miRNAs in the initiation and progression of chordoma.
Since the majority of miRNAs are considered to be
involved in carcinogenesis by inhibiting target mRNAs, targets of
these 42 altered miRNAs were predicted in the present study. In
total, 10,292 putative target genes were predicted by the
intersection of three databases (Targetscan, microRNA.org and PITA). Further GO analyses revealed
that the majority of predicted target genes functioned in
regulation of transcription, signal transduction and multicellular
organismal development. These functions are known to be associated
with human tumorigenesis and cancer progression.
Signaling pathways regulated by validated targets of
the dysregulated miRNAs were assessed using KEGG pathway analysis.
In the present study, the most significant pathways were associated
with tumor pathogenesis, including signaling pathways in cancer,
MAPK signaling pathway, regulation of actin cytoskeleton, focal
adhesion and endocytosis. Consistent with previous studies, the
MAPK signaling pathway was the most highly overrepresented genetic
pathway (17,18). Additionally, our previous study
demonstrated that Raf-1 and extracellular-signal-regulated kinase
1/2, which are important members of the MAPK signaling pathway,
were overexpressed in chordoma and associated with tumor
progression (19). However, further
study was required to evaluate the exact role of the MAPK signaling
pathway in the development and progression of chordoma.
In particular, the Wnt signaling pathway was
demonstrated to be important in chordoma development. Previous
studies have indicated that the Wnt signaling pathway serves an
important role in cell proliferation, differentiation, survival and
apoptosis, and is implicated in various tumor types (20,21). In
the present study, it was observed that hsa-miR-185-5p was
significantly dysregulated in chordoma tissues and had the
potential to target the Wnt signaling pathway. Li et al
(22) confirmed that miR-185-3p
contributed to the radioresistance of nasopharyngeal carcinoma via
modulation of WNT2B expression in vitro. Liu et al
(23) confirmed that miR-185 was a
negative regulator of RhoA and cell division cycle 42, and their
cellular activities, and was able to inhibit proliferation and
invasion of colorectal cancer cells. Fu et al (24) demonstrated that miR-185 inhibited the
proliferation of breast cancer cells by regulating the expression
of c-Met, indicating its potential as a therapeutic target for
breast cancer. Therefore, elucidating oncogenic mechanisms by which
miRNAs regulate the Wnt signaling pathway represents a promising
strategy for identifying novel therapeutic targets in chordoma.
In conclusion, 42 significantly dysregulated miRNAs
were discovered in chordoma, including 19 upregulated and 23
downregulated miRNAs. In addition, 10,292 potential target
transcripts were predicted for these 42 miRNAs. Integrated GO and
KEGG pathway analyses indicated that the MAPK signaling pathway may
serve an important role in chordoma development. In particular, the
Wnt signaling pathway may have a vital functional effect on
chordoma tumor pathogenesis, and hsa-miR-185-5p was revealed as a
potential key oncogenic miRNA in chordoma development via the Wnt
signaling pathway. These results indicate that dysregulated
expression of miRNAs and their potential target mRNAs are
prominently involved in the pathogenesis of chordoma, which
suggests that dysregulated miRNAs may serve as potential biomarkers
and therapeutic targets in patients with chordoma.
Acknowledgements
The present study was funded by the Jiangsu
Provincial Special Program of Medical Science (grant no.
BL2012004), the Natural Science Foundation of the Colleges and
Universities in Jiangsu Province (grant no. 16KJD320004) and the
Suzhou Civic ‘Science and Education Guardian’ Youth Science and
Technology Project (grant no. KJXW2014009).
References
|
1
|
Casali PG, Stacchiotti S, Sangalli C, Olmi
P and Gronchi A: Chordoma. Curr Opin Oncol. 19:367–370. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McMaster ML, Goldstein AM, Bromley CM,
Ishibe N and Parry DM: Chordoma: Incidence and survival patterns in
the United States, 1973–1995. Cancer Causes Control. 12:1–11. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fuchs B, Dickey ID, Yaszemski MJ, Inwards
CY and Sim FH: Operative management of sacral chordoma. J Bone
Joint Surg Am. 87:2211–2216. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sciubba DM, Cheng JJ, Petteys RJ, Weber
KL, Frassica DA and Gokaslan ZL: Chordoma of the sacrum and
vertebral bodies. J Am Acad Orthop Surg. 17:708–717. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chugh R, Tawbi H, Lucas DR, Biermann JS,
Schuetze SM and Baker LH: Chordoma: The nonsarcoma primary bone
tumor. Oncologist. 12:1344–1350. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kong YW, Ferland-McCollough D, Jackson TJ
and Bushell M: microRNAs in cancer management. Lancet Oncol.
13:e249–e258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Duan Z, Choy E, Nielsen GP, Rosenberg A,
Iafrate J, Yang C, Schwab J, Mankin H, Xavier R and Hornicek FJ:
Differential expression of microRNA (miRNA) in chordoma reveals a
role for miRNA-1 in Met expression. J Orthop Res. 28:746–752.
2010.PubMed/NCBI
|
|
9
|
Bayrak OF, Gulluoglu S, Aydemir E, Ture U,
Acar H, Atalay B, Demir Z, Sevli S, Creighton CJ, Ittmann M, et al:
MicroRNA expression profiling reveals the potential function of
microRNA-31 in chordomas. J Neurooncol. 115:143–151. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y, Schiff D, Park D and Abounader R:
MicroRNA-608 and microRNA-34a regulate chordoma malignancy by
targeting EGFR, Bcl-xL and MET. PLoS One. 9:e915462014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vujovic S, Henderson S, Presneau N, Odell
E, Jacques TS, Tirabosco R, Boshoff C and Flanagan AM: Brachyury, a
crucial regulator of notochordal development, is a novel biomarker
for chordomas. J Pathol. 209:157–165. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shen J, Shi Q, Lu J, Wang DL, Zou TM, Yang
HL and Zhu GQ: Histological study of chordomaorigin from fetal
notochordal cell rests. Spine (Phila Pa 1976). 38:2165–2170. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ziyan W, Shuhua Y, Xiufang W and Xiaoyun
L: MicroRNA-21 is involved in osteosarcoma cell invasion and
migration. Med Oncol. 28:1469–1474. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang C, Liu K, Li T, Fang J, Ding Y, Sun
L, Tu T, Jiang X, Du S, Hu J, et al: miR-21: A gene of dual
regulation in breast cancer. Int J Oncol. 48:161–172. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Basati G, Razavi Emami A, Abdi S and
Mirzaei A: Elevated level of microRNA-21 in the serum of patients
with colorectal cancer. Med Oncol. 31:2052014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Long C, Jiang L, Wei F, Ma C, Zhou H, Yang
S, Liu X and Liu Z: Integrated miRNA-mRNA analysis revealing the
potential roles of miRNAs in chordomas. PLoS One. 8:e666762013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tamborini E, Virdis E, Negri T, Orsenigo
M, Brich S, Conca E, Gronchi A, Stacchiotti S, Manenti G, Casali
PG, et al: Analysis of receptor tyrosine kinases (RTKs) and
downstream pathways in chordomas. Neuro Oncol. 12:776–789. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang K, Chen H, Zhang B, Sun J, Lu J,
Chen K and Yang H: Overexpression of Raf-1 and ERK1/2 in sacral
chordoma and association with tumor recurrence. Int J Clin Exp
Pathol. 8:608–614. 2015.PubMed/NCBI
|
|
20
|
Tian J, He H and Lei G: Wnt/β-catenin
pathway in bone cancers. Tumour Biol. 35:9439–9445. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song JL, Nigam P, Tektas SS and Selva E:
microRNA regulation of Wnt signaling pathways in development and
disease. Cell Signal. 27:1380–1391. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li G, Wang Y, Liu Y, Su Z, Liu C, Ren S,
Deng T, Huang D, Tian Y and Qiu Y: miR-185-3p regulates
nasopharyngeal carcinoma radioresistance by targeting WNT2B in
vitro. Cancer Sci. 105:1560–1568. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu M, Lang N, Chen X, Tang Q, Liu S,
Huang J, Zheng Y and Bi F: miR-185 targets RhoA and Cdc42
expression and inhibits the proliferation potential of human
colorectal cells. Cancer Lett. 301:151–160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fu P, Du F, Yao M, Lv K and Liu Y:
MicroRNA-185 inhibits proliferation by targeting c-Met in human
breast cancer cells. Exp Ther Med. 8:1879–1883. 2014.PubMed/NCBI
|