Introduction
Colorectal carcinoma (CRC) is the third most common
type of cancer worldwide; 40–50% of newly diagnosed patients have
already progressed to metastasis and are therefore resistant to
conventional therapy, with an increased chance of recurrence
(1,2).
Despite advances in CRC therapy, the prognosis of patients with
metastatic CRC (mCRC) remains poor, with a median overall survival
(OS) time of 18–21 months (3). Thus,
the major risk factor for patients with CRC is the development of
metastasis.
Understanding the molecular mechanisms that drive
CRC progression and metastatic processes may facilitate the
development of better treatment strategies to improve patient
prognosis and disease management, and aid in the identification of
novel molecular prognostic factors and therapeutic targets. One
candidate molecule with potential as a prognostic marker or
therapeutic target is macrophage-capping protein (CapG). CapG is a
ubiquitous actin-binding protein of the gelsolin/villin superfamily
that is associated with cell motility (4). A previous study demonstrated that bone
marrow-derived macrophages from CapG-deficient mice exhibited
distinct motility defects and the inhibition of receptor-mediated
ruffling, suggesting that CapG is associated with motility
(4). Furthermore, the overexpression
of CapG has been detected in a range of types of cancer, including
pancreatic, breast and ovarian carcinoma, in which the contribution
of CapG expression to cancer cell metastatic behavior is validated
(5). However, there is limited
information regarding the role of CapG in CRC.
Therefore, the present study investigated the role
of CapG in CRC development and progression, with potential novel
insights for CRC diagnosis, treatment and prognosis.
Materials and methods
Human tissue sample collection
Between October 2014 to March 2015, fresh tissues
were obtained from 84 patients with CRC (49 males, 35 females) that
underwent CRC resection without neoadjuvant treatment at the
Zhongnan Hospital of Wuhan University (Wuhan, China). The mean age
was 59.3 (range, 29–85) years. the study was approved by the
Zhongnan Hospital of Wuhan University Ethics Committee. Informed,
written consent was obtained from all participants in the study.
The neoplastic tissues were collected from 84 patients, whereas
additional non-neoplastic epithelial tissue samples (~5 cm from the
border of the main tumor lesion) were collected from 19 of the
patients. The tissue samples were formalin-fixed and
paraffin-embedded. Data regarding the clinicopathological features
of the patients, including sex, age, tumor location, tumor
differentiation, lymph node metastasis (LNM) status and clinical
stage determined according to the Dukes system (6) for CRC staging were extracted from
patient records. Patients that had received prior treatment or that
exhibited metaplasia, dysplasia or atypical hyperplasia were
excluded from the study.
Immunohistochemistry (IHC)
For IHC, formalin-fixed, paraffin-embedded CRC and
non-neoplastic epithelial tissues were cut into 4-µm sections. The
sections were deparaffinized in xylene and rehydrated in a
descending series of ethanol concentrations. For antigen retrieval,
sections were immersed in antigen-unmasking solution (BOND Epitope
Retrieval 1; Leica Microsystems, Inc., Buffalo Grove, IL, United
States; cat. no. AR9961, pH 6.0, 10 min, 100°C) and boiled in a
microwave oven for 15 sec. Tissue sections were incubated with
anti-CapG antibodies (no. 10194–1-AP; dilution, 1:500; ProteinTech
Group, Inc., Chicago, IL, USA) at room temperature for 60 min. A
S-P immunohistochemical kit (Fuzhou Maixin Biological Technology,
Ltd., Fuzhou, China) was then applied according to the
manufacturer's protocol. Immunohistochemical reactions were
developed with 3,3′-diaminobenzidine tetrahydrochloride (Fuzhou
Maixin Biological Technology, Ltd.) and counterstained with
hematoxylin for 30 sec, prior to mounting.
Immunostaining was blindly evaluated by two
independent experienced pathologists in an effort to provide a
consensus on staining patterns. The number of positive cells per
core were counted at ×200 and ×400 magnification. A total of 5
cores were taken per sample and the diameter of each core was 4 cm.
The grade of each section was judged by the staining intensity and
percentage of positive cells. The scores for staining intensity
were 0 (no staining), 1 (weak staining), 2 (moderate staining) or 3
(strong staining); the scores for the percentage of positive cells
were 0 (<5%), 1 (≥5% and <25%), 2 (≥25% and <50%), 3 (≥50%
and <75%) or 4 (≥75%). The combined IHC score was the staining
intensity score multiplied by the positive percentage score:
Negative (combined score, ≤1) was designated with ‘−’, weak
positive (combined score, 2–4) with ‘+’, moderate positive
(combined score, 6–8) with ‘++’ and strong positive (combined
score, 9–12) with ‘+++’, as per a previously described method
(7).
Cell culture and small interfering
(si)RNA
HCT116 CRC cells were obtained from the Scientific
Research Center of the Zhongnan Hospital of Wuhan University and
cultured in HyClone RPMI-1640 medium (GE Healthcare Life Sciences,
Logan, UT, USA) with 10% fetal bovine serum (FBS; Zhejiang Tianhang
Biotechnology Co., Ltd., Luohe, China). The cells were incubated at
37°C in a humidified atmosphere with 5% CO2 throughout
the study. siRNA oligo duplexes were produced by Invitrogen (Thermo
Fisher Scientific, Inc.). The target sequences for CapG-siRNA were
forward, 5′-GUGUGGAGUCAGCAUUUCAdTdT-3′ and reverse,
3′-dTdTCACACCUCAGUCGUAAAGU-5′; and the negative control sequences
were forward, 5′-UUCUCCGAACGUGUCACGUdTdT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAAdTdT-3′. The CapG or negative control siRNA
was transfected into HCT116 cells with Lipofectamine 2000
transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Following incubation for
48 h, the cells were harvested and the efficacy of RNA interference
(RNAi) was confirmed by western blotting.
Western blotting
The HCT116 cells were lysed with 100–300 µl RIPA
buffer supplemented with protease inhibitors (Roche Diagnostics,
Basel, Switzerland). The protein concentration was measured with
the BCA Protein assay (Bio-Rad Laboratories, Inc., Hercules, CA,
USA). From each sample, 20 µg total protein per lane was separated
by SDS-PAGE (10% gel). Proteins were electroblotted onto a
polyvinylidene fluoride membrane and blocked overnight in 0.05%
Tween-20 in PBS with 5% dried skimmed milk at 4°C. Primary
antibodies against CapG (no. 10194-1-AP; dilution, 1:1,000;
ProteinTech Group, Inc.) and GAPDH (no. BM1985; 1:1,000; Wuhan
Boster Biological Technology Ltd., Wuhan, China) were incubated
with the membrane for 1 h at room temperature. Membranes were then
washed with 0.05% Tween-20 in PBS and incubated with secondary
antibodies, including peroxidase-conjugated anti-rabbit (no.
sc2357; 1:3,000; against CapG antibodies) and anti-mouse (no.
sc516142; 1:3,000; against GAPDH antibodies) (both from Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) antibodies, for 45 min at
room temperature. The membranes were visualized using an enhanced
chemiluminescence detection kit (Santa Cruz Biotechnology, Inc.).
GAPDH was used as a control for protein loading. Quantification of
the intensity of immunoblots was performed by Bio-Rad Quantity One
software (version 4.1; Bio-Rad Laboratories, Inc.).
Wound healing assay
HCT116 cells were treated with siRNA as previously
described. Following incubation for 24 h, the cells were removed by
trypsinization, counted and plated at 4×105 cells/ml in
6-well plates. Cells were incubated overnight to yield confluent
monolayers. A wound in the monolayer was produced with a pipette
tip and images were captured at 0 and 24 h after wounding. Wound
closure (%) was determined as the wound width at 24 h relative to
the width at 0 h. Experiments were performed in triplicate and
repeated ≥3 times.
Transwell migration assay
In vitro tumor cell migration was measured
using Transwell chambers without matrigel. (BD Biosciences,
Bedford, MA, USA) according to the manufacturer's protocol. In
brief, 2×105 cells with/without small interfering RNA in
RPMI-1640 medium with 2% FBS were plated in the upper chamber, with
RPMI-1640 medium containing 10% FBS in the bottom chamber. The
cells were incubated for 24 h. The cells on the bottom surface were
fixed in 4% formalin for 15 min at room temperate, washed with PBS
twice, stained with 0.1% crystal violet for 15 min at room
temperate, and were counted. Cell counting was manually performed
in 5 areas per membrane with an optical microscope at ×200
magnification.
Statistical analysis
The CRC mRNA expression profiles GSE14333 and
GSE39582 were downloaded from the Gene Expression Omnibus
(http://www.ncbi.nlm.nih.gov/geo).
Disease-free survival (DFS) analyses were performed with the
Kaplan-Meier method and the results were compared by the log-rank
test. The median (for GSE14333, the median is 8.83; for GSE39582,
the median is 8.87) was used as the cut-off value in the
Kaplan-Meier analysis to define the low and high expression groups.
Associations between CapG expression and clinicopathological
parameters were assessed by the χ2 test. The wound
healing and Transwell assay results were analyzed by the t-test.
Data are expressed as the mean (n=3) ± standard deviation.
P<0.05 was considered to represent a statistically significant
difference. Statistical analyses were performed using GraphPad
Prism software (version 6.0; GraphPad Software, Inc., La Jolla, CA,
USA).
Results
CapG expression is higher in CRC
tissue than in non-tumor tissue samples
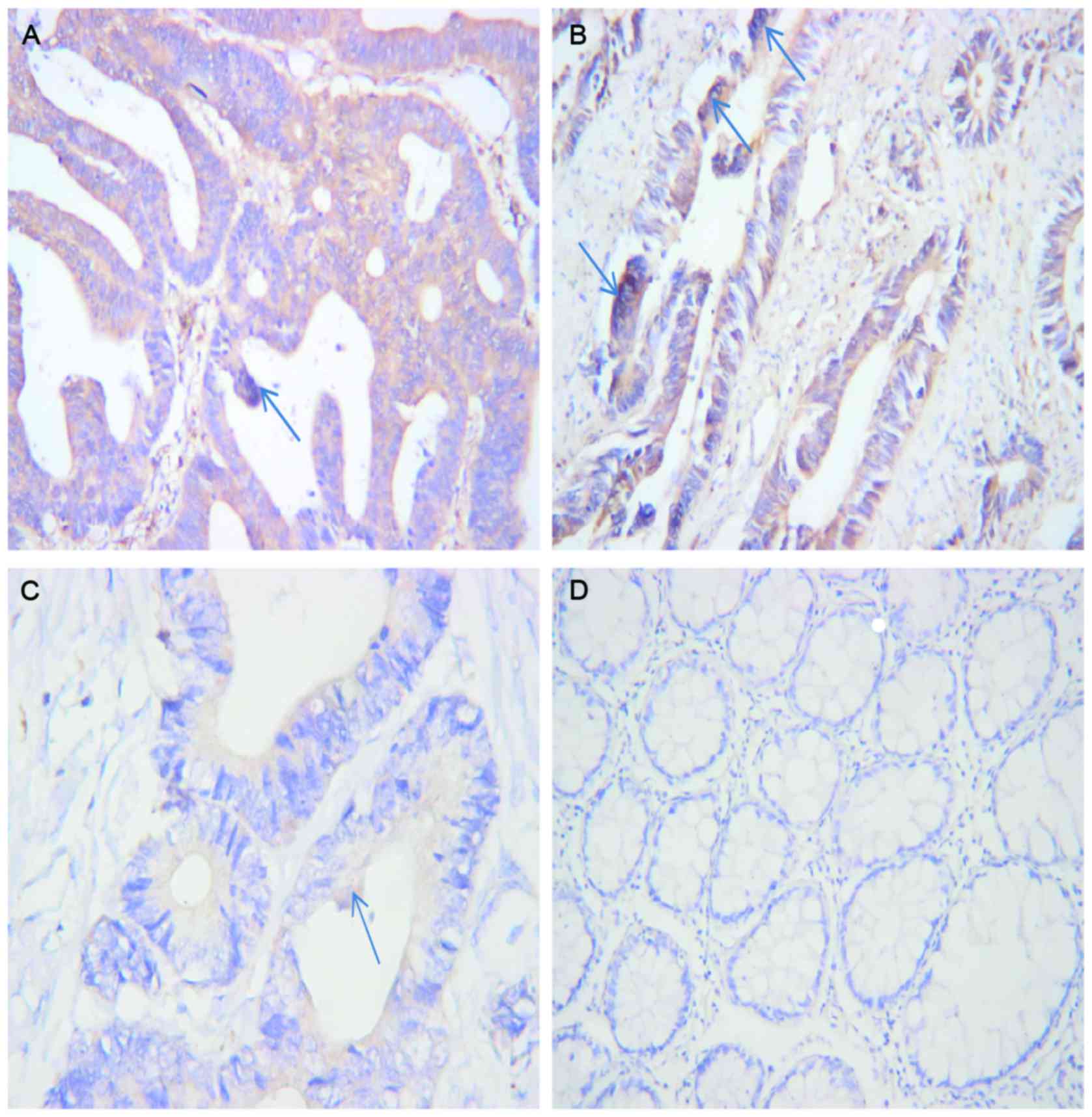
The IHC results revealed that CapG expression in the
CRC tissue was higher than in the non-tumor tissue. The positive
CapG expression rate was 84% (16/19) for CRC tissue and 26% (5/19)
for non-tumor tissue. Compared with the non-tumor tissue, CRC
tissue exhibited a significantly increased rate of immunoreactivity
for CapG (P<0.001; Table I). CapG
positivity was identified primarily in the cytoplasm and nucleus of
the CRC cells (Fig. 1).
 | Table I.CapG IHC scores in CRC tissue compared
with non-tumor tissue samples. |
Table I.
CapG IHC scores in CRC tissue compared
with non-tumor tissue samples.
|
| CapG IHC scores |
|
|
|---|
|
|
|
|
|
|---|
| Tissue type | Total | − | + | ++ | +++ | χ2 | P-value |
|---|
| CRC tissue | 19 | 3 | 2 | 10 | 4 | 20.618 | <0.001 |
| Non-tumor tissue | 19 | 14 | 5 | 0 | 0 |
|
|
CapG overexpression is associated with
risk-associated prognostic factors and the progression of CRC
Potential associations between CapG expression and
clinicopathological parameters are summarized in Table II. The results indicated that the
positive expression of CapG was significantly associated with tumor
site, LNM status, tumor differentiation and clinical stage
(P=0.021, P=0.036, P=0.012 and P=0.009, respectively); however,
there was no significant difference in expression associated with
sex or age (P=0.366 and P=0.789, respectively). These results may
suggest that CapG functions as an oncogene in CRC, and CapG may
represent a novel prognostic factor for CRC following curative
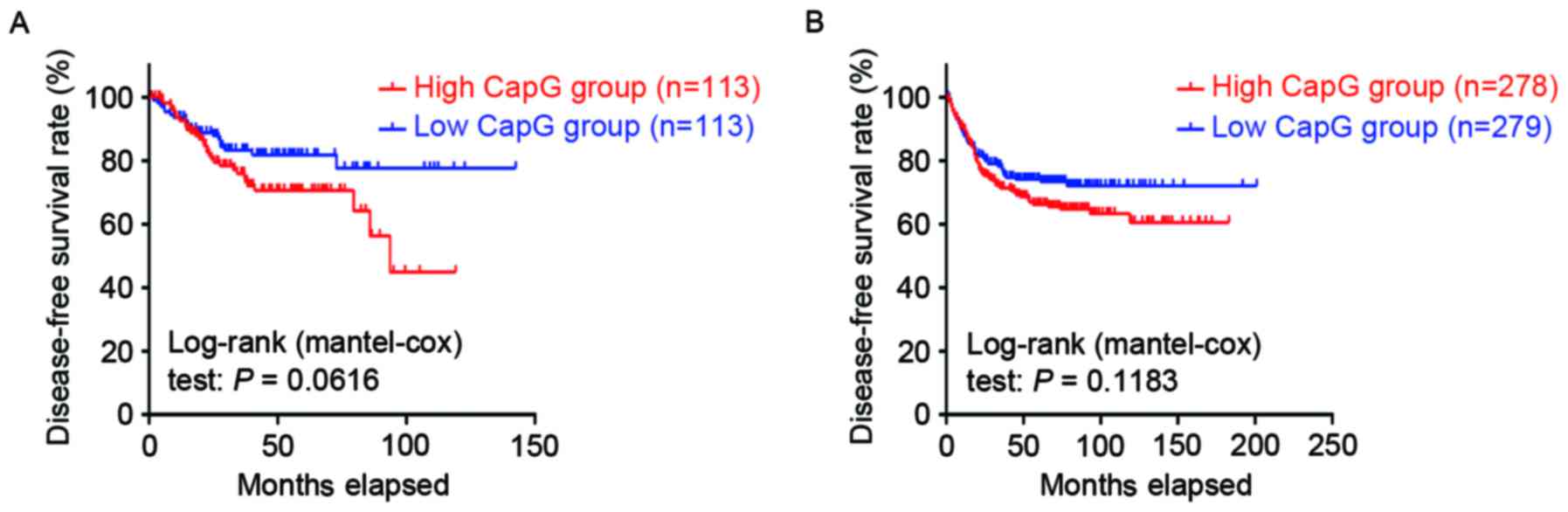
colorectal resection. However, Kaplan-Meier survival analysis of
the GSE14333 and GSE39582 expression profiles demonstrated that DFS
time did not differ significantly between the patients with CRC
with tumors with high and low CapG expression levels (GSE14333;
P=0.0616, Fig. 2A; GSE39582,
P=0.1183, Fig. 2B, respectively).
 | Table II.Association of the clinical features
of patients with CRC with CapG IHC scores. |
Table II.
Association of the clinical features
of patients with CRC with CapG IHC scores.
|
| CapG IHC scores |
|
|
|---|
|
|
|
|
|
|---|
| Clinical
features | Total | − | + | ++ | +++ | χ2 | P-value |
|---|
| All patients | 84 | 4 | 22 | 31 | 27 |
|
|
| Sex |
|
|
|
|
| 3.17 | 0.366 |
|
Male | 49 | 1 | 12 | 21 | 15 |
|
|
|
Female | 35 | 3 | 10 | 10 | 12 |
|
|
| Age |
|
|
|
|
|
1.05 | 0.789 |
|
≥65 | 30 | 2 | 9 | 11 | 8 |
|
|
|
<65 | 54 | 2 | 13 | 20 | 19 |
|
|
| Location |
|
|
|
|
| 14.96 | 0.021 |
|
Rectum | 28 | 2 | 14 | 7 | 5 |
|
|
| Left
colon | 32 | 1 | 3 | 15 | 13 |
|
|
| Right
colon | 24 | 1 | 5 | 9 | 9 |
|
|
| Lymph node
metastasis |
|
|
|
|
| 8.53 | 0.036 |
|
Yes | 34 | 1 | 6 | 10 | 17 |
|
|
| No | 50 | 3 | 16 | 21 | 10 |
|
|
| Tumor
differentiation |
|
|
|
|
| 16.43 | 0.012 |
|
High | 15 | 2 | 7 | 4 | 2 |
|
|
|
Moderate | 52 | 1 | 15 | 21 | 15 |
|
|
|
Low | 17 | 1 | 0 | 6 | 10 |
|
|
| Stage |
|
|
|
|
| 11.64 | 0.009 |
|
A/B | 41 | 4 | 15 | 14 | 8 |
|
|
|
C/D | 43 | 0 | 7 | 17 | 19 |
|
|
Reduction of CapG significantly
inhibits the in vitro motility of HCT116 cells
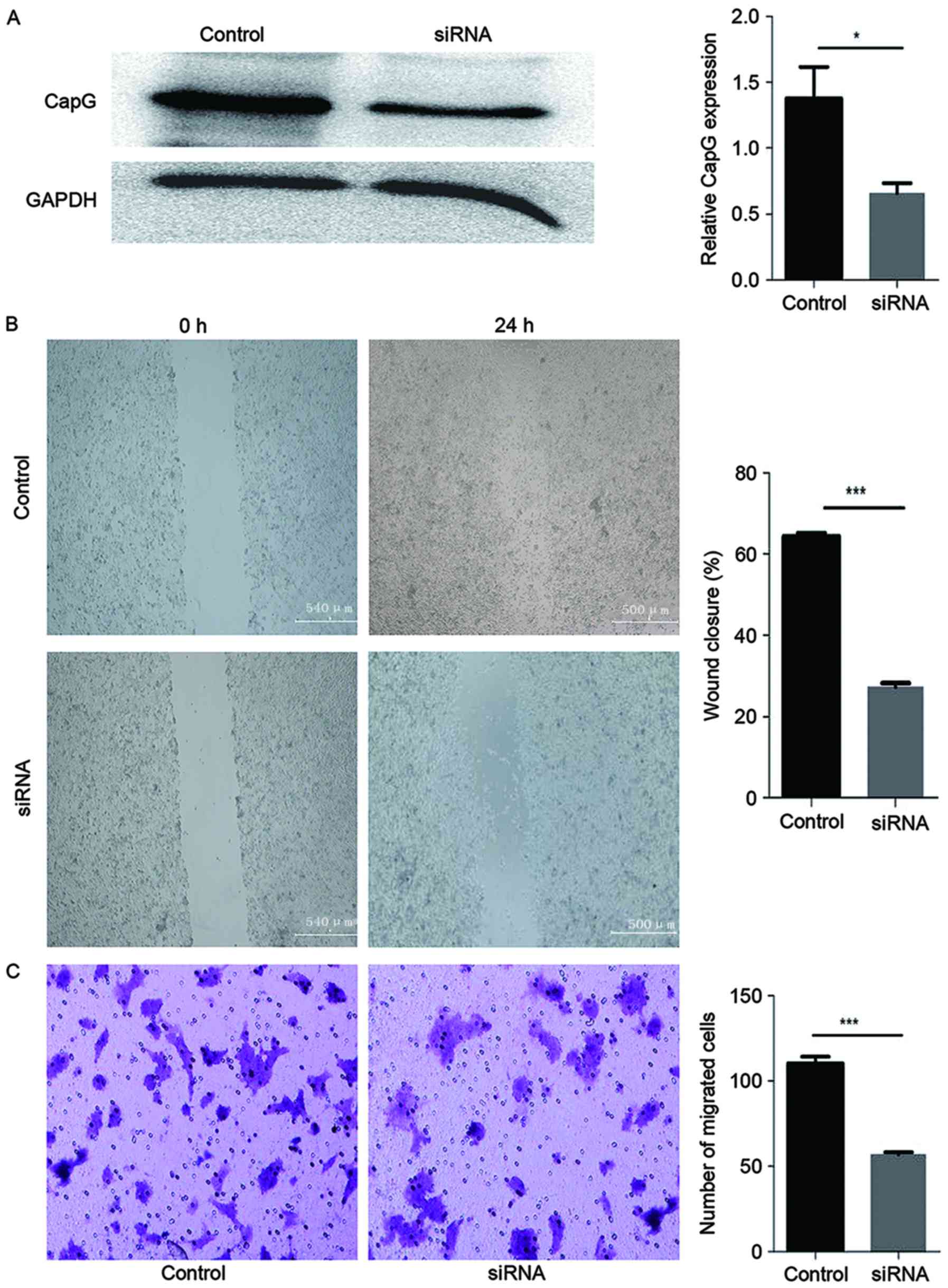
RNAi was effective in reducing the expression level
of CapG protein in HCT116 cells (Fig
3A). CapG levels were diminished in HCT116 cells at 48 h after
transfection with CapG-specific siRNA compared with cells
transfected with the control siRNA.
A migration assay and an in vitro
wound-healing assay were used to assess the effect of CapG
knockdown on cell motility. In the wound healing assay, a wound in
a monolayer of cells transfected with the control siRNA exhibited
>65% wound closure. In contrast, cells transfected with the
CapG-targeting siRNA exhibited <27% wound closure. This
demonstrated that the capacity for cell migration significantly
decreased when CapG expression was suppressed (P<0.001; Fig. 3B).
The result of the cell migration assay was
consistent with the result of the wound healing assay. Transfection
of the HCT116 cells with the siRNA against CapG resulted in a
reduced extent of motility compared with corresponding cells
treated with control siRNA, as demonstrated by the reduced rate of
migration (P<0.001; Fig. 3C).
Discussion
CRC is the third most common type of cancer
worldwide (8). The 5-year survival
rate is ~90% for patients with early stage CRC, but decreases to
<10% in patients with distant metastasis (9). Therefore, it is necessary to identify
mCRC risk-associated biomarkers.
CapG, a 348-amino acid protein, belongs to the
actin-binding protein family, is ubiquitously expressed in normal
tissue, with particular abundance in macrophages (10,11), and
is associated with cell signaling, receptor-mediated membrane
ruffling, phagocytosis and motility (12,13). It
has been reported to modulate the motility of cells by interacting
differentially with the actin cytoskeleton (14). CapG was originally isolated from the
cytoplasm of alveolar macrophages and has been demonstrated to be
involved in the control of actin-based cell motility and membrane
ruffling (phagocytosis) of non-muscle cells; it may also function
as a nuclear actin-binding protein to prevent nuclear actin from
polymerizing and maintain it in a monomeric globular or short
oligomeric form (14). Alteration to
CapG can change the cell morphology and reduce motility (15), particularly important features of
cancer cells during invasion and metastasis.
It has been reported that CapG is associated with
invasion and metastasis in various types of malignancy (15–17).
However, to the best of our knowledge, the effect of CapG in
primary CRC was not investigated prior to the present study. In the
present study, the expression and function of CapG in CRC were
investigated, and the results demonstrated that the expression
level of CapG protein in CRC tissue was significantly higher than
in non-tumor tissue. This observation was consistent with the
results of studies regarding other types of cancer, including oral
(18), pancreatic (19), ovarian (20–22) and
breast cancer (23). In the present
study, the positive expression of CapG in the cytoplasm and nucleus
was significantly associated with CRC location, differentiation,
LMN status and clinical stage. Other clinical studies of
nasopharyngeal carcinoma (24),
non-small cell lung cancer (25) and
cholangiocarcinoma (26) concluded
that high CapG expression was associated with poor differentiation
and clinical stage, and that the patients with CapG-positive tumors
exhibited a worse prognosis. Ichikawa et al (15) performed two-dimensional gel
electrophoresis to obtain protein expression profiles for 3,228
proteins, and identified that CapG was upregulated in the tumor
tissues of patients with LNM. This is consistent with the result of
the present study. Furthermore, although there was no identified
statistical significance, the expression of CapG trended towards an
association with DFS time, which implies that CapG may be useful in
establishing a prognosis in CRC.
Clinical studies of pancreatic ductal and lung
adenocarcinoma have also demonstrated that high CapG expression was
associated with an increased tumor size (19,25).
Morofuji et al studied the proteomic profile of
cholangiocarcinoma and identified CapG expression as a novel
biomarker for predicting the response to gemcitabine treatment, and
as a prognostic indicator in cholangiocarcinoma (26). However, further validation studies are
required to establish whether CapG may exhibit similar functions in
CRC.
The IHC results revealed that the positive
expression of CapG in the cytoplasm and nucleus was significantly
associated with the location of the CRC tumor; the highest
expression of CapG was identified in tumors from the left side of
the colon. Compared with CRC from the right side of the colon,
left-sided colon cancer is more likely to metastasize (27,28). An
additional study identified that CRC that metastasized to the liver
had a higher expression of CapG (29). This result may support the conclusion
from the present study that CapG in CRC was significantly
associated with the tumor site, as CRC from different locations is
associated with different rates of liver metastasis (28).
When CapG expression was suppressed in the present
study, the motility of CRC cells was reduced. In the migration and
in vitro wound-healing assays, cell migration was
significantly inhibited following the transfection of HCT116 cells
with siRNA against CapG. This suggests that CapG contributes to the
motility of CRC cells. Other studies have reported that the
knockdown of CapG in hepatocellular carcinoma (17) and pancreatic cancer (19) cells can attenuate cancer cell
invasion, motility and aggression. However, Watari et al
(30) identified that CapG may act as
a tumor suppressor in stomach cancer, lung cancer and melanoma.
Therefore, the role of CapG in tumor cells may depend on the cell
type.
The nuclear import of CapG is energy dependent and
requires the cytosolic receptor importin β (31). It has been reported that the
overexpression of nuclear CapG, but not cytoplasmic CapG,
predominantly contributes to cell invasion (31). Whether cytoplasmic or nuclear CapG
expression promote cell invasion specifically in CRC may require
further study.
Taken together, the results of the present study
demonstrated that CapG expression was increased in CRC tissue and
associated with poor prognostic risk factors. The observation that
the downregulation of CapG in CRC cells diminished their motility
may imply the involvement of CapG in the motility, and consequently
the dissemination, of CRC cells. These findings may provide novel
insights to understanding the molecular mechanisms of CRC
metastasis, and CapG may be a potential biomarker for predicting
the prognosis of CRC.
Acknowledgements
The authors would like to thank the Department of
Gastroenterology, Zhongnan Hospital of Wuhan University, the
Clinical Center for Intestinal and Colorectal Diseases of Hubei
Province, the Key Laboratory of Intestinal and Colorectal Diseases
of Hubei Province, and the Institute of Virology, Wuhan University,
for technical support and guidance during this study.
References
|
1
|
Ferlay J, Autier P, Boniol M, Heanue M,
Colombet M and Boyle P: Estimates of the cancer incidence and
mortality in Europe in 2006. Ann Oncol. 18:581–592. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Manfredi S, Lepage C, Hatem C, Coatmeur O,
Faivre J and Bouvier AM: Epidemiology and management of liver
metastases from colorectal cancer. Ann Surg. 244:254–259. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Poston GJ, Figueras J, Giuliante F, Nuzzo
G, Sobrero AF, Gigot JF, Nordlinger B, Adam R, Gruenberger T, Choti
MA, et al: Urgent need for a new staging system in advanced
colorectal cancer. J Clin Oncol. 26:4828–4833. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Witke W, Li W, Kwiatkowski DJ and
Southwick FS: Comparisons of CapG and gelsolin-null macrophages:
Demonstration of a unique role for CapG in receptor-mediated
ruffling, phagocytosis and vesicle rocketing. J Cell Biol.
154:775–784. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dahl E, Sadr-Nabavi A, Klopocki E, Betz B,
Grube S, Kreutzfeld R, Himmelfarb M, An HX, Gelling S, Klaman I, et
al: Systematic identification and molecular characterization of
genes differentially expressed in breast and ovarian cancer. J
Pathol. 205:21–28. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Harpaz N and Saxena R: Modern surgical
pathologyGastrointestinal Tract, Large Intestine. Weidner N, Cote
RJ, Suster S and Weiss LM: 1. 1st. Saunders, Philadelphia: pp.
749–852. 2003
|
|
7
|
Weber A, Kristiansen I, Johannsen M,
Oelrich B, Scholmann K, Gunia S, May M, Meyer HA, Behnke S, Moch H
and Kristiansen G: The FUSE binding proteins FBP1 and FBP3 are
potential c-myc regulators in renal, but not in prostate and
bladder cancer. BMC Cancer. 8:3692008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Harrison S and Benziger H: The molecular
biology of colorectal carcinoma and its implications: A review.
Surgeon. 9:200–210. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Miyoshi N, Ohue M, Shingai T, Noura S,
Sugimura K, Akita H, Gotoh K, Motoori M, Takahashi H, Kishi K, et
al: Clinicopathological characteristics and prognosis of stage IV
colorectal cancer. Mol Clin Oncol. 3:1093–1098. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gau DM, Lesnock JL, Hood BL, Bhargava R,
Sun M, Darcy K, Luthra S, Chandran U, Conrads TP, Edwards RP, et
al: BRCA1 deficiency in ovarian cancer is associated with
alteration in expression of several key regulators of cell
motility-A proteomics study. Cell Cycle. 14:1884–1892. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kobayashi M, Nagashio R, Ryuge S, Murakami
Y, Yanagita K, Nakashima H, Matsumoto T, Jiang SX, Saegusa M, Satoh
Y, et al: Acquisition of useful sero-diagnostic autoantibodies
using the same patients'sera and tumor tissues. Biomed Res.
35:133–143. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Morelli M, Scumaci D, Di Cello A,
Venturella R, Donato G, Faniello MC, Quaresima B, Cuda G, Zullo F
and Costanzo F: DJ-1 in endometrial cancer: A possible biomarker to
improve differential diagnosis between subtypes. Int J Gynecol
Cancer. 24:649–658. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Da Costa GG, Gomig TH, Kaviski R, Sousa
Santos K, Kukolj C, De Lima RS, De Andrade Urban C, Cavalli IJ and
Ribeiro EM: Comparative proteomics of tumor and paired normal
breast tissue highlights potential biomarkers in breast cancer.
Cancer Genomics Proteomics. 12:251–261. 2015.PubMed/NCBI
|
|
14
|
Renz M and Langowski J: Dynamics of the
CapG actin-binding protein in the cell nucleus studied by FRAP and
FCS. Chromosome Res. 16:427–437. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ichikawa H, Kanda T, Kosugi S, Kawachi Y,
Sasaki H, Wakai T and Kondo T: Laser microdissection and
two-dimensional difference gel electrophoresis reveal the role of a
novel macrophage-capping protein in lymph node metastasis in
gastric cancer. J Proteome Res. 12:3780–3791. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Glaser J, Neumann MH, Mei Q, Betz B, Seier
N, Beyer I, Fehm T, Neubauer H, Niederacher D and Fleisch MC:
Macrophage capping protein CapG is a putative oncogene involved in
migration and invasiveness in ovarian carcinoma. Biomed Res Int.
2014:3798472014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kimura K, Ojima H, Kubota D, Sakumoto M,
Nakamura Y, Tomonaga T, Kosuge T and Kondo T: Proteomic
identification of the macrophage-capping protein as a protein
contributing to the malignant features of hepatocellular carcinoma.
J Proteomics. 78:362–373. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nomura H, Uzawa K, Ishigami T, Kouzu Y,
Koike H, Ogawara K, Siiba M, Bukawa H, Yokoe H, Kubosawa H and
Tanzawa H: Clinical significance of gelsolin-like actin-capping
protein expression in oral carcinogenesis: An immunohistochemical
study of premalignant and malignant lesions of the oral cavity. BMC
Cancer. 8:392008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thompson CC, Ashcroft FJ, Patel S, Saraga
G, Vimalachandran D, Prime W, Campbell F, Dodson A, Jenkins RE,
Lemoine NR, et al: Pancreatic cancer cells overexpress gelsolin
family-capping proteins, which contribute to their cell motility.
Gut. 56:95–1067. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ouellet V, Provencher DM, Maugard CM, Le
Page C, Ren F, Lussier C, Novak J, Ge B, Hudson TJ, Tonin PN and
Mes-Masson AM: Discrimination between serous low malignant
potential and invasive epithelial ovarian tumors using molecular
profiling. Oncogene. 24:4672–4687. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Partheen K, Levan K, Osterberg L, Claesson
I, Fallenius G, Sundfeldt K and Horvath G: Four potential
biomarkers as prognostic factors in stage III serous ovarian
adenocarcinomas. Int J Cancer. 123:2130–2137. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang F, Li C, Liu H, Wang Y, Chen Y and
Wu X: The functional proteomics analysis of VEGF-treated human
epithelial ovarian cancer cells. Tumour Biol. 35:12379–12387. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kang S, Kim MJ, An H, Kim BG, Choi YP,
Kang KS, Gao MQ, Park H, Na HJ, Kim HK, et al: Proteomic molecular
portrait of interface zone in breast cancer. J Proteome Res.
9:5638–5645. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li MX, Xiao ZQ, Chen YH, Peng F, Li C,
Zhang PF, Li MY, Li F, Duan CJ, Li DJ, et al: Proteomic analysis of
the stroma-related proteins in nasopharyngeal carcinoma and normal
nasopharyngeal epithelial tissues. Med Oncol. 27:134–144. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu WY, Hunag YY, Liu XG, He JY, Chen DD,
Zeng F, Zhou JH and Zhang YK: Prognostic evaluation of CapG,
gelsolin, P-gp, GSTP1, and Topo-II proteins in non-small cell lung
cancer. Anat Rec (Hoboken). 295:208–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Morofuji N, Ojima H, Onaya H, Okusaka T,
Shimada K, Sakamoto Y, Esaki M, Nara S, Kosuge T, Asahina D, et al:
Macrophage-capping protein as a tissue biomarker for prediction of
response to gemcitabine treatment and prognosis in
cholangiocarcinoma. J Proteomics. 75:1577–1589. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Komuro K, Tada M, Tamoto E, Kawakami A,
Matsunaga A, Teramoto K, Shindoh G, Takada M, Murakawa K, Kanai M,
et al: Right- and left-sided colorectal cancers display distinct
expression profiles and the anatomical stratification allows a high
accuracy prediction of lymph node metastasis. J Surg Res.
124:216–224. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Konopke R, Distler M, Ludwig S and
Kersting S: Location of liver metastases reflects the site of the
primary colorectal carcinoma. Scand J Gastroenterol. 43:192–195.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kang B, Hao C, Wang H, Zhang J, Xing R,
Shao J, Li W, Xu N, Lu Y and Liu S: Evaluation of
hepatic-metastasis risk of colorectal cancer upon the protein
signature of PI3K/AKT pathway. J Proteome Res. 7:3507–3515. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Watari A, Takaki K, Higashiyama S, Li Y,
Satomi Y, Takao T, Tanemura A, Yamaguchi Y, Katayama I, Shimakage
M, et al: Suppression of tumorigenicity, but not anchorage
independence, of human cancer cells by new candidate tumor
suppressor gene CapG. Oncogene. 25:7373–7380. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
De Corte V, Van Impe K, Bruyneel E,
Boucherie C, Mareel M, Vandekerckhove J and Gettemans J: Increased
importin-beta-dependent nuclear import of the actin modulating
protein CapG promotes cell invasion. J Cell Sci. 117:5283–5292.
2004. View Article : Google Scholar : PubMed/NCBI
|