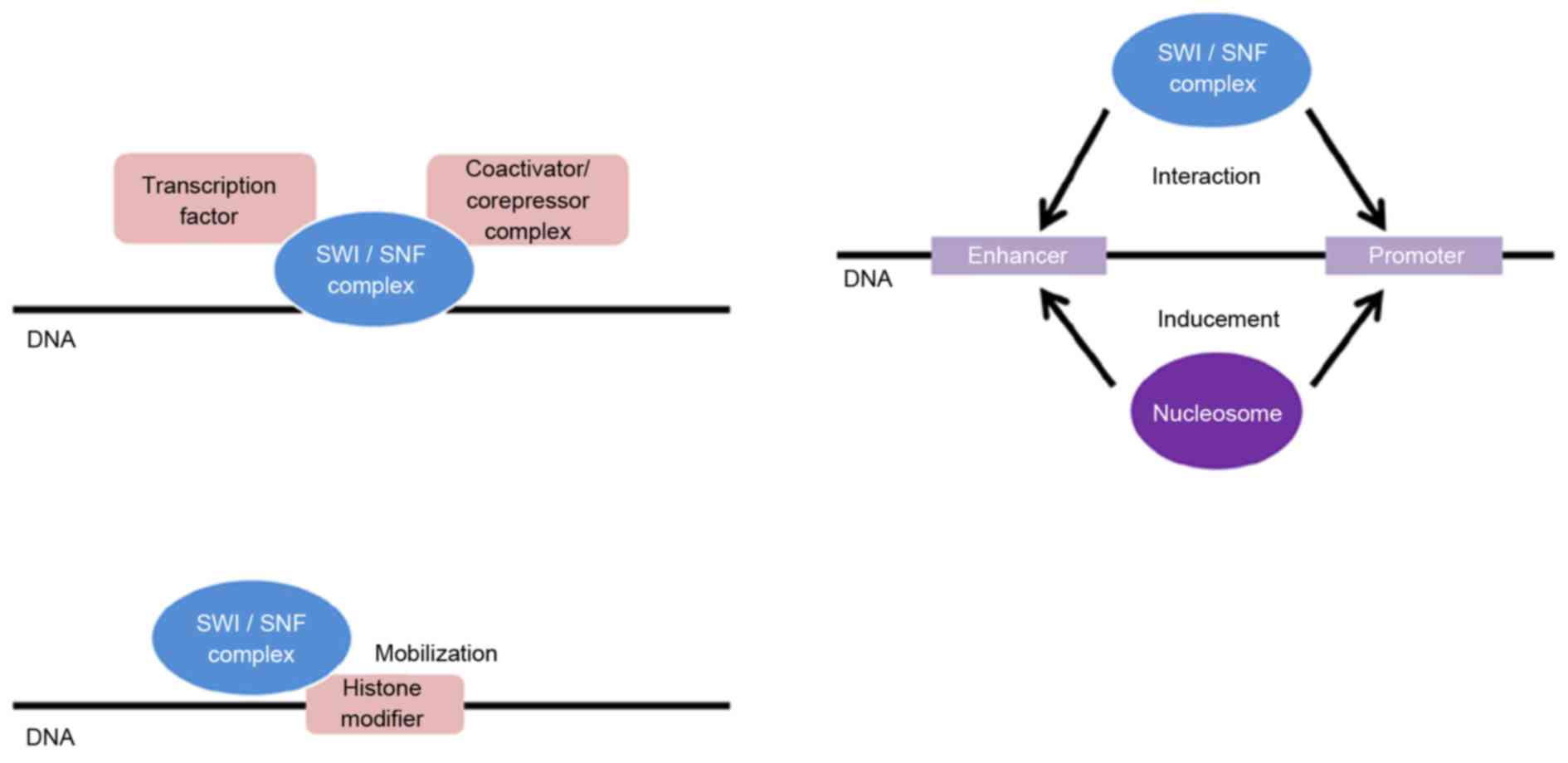
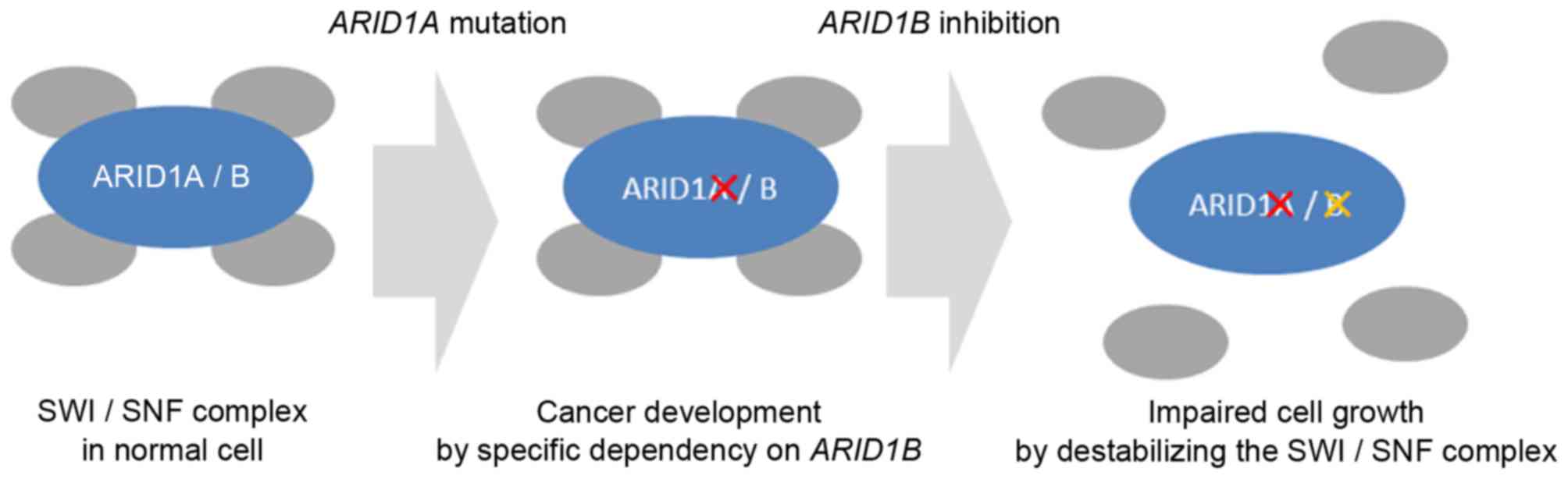
|
1
|
Weaver IC, Korgan AC, Lee K, Wheeler RV,
Hundert AS and Goguen D: Stress and the emerging roles of chromatin
remodeling in signal integration and stable transmission of
reversible phenotypes. Front Behav Neurosci. 11:412017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Takeda T, Banno K, Okawa R, Yanokura M,
Iijima M, Irie-Kunitomi H, Nakamura K, Iida M, Adachi M, Umene K,
et al: ARID1A gene mutation in ovarian and endometrial cancers
(Review). Oncol Rep. 35:607–613. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Clapier CR and Cairns BR: The biology of
chromatin remodeling complexes. Annu Rev Biochem. 78:273–304. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ronan JL, Wu W and Crabtree GR: From
neural development to cognition: Unexpected roles for chromatin.
Nat Rev Genet. 14:347–359. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang B, Jiang C and Zhang R: Epigenetics:
The language of the cell? Epigenomics. 6:73–88. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Alberts B, Johnson A, Lewis J, Raff M,
Roberts K and Walter P: Molecular biology of the cell, 5th edition.
Science. 215–216. 2008.PubMed/NCBI
|
|
7
|
Wilson BG and Roberts CW: SWI/SNF
nucleosome remodellers and cancer. Nat Rev Cancer. 11:481–492.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Oike T, Ogiwara H, Nakano T, Yokota J and
Kohno T: Inactivating mutations in SWI/SNF chromatin remodeling
genes in human cancer. Jpn J Clin Oncol. 43:849–855. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jones S, Wang TL, Shih IeM, Mao TL,
Nakayama K, Roden R, Glas R, Slamon D, Diaz LA Jr, Vogelstein B, et
al: Frequent mutations of chromatin remodeling gene ARID1A in
ovarian clear cell carcinoma. Science. 330:228–231. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mayes K, Qiu Z, Alhazmi A and Landry JW:
ATP-dependent chromatin remodeling complexes as novel targets for
cancer therapy. Adv Cancer Res. 121:183–233. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li WD, Li QR, Xu SN, Wei FJ, Ye ZJ, Cheng
JK and Chen JP: Exome sequencing identifies an MLL3 gene germ line
mutation in a pedigree of colorectal cancer and acute myeloid
leukemia. Blood. 121:1478–1479. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fujimoto A, Totoki Y, Abe T, Boroevich KA,
Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, et al:
Whole-genome sequencing of liver cancers identifies etiological
influences on mutation patterns and recurrent mutations in
chromatin regulators. Nat Genet. 44:760–764. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao
S, Wu R, Chen C, Li X, Zhou L, et al: Frequent mutations of
chromatin remodeling genes in transitional cell carcinoma of the
bladder. Nat Genet. 43:875–878. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lindberg J, Mills IG, Klevebring D, Liu W,
Neiman M, Xu J, Wikström P, Wiklund P, Wiklund F, Egevad L and
Grönberg H: The mitochondrial and autosomal mutation landscapes of
prostate cancer. Eur Urol. 63:702–708. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Watanabe Y, Castoro RJ, Kim HS, North B,
Oikawa R, Hiraishi T, Ahmed SS, Chung W, Cho MY, Toyota M, et al:
Frequent alteration of MLL3 frameshift mutations in microsatellite
deficient colorectal cancer. PLoS One. 6:e233202011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zang ZJ, Cutcutache I, Poon SL, Zhang SL,
McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, et al:
Exome sequencing of gastric adenocarcinoma identifies recurrent
somatic mutations in cell adhesion and chromatin remodeling genes.
Nat Genet. 44:570–574. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu P, Morrison C, Wang L, Xiong D, Vedell
P, Cui P, Hua X, Ding F, Lu Y, James M, et al: Identification of
somatic mutations in non-small cell lung carcinomas using
whole-exome sequencing. Carcinogenesis. 33:1270–1276. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ellis MJ, Ding L, Shen D, Luo J, Suman VJ,
Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, et al:
Whole-genome analysis informs breast cancer response to aromatase
inhibition. Nature. 486:353–360. 2012.PubMed/NCBI
|
|
19
|
Biankin AV, Waddell N, Kassahn KS, Gingras
MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J,
et al: Pancreatic cancer genomes reveal aberrations in axon
guidance pathway genes. Nature. 491:399–405. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Je EM and Lee SH, Yoo NJ and Lee SH:
Mutational and expressional analysis of MLL genes in gastric and
colorectal cancers with microsatellite instability. Neoplasma.
60:188–195. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu JN and Roberts CW: ARID1A mutations in
cancer: Another epigenetic tumor suppressor? Cancer Discov.
3:35–43. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Trotter KW, Fan HY, Ivey ML, Kingston RE
and Archer TK: The HSA domain of BRG1 mediates critical
interactions required for glucocorticoid receptor-dependent
transcriptional activation in vivo. Mol Cell Biol. 28:1413–1426.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Inoue H, Furukawa T, Giannakopoulos S,
Zhou S, King DS and Tanese N: Largest subunits of the human SWI/SNF
chromatin-remodeling complex promote transcriptional activation by
steroid hormone receptors. J Biol Chem. 277:41674–41685. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang HN, Lin MC, Huang WC, Chiang YC and
Kuo KT: Loss of ARID1A expression and its relationship with
PI3K-Akt pathway alterations and ZNF217 amplification in ovarian
clear cell carcinoma. Mod Pathol. 27:983–990. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Biegel JA, Busse TM and Weissman BE:
SWI/SNF chromatin remodeling complexes and cancer. Am J Med Genet C
Semin Med Genet. 166C:350–366. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Reisman D, Glaros S and Thompson EA: The
SWI/SNF complex and cancer. Oncogene. 28:1653–1668. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yamada M, Sato N, Ikeda S, Arai T, Sawabe
M, Mori S, Yamada Y, Muramatsu M and Tanaka M: Association of the
chromodomain helicase DNA-binding protein 4 (CHD4) missense
variation p.D140E with cancer: Potential interaction with smoking.
Genes Chromosomes Cancer. 54:122–128. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao S, Choi M, Overton JD, Bellone S,
Roque DM, Cocco E, Guzzo F, English DP, Varughese J, Gasparrini S,
et al: Landscape of somatic single-nucleotide and copy-number
mutations in uterine serous carcinoma. Proc Natl Acad Sci USA.
110:2916–2921. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Le Gallo M, O'Hara AJ, Rudd ML, Urick ME,
Hansen NF, O'Neil NJ, Price JC, Zhang S, England BM, Godwin AK, et
al: Exome sequencing of serous endometrial tumors identifies
recurrent somatic mutations in chromatin-remodeling and ubiquitin
ligase complex genes. Nat Genet. 44:1310–1315. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim MS, Chung NG, Kang MR, Yoo NJ and Lee
SH: Genetic and expressional alterations of CHD genes in gastric
and colorectal cancers. Histopathology. 58:660–668. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sasaki MM, Skol AD, Bao R, Rhodes LV,
Chambers R, Vokes EE, Cohen EE and Onel K: Integrated genomic
analysis suggests MLL3 is a novel candidate susceptibility gene for
familial nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers
Prev. 24:1222–1228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Villacis RA, Miranda PM, Gomy I, Santos
EM, Carraro DM, Achatz MI, Rossi BM and Rogatto SR: Contribution of
rare germline copy number variations and common susceptibility loci
in Lynch syndrome patients negative for mutations in the mismatch
repair genes. Int J Cancer. 138:1928–1935. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim KH and Roberts CW: Targeting EZH2 in
cancer. Nat Med. 22:128–134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang Q, Laknaur A, Elam L, Ismail N,
Gavrilova-Jordan L, Lue J, Diamond MP and Al-Hendy A:
Identification of polycomb group protein EZH2-mediated DNA mismatch
repair gene MSH2 in human uterine fibroids. Reprod Sci.
23:1314–1325. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cai L, Wang Z and Liu D: Interference with
endogenous EZH2 reverses the chemotherapy drug resistance in
cervical cancer cells partly by up-regulating Dicer expression.
Tumour Biol. 37:6359–6369. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lu C, Han HD, Mangala LS, Ali-Fehmi R,
Newton CS, Ozbun L, Armaiz-Pena GN, Hu W, Stone RL, Munkarah A, et
al: Regulation of tumor angiogenesis by EZH2. Cancer Cell.
18:185–197. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Crotzer DR, Sun CC, Coleman RL, Wolf JK,
Levenback CF and Gershenson DM: Lack of effective systemic therapy
for recurrent clear cell carcinoma of the ovary. Gynecol Oncol.
105:404–408. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Viganó P, Somigliana E, Chiodo I, Abbiati
A and Vercellini P: Molecular mechanisms and biological
plausibility underlying the malignant transformation of
endometriosis: A critical analysis. Hum Reprod Update. 12:77–89.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nishikimi K, Kiyokawa T, Tate S, Iwamoto M
and Shozu M: ARID1A expression in ovarian clear cell carcinoma with
an adenofibromatous component. Histopathology. 67:866–871. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nagl NG Jr, Wang X, Patsialou A, Van Scoy
M and Moran E: Distinct mammalian SWI/SNF chromatin remodeling
complexes with opposing roles in cell-cycle control. EMBO J.
26:752–763. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Weissman B and Knudsen KE: Hijacking the
chromatin remodeling machinery: Impact of SWI/SNF perturbations in
cancer. Cancer Res. 69:8223–8230. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wiegand KC, Shah SP, Al-Agha OM, Zhao Y,
Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, et
al: ARID1A mutations in endometriosis-associated ovarian
carcinomas. N Engl J Med. 363:1532–1543. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Itamochi H, Oumi N, Oishi T, Shoji T,
Fujiwara H, Sugiyama T, Suzuki M, Kigawa J and Harada T: Loss of
ARID1A expression is associated with poor prognosis in patients
with stage I/II clear cell carcinoma of the ovary. Int J Clin
Oncol. 20:967–973. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Conlon N, Silva A, Guerra E, Jelinic P,
Schlappe BA, Olvera N, Mueller JJ, Tornos C, Jungbluth AA, Young
RH, et al: Loss of SMARCA4 expression is both sensitive and
specific for the diagnosis of small cell carcinoma of ovary,
hypercalcemic type. Am J Surg Pathol. 40:395–403. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jelinic P, Mueller JJ, Olvera N, Dao F,
Scott SN, Shah R, Gao J, Schultz N, Gonen M, Soslow RA, et al:
Recurrent SMARCA4 mutations in small cell carcinoma of the ovary.
Nat Genet. 46:424–426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ramos P, Karnezis AN, Craig DW, Sekulic A,
Russell ML, Hendricks WP, Corneveaux JJ, Barrett MT, Shumansky K,
Yang Y, et al: Small cell carcinoma of the ovary, hypercalcemic
type, displays frequent inactivating germline and somatic mutations
in SMARCA4. Nat Genet. 46:427–429. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Witkowski L, Carrot-Zhang J, Albrecht S,
Fahiminiya S, Hamel N, Tomiak E, Grynspan D, Saloustros E, Nadaf J,
Rivera B, et al: Germline and somatic SMARCA4 mutations
characterize small cell carcinoma of the ovary, hypercalcemic type.
Nat Genet. 46:438–443. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Schneppenheim R, Frühwald MC, Gesk S,
Hasselblatt M, Jeibmann A, Kordes U, Kreuz M, Leuschner I, Subero
Martin JI, Obser T, et al: Germline nonsense mutation and somatic
inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor
predisposition syndrome. Am J Hum Genet. 86:279–284. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yoshimoto T, Matsubara D, Nakano T, Tamura
T, Endo S, Sugiyama Y and Niki T: Frequent loss of the expression
of multiple subunits of the SWI/SNF complex in large cell carcinoma
and pleomorphic carcinoma of the lung. Pathol Int. 65:595–602.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Helming KC, Wang X and Roberts CW:
Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer Cell.
26:309–317. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sherman ME: Theories of endometrial
carcinogenesis: A multidisciplinary approach. Mod Pathol.
13:295–308. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Polo SE, Kaidi A, Baskcomb L, Galanty Y
and Jackson SP: Regulation of DNA-damage responses and cell-cycle
progression by the chromatin remodelling factor CHD4. EMBO J.
29:3130–3139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Duman BB, Kara IO, Günaldi M and Ercolak
V: Malignant mixed Mullerian tumor of the ovary with two cases and
review of the literature. Arch Gynecol Obstet. 283:1363–1368. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sharma NK, Sorosky JI, Bender D, Fletcher
MS and Sood AK: Malignant mixed mullerian tumor (MMMT) of the
cervix. Gynecol Oncol. 97:442–445. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ahuja A, Safaya R, Prakash G, Kumar L and
Shukla NK: Primary mixed mullerian tumor of the vagina - a case
report with review of the literature. Pathol Res Pract.
207:253–255. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
George EM, Herzog TJ, Neugut AI, Lu YS,
Burke WM, Lewin SN, Hershman DL and Wright JD: Carcinosarcoma of
the ovary: Natural history, patterns of treatment, and outcome.
Gynecol Oncol. 131:42–45. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Jones S, Stransky N, McCord CL, Cerami E,
Lagowski J, Kelly D, Angiuoli SV, Sausen M, Kann L, Shukla M, et
al: Genomic analyses of gynaecologic carcinosarcomas reveal
frequent mutations in chromatin remodelling genes. Nat Commun.
5:50062014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lee J, Kim DH, Lee S, Yang QH, Lee DK, Lee
SK, Roeder RG and Lee JW: A tumor suppressive coactivator complex
of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase
MLL3 or its paralogue MLL4. Proc Natl Acad Sci USA. 106:8513–8518.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kwon JE, La M, Oh KH, Oh YM, Kim GR, Seol
JH, Baek SH, Chiba T, Tanaka K, Bang OS, et al: BTB
domain-containing speckle-type POZ protein (SPOP) serves as an
adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase.
J Biol Chem. 281:12664–12672. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Samartzis EP, Gutsche K, Dedes KJ, Fink D,
Stucki M and Imesch P: Loss of ARID1A expression sensitizes cancer
cells to PI3K- and AKT-inhibition. Oncotarget. 5:5295–5303. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bitler BG, Aird KM, Garipov A, Li H,
Amatangelo M, Kossenkov AV, Schultz DC, Liu Q, Shih IeM,
Conejo-Garcia JR, et al: Synthetic lethality by targeting EZH2
methyltransferase activity in ARID1A-mutated cancers. Nat Med.
21:231–238. 2015.PubMed/NCBI
|
|
62
|
Guan B, Gao M, Wu CH, Wang TL and Shih
IeM: Functional analysis of in-frame indel ARID1A mutations reveals
new regulatory mechanisms of its tumor suppressor functions.
Neoplasia. 14:986–993. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Helming KC, Wang X, Wilson BG, Vazquez F,
Haswell JR, Manchester HE, Kim Y, Kryukov GV, Ghandi M, Aguirre AJ,
et al: ARID1B is a specific vulnerability in ARID1A-mutant cancers.
Nat Med. 20:251–254. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Nagymanyoki Z, Mutter GL, Hornick JL and
Cibas ES: ARID1A is a useful marker of malignancy in peritoneal
washings for endometrial carcinoma. Cancer Cytopathol. 123:253–257.
2015. View Article : Google Scholar : PubMed/NCBI
|