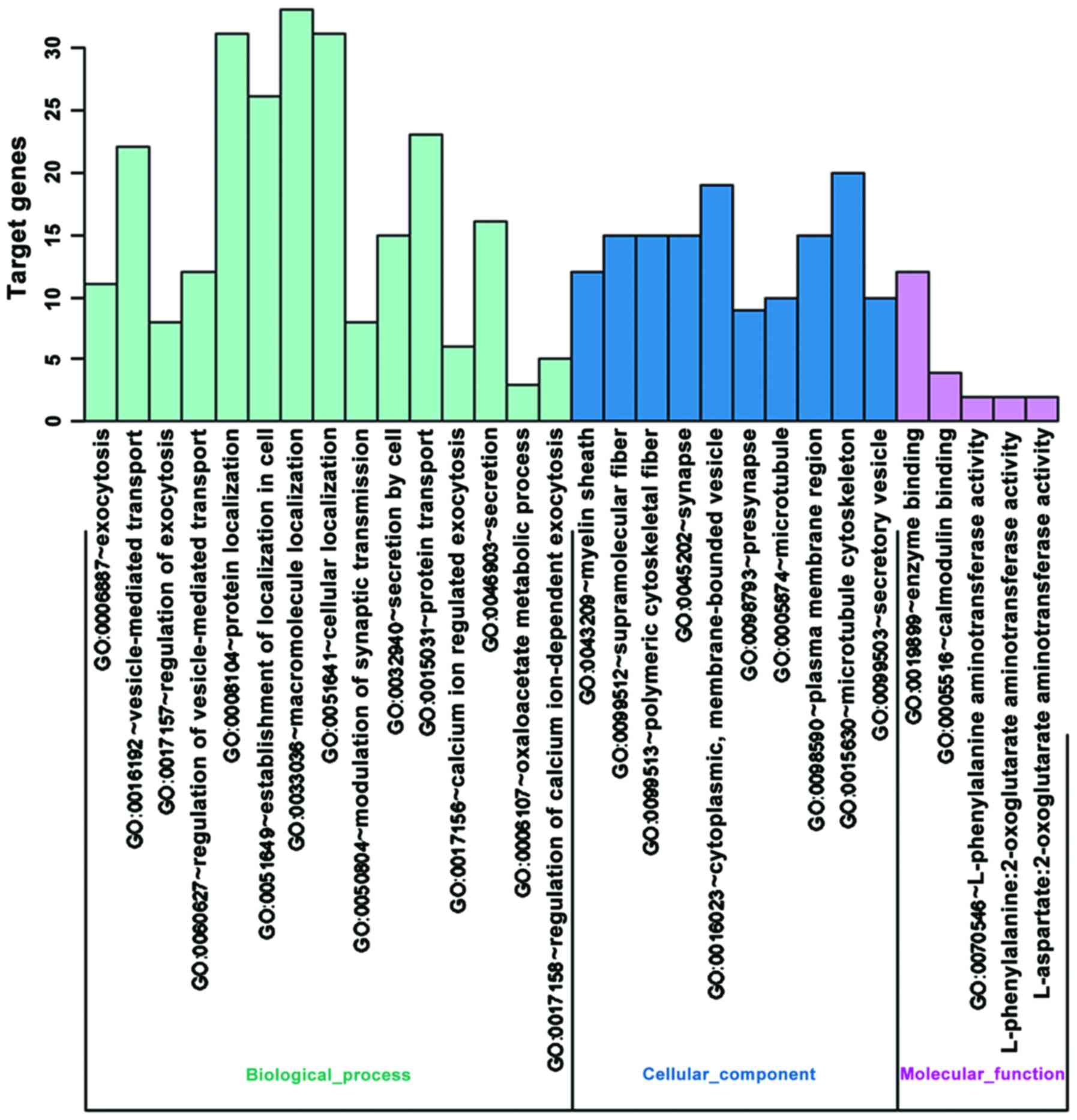
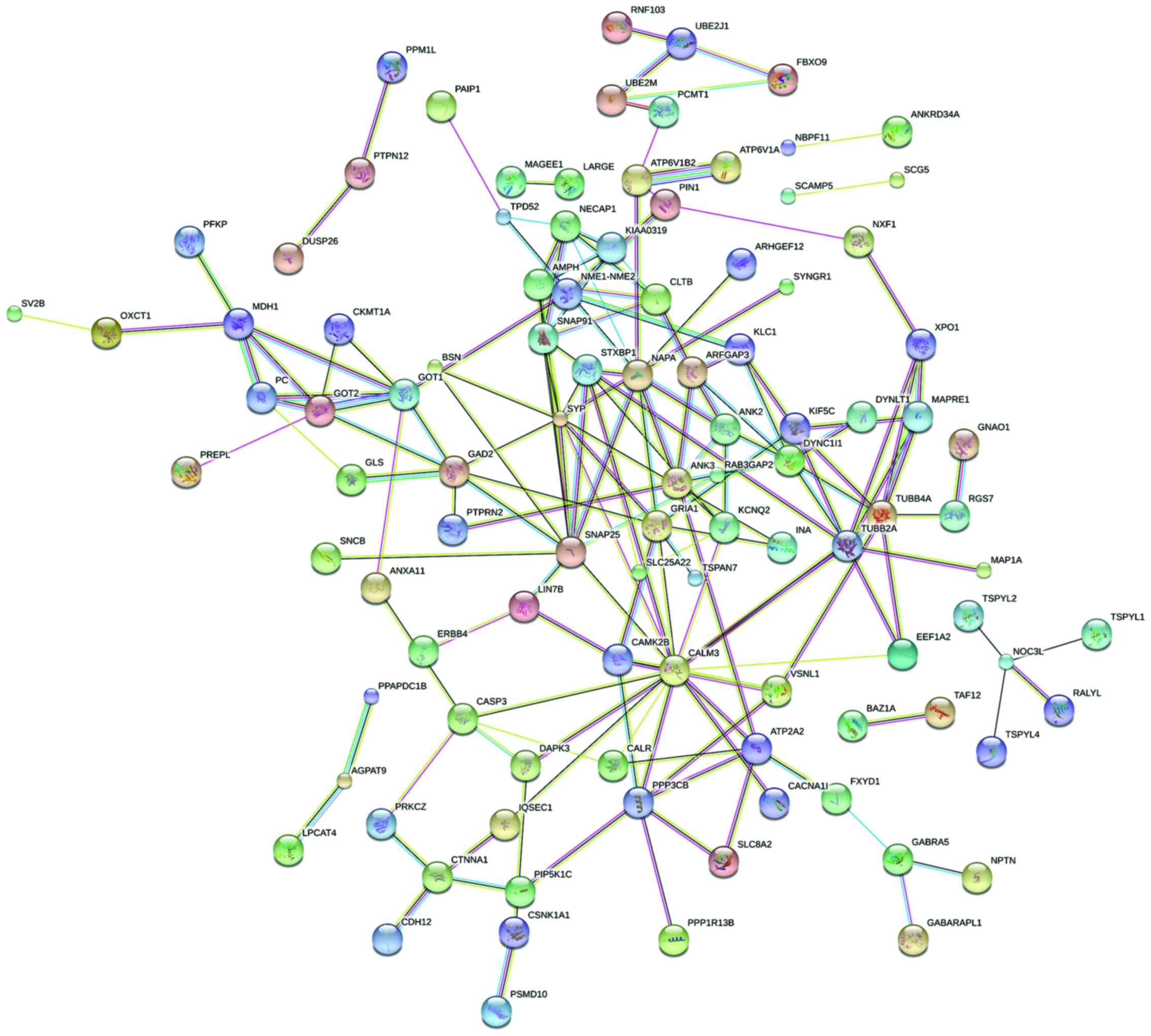
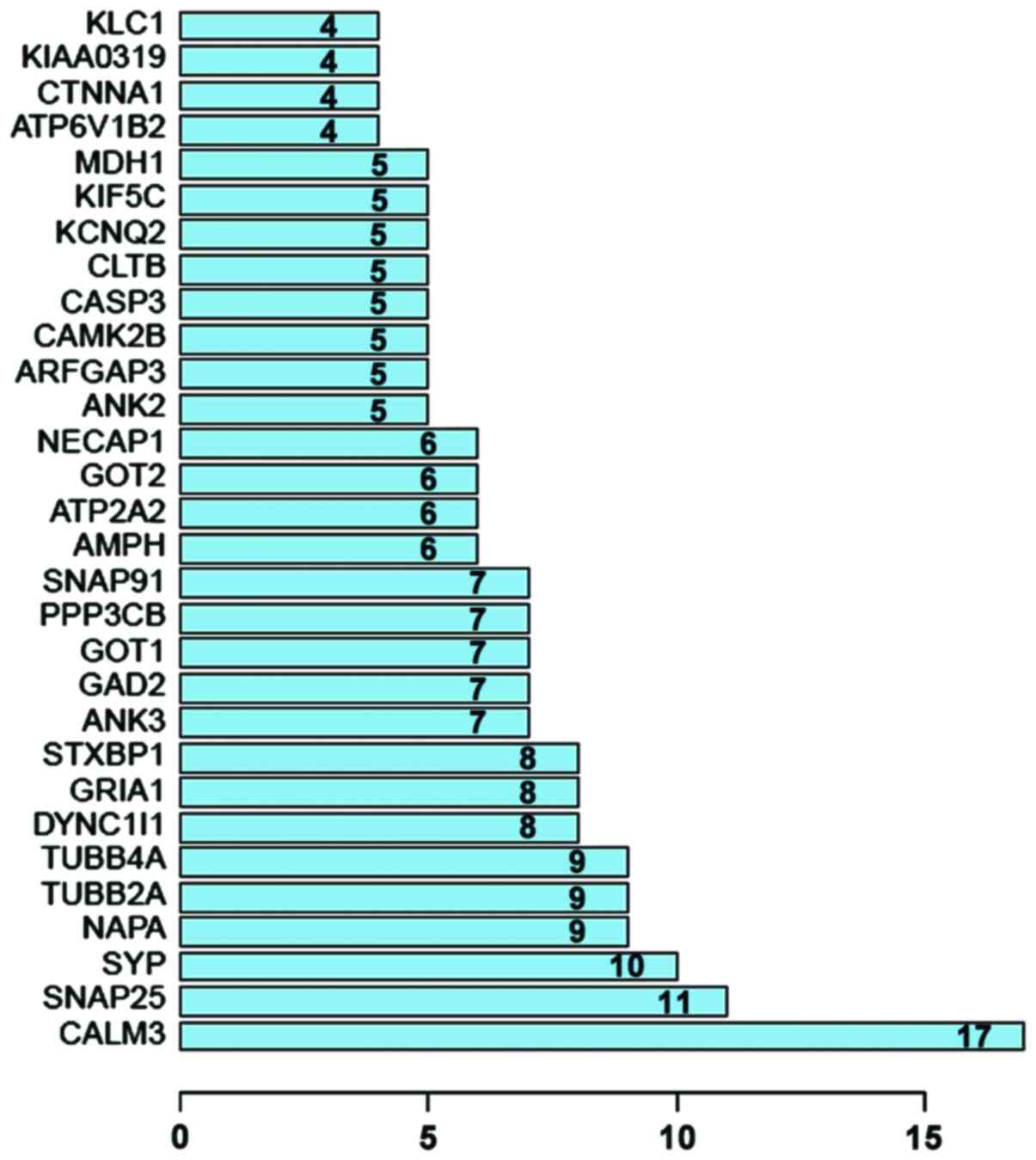
|
1
|
Lathia JD, Mack SC, Mulkearns-Hubert EE,
Valentim CL and Rich JN: Cancer stem cells in glioblastoma. Genes
Dev. 29:1203–1217. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jin Z, Jin RH, Ma C, Li HS and Xu HY:
Serum expression level of miR-504 can differentiate between
glioblastoma multiforme and solitary brain metastasis of non-small
cell lung carcinoma. J BUON. 22:474–480. 2017.PubMed/NCBI
|
|
3
|
Sharma V, Dixit D, Koul N, Mehta VS and
Sen E: Ras regulates interleukin-1β-induced HIF-1α transcriptional
activity in glioblastoma. J Mol Med (Berl). 89:123–136. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Reed GJ, Boczek NJ, Etheridge SP and
Ackerman MJ: CALM3 mutation associated with long QT syndrome. Heart
Rhythm. 12:419–422. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Olar A and Aldape KD: Using the molecular
classification of glioblastoma to inform personalized treatment. J
Pathol. 232:165–177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lun M, Lok E, Gautam S, Wu E and Wong ET:
The natural history of extracranial metastasis from glioblastoma
multiforme. J Neurooncol. 105:261–273. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Goodenberger ML and Jenkins RB: Genetics
of adult glioma. Cancer Genet. 205:613–621. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jones TS and Holland EC: Standard of care
therapy for malignant glioma and its effect on tumor and stromal
cells. Oncogene. 31:1995–2006. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stupp R, Hegi ME, Mason WP, van den Bent
MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B,
Belanger K, et al: European Organisation for Research and Treatment
of Cancer Brain Tumour and Radiation Oncology Groups; National
Cancer Institute of Canada Clinical Trials Group: Effects of
radiotherapy with concomitant and adjuvant temozolomide versus
radiotherapy alone on survival in glioblastoma in a randomised
phase III study: Five-year analysis of the EORTC-NCIC trial. Lancet
Oncol. 10:459–466. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bo LJ, Wei B, Li ZH, Wang ZF, Gao Z and
Miao Z: Bioinformatics analysis of miRNA expression profile between
primary and recurrent glioblastoma. Eur Rev Med Pharmacol Sci.
19:3579–3586. 2015.PubMed/NCBI
|
|
11
|
Carro MS, Lim WK, Alvarez MJ, Bollo RJ,
Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al:
The transcriptional network for mesenchymal transformation of brain
tumours. Nature. 463:318–325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lathia JD, Gallagher J, Myers JT, Li M,
Vasanji A, McLendon RE, Hjelmeland AB, Huang AY and Rich JN: Direct
in vivo evidence for tumor propagation by glioblastoma cancer stem
cells. PLoS One. 6:e248072011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xie Q, Mittal S and Berens ME: Targeting
adaptive glioblastoma: An overview of proliferation and invasion.
Neuro-oncol. 16:1575–1584. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Batchelor TT, Reardon DA, de Groot JF,
Wick W and Weller M: Antiangiogenic therapy for glioblastoma:
Current status and future prospects. Clin Cancer Res. 20:5612–5619.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xie Y, Bergström T, Jiang Y, Johansson P,
Marinescu VD, Lindberg N, Segerman A, Wicher G, Niklasson M,
Baskaran S, et al: The human glioblastoma cell culture resource:
Validated cell models representing all molecular subtypes.
EBioMedicine. 2:1351–1363. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo E, Wang Z and Wang S: miR-200c and
miR-141 inhibit ZEB1 synergistically and suppress glioma cell
growth and migration. Eur Rev Med Pharmacol Sci. 20:3385–3391.
2016.PubMed/NCBI
|
|
17
|
Joo KM, Kim J, Jin J, Kim M, Seol HJ,
Muradov J, Yang H, Choi YL, Park WY, Kong DS, et al:
Patient-specific orthotopic glioblastoma xenograft models
recapitulate the histopathology and biology of human glioblastomas
in situ. Cell Rep. 3:260–273. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meyer M, Reimand J, Lan X, Head R, Zhu X,
Kushida M, Bayani J, Pressey JC, Lionel AC, Clarke ID, et al:
Single cell-derived clonal analysis of human glioblastoma links
functional and genomic heterogeneity. Proc Natl Acad Sci USA.
112:851–856. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ogden AT, Waziri AE, Lochhead RA, Fusco D,
Lopez K, Ellis JA, Kang J, Assanah M, McKhann GM, Sisti MB, et al:
Identification of A2B5+CD133−
tumor-initiating cells in adult human gliomas. Neurosurgery.
62:505–515. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Savary K, Caglayan D, Caja L, Tzavlaki K,
Bin Nayeem S, Bergström T, Jiang Y, Uhrbom L, Forsberg-Nilsson K,
Westermark B, et al: Snail depletes the tumorigenic potential of
glioblastoma. Oncogene. 32:5409–5420. 2013. View Article : Google Scholar : PubMed/NCBI
|