Introduction
Cervical cancer (CC), which is the third most common
cause of cancer in women in the world, is mainly associated with
infection by human papillomavirus (HPV), which is a small,
circular, double-stranded DNA virus that is also associated with
cervical neoplasia, anogenital warts and other anogenital cancers
(1,2).
According to previous reports from the International Agency for
Research on Cancer, HPV could be classified into high-risk (HR)
types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73 and
82), probable HR types (26, 53 and 66) and low-risk types (6, 11,
40, 42, 43, 44, 54, 61, 70, 72, 81 and CP6108) (3).
HR HPV, which is considered to be a necessary
biological factor for CC, can express specific viral genes such as
E6 and E7, which have been observed in CC cell lines and biopsies
(4,5).
These specific viral genes express oncoproteins (such as E6 and E7
proteins) that can be inserted into the host's cell genome
(6). Therefore, E6 and E7 proteins
may be critical for identifying HR HPV, and consequently could be a
potential new biomarker for CC diagnosis.
Currently, the established methods for the screening
of CC in clinical trials include Pap test, ThinPrep cytological
test (TCT), HPV DNA determination by Hybrid Capture 2 (HC2) test,
HPV messenger RNA (mRNA) determination by PreTect HPV-Proofer and
colposcopy (7). However, due to the
low sensitivity of the aforementioned methods, females with high
risk of developing CC require regular re-tests to confirm the
accuracy of the results (7). Among
patients with negative cytology results but positive HPV DNA test
results, a small percentage will develop CC (8). Therefore, an efficient diagnostic method
is required for these particular patients
(TCT−/HPV+).
The present study aimed to evaluate E6/E7 proteins
as the main biomarkers of pre-CC lesions, and to determine the
suitability of E6/E7 protein detection as a potential screening
method for CC diagnosis.
Materials and methods
Subjects
A total of 450 samples from female patients with
suspected cervical intraepithelial neoplasia (CIN) (positive in ≥1
indicator of TCT and HC2 test, excluding carcinoma patients)
according to routine physical examination were collected from the
Cervical Department (International Peace Maternity and Child Health
Hospital, Shanghai, China) from March 2014 to February 2015. Of
these patients, 348 were diagnosed as CIN2+ and the rest
were diagnosed as CIN2−. The patients were aged from 18
to 45 years old, and had not been subjected to any particular
treatment, including hysterectomy, radiotherapy or chemotherapy.
Each sample was analyzed by cytological test, HPV DNA detection by
polymerase chain reaction and E6/E7 protein expression detection by
western blotting (9,10). In addition, histologic diagnosis was
applied to determine the stage of CIN (11). Since histopathology is recognized as
the gold standard for tumor diagnosis, the results of E6/E7 protein
detection, HPV DNA evaluation and TCT were compared with the
histopathologic results in terms of their sensitivity, specificity,
positive predictive value (PPV) and negative predictive value (NPV)
(11).
Preparation and preservation of
samples
Cervical TCT samples obtained from biopsy were
prepared in two formats, either using PreservCyt®
Solution (Hologic Inc., Bedford, MA, USA) or SurePath™
Preservative Solution (TriPath Imaging Inc., Burlington, NC, USA).
A total of 2 ml cervical specimen preparation (containing ~75 µl
cell pellets per 1 ml solution) were collected in 20 ml
PreservCyt® Solution as specimens for TCT. All samples
were maintained at −20°C prior to use, and were tested within 1–2
weeks after collection. Specimens with abnormal TCT findings were
rinsed into Specimen Transport Medium (Digene Corporation,
Gaithersburg, MD, USA) to detect HPV DNA by Hybrid Capture Tube
test (HCT; Digene Corporation) following the manufacturer's
protocol for PreservCyt® Solution-containing specimens.
The specimens were stored at 4°C. The threshold of positive HPV
detection was a relative light unit/cut-off ratio (RLU/CO) of ≥1.0.
Samples containing <5 µl cell precipitate were discarded.
TCT
TCT was used to identify high-grade squamous
intraepithelial lesions (HSILs) (7).
A liquid based cell plastic brush was inserted into the cervix at
the squamo-columnar junction, and was rotated 5–8 circles. The
exfoliated cells were stored in liquid-based cell preservation
solution purchased from Beijing TCT Medical Technology Co., Ltd.
(Beijing, China). The ultra-thin coating was made by ThinPrep 2000
system (Cytyc Corporation, Marlborough, MA, USA). The cells were
fixed with 95% alcohol aqueous solution for 15 min at room
temperature and pap stained for 3 min at room temperature, and then
observed under the 15JF microscope (magnification, ×40) from
Shanghai CSOIF Co., Ltd. (Shanghai, China). Liquid-based cell
plastic brushes and liquid-based cell preservation solution were
purchased from Beijing TCT Medical Technology Co., Ltd. (Beijing,
China). All steps (sample collection, preparation and final
staining) were followed according to the manufacturer's protocol
(Beijing TCT Medical Technology Co., Ltd.). The samples were
classified into five grades according to The Bethesda System:
Within normal limitation (normal); atypical squamous cells with
undetermined significance; low grade squamous intraepithelial
lesion (LSIL); HSIL; and ASCs where HSIL cannot be excluded
(12).
HC2 test
HC2 test (Qiagen Sciences, Inc., Gaithersburg, MD,
USA), a signal amplification test based on the hybridization of a
RNA probe cocktail for 13 high-risk oncogenic types with the target
DNA, and capture and detection of the DNA-RNA hybrid by
chemiluminescence, was applied to quantitate the level of HPV DNA
for its standardization approved by the USA Food and Drug
Administration (13). This test was
performed with 4 ml of PreservCyt samples, and then mixed with 400
µl transfer buffer (digeneHC2HPVDNA detection kit; Qiagen Sciences,
Inc.). Following centrifugation (135 × g for 15 min at 25°C), 75 µl
of the supernatant was added to the microwell plate in a water bath
at 65°C for 15 min. The solution was mixed for 30 sec, and then
kept in a water bath at 65°Cfor 15 min. Denaturant (150 µl;
digeneHC2HPVDNA detection kit; Qiagen Sciences, Inc.) was then
added to the mixture. Following TCT, the residual samples were
denatured by asymmetric PCR (Piko® Thermal Cycler
96-well system; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
to obtain single-stranded DNA (13),
which was then mixed and reacted with an RNA probe cocktail (BD
Pharmingen; BD Biosciences, San Jose, CA, USA) for 13 HR-HPV
oncogenic types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and
68) to capture oncogenic HPV subtypes using DML-2000 gene
hybridization amplifiers (Digene Corporation). The type of positive
point HPV was determined according to the distribution pattern of
HPV. RLU/CO values of ≥1.0 were determined as positive. All samples
were evaluated in triplicate.
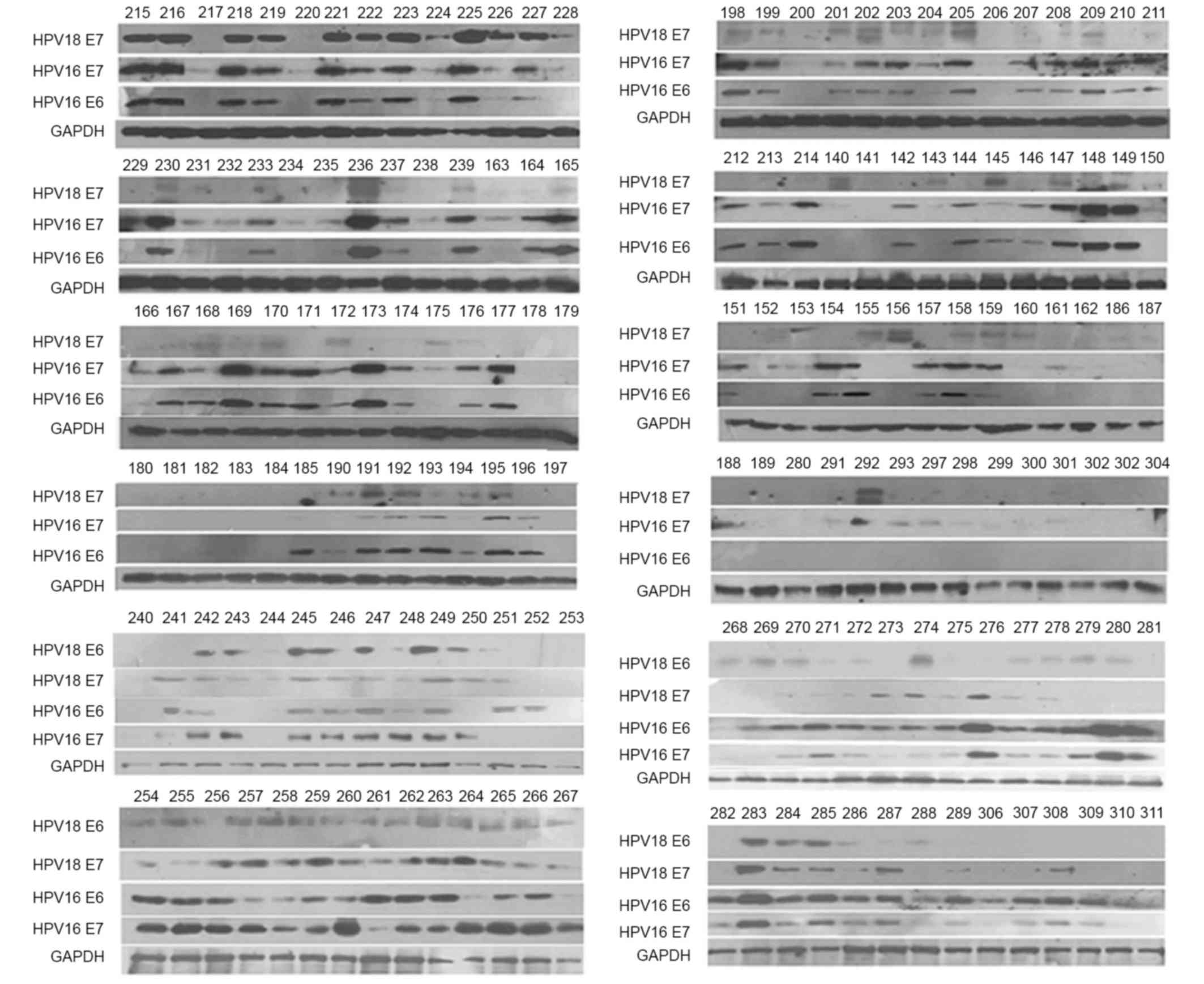
Western blot assay
The human cervical cancer cells obtained from biopsy
were frozen for 2 days. Protein extraction was performed from
frozen cells by adding RIPA Rapid Cell Lysates (catalog no.
BYL40825; Shanghai Jierdun Biotech Co., Ltd., Shanghai, China),
which contained Halt Protease and Phosphatase Inhibitor Cocktail
(Thermo Fisher Scientific, Inc.) as previously described (14). The protein concentration in the cell
lysates was determined with Pierce BCA Protein Assay kit (catalog
no. PICPI23223; Thermo Fisher Scientific, Inc.). A total of 25 µg
protein was loaded on 15% polyacrylamide gels, according to the
molecular weight of E6/E7 proteins (HPV16 E6/E7 and HPV18 E6, 17
kDa; and HPV18 E7, 12 kDa), for electrophoresis as described
previously (15). GAPDH was applied
as an internal control to confirm equal loading of cell lysates.
Upon electrophoresis, samples were transferred to polyvinylidene
fluoride membranes and blocked with 5% skimmed milk powder diluted
in PBS-Tween-20 buffer. Next, primary antibodies (C1P5; catalog
no., sc-460) against the E6/E7 proteins of HPV16 (HPV16 E6
dilution, 1:200; and HPV16 E7 dilution, 1:1,500), antibodies (BF7;
catalog no., ab20192) against the E6 proteins of HPV18 or
antibodies (8E2; catalog no., ab100953) against the E7 proteins of
HPV18 (HPV18 E6; dilution, 1:600; and HPV18 E7 dilution, 1:1,000)
for 1 h at room temperature were added, followed by addition of
horseradish peroxidase-labeled secondary antibodies (dilution
1:1,000) incubated for 1 h at room temperature. All antibodies,
together with anti-GAPDH antibody (6C5; catalog no., ab8245) were
purchased from Abcam (Cambridge, UK), with the exception of
anti-HPV16 E6 antibody, which was purchased from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA). Subsequently, samples were
subjected to enhanced chemiluminescence detection (GE Healthcare,
Chicago, IL, USA). Finally, the representative gray value of
protein expression was scanned (Fig.
1) and analyzed by Quantity One 1-D analysis software version
4.6.9 (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Based on the
positive/negative value of 1,393 volunteers from the Pap test
between April 2012 and February 2015 in south China, the E6/E7 for
the western blot analysis with an optical density under 450 nm
value of ≥0.4±0.005 were considered (16). The test was used to observe the
exfoliated cells from the cervix fixed with physiological saline
under the microscope.
Histological diagnosis
Histologic diagnosis, which is deemed as the gold
standard for the diagnosis of CC, was used in the present study to
clarify the stage of CIN with CIN2+ serving as the disease endpoint
(17). The main steps were performed
according to the procedure described by Ratnam et al
(17). Cervical biopsy results read
by one or more pathologists were obtained from participating
centers and accepted as the disease endpoint for the study
purposes. Pathologists were blinded to HPV results.
Statistical analysis
Categorical variables were assessed in the form of
percentages. Comparison among groups based on percentage values was
assessed for statistical significance. Differences in the
quantitative values (such as age) between groups were analyzed by
the Student's t-test. Statistical analysis was performed by
SPSS13.0 for Windows (SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Results of histological diagnosis
The results from all four methods are shown in
Table I. The proportion of E6/E7
protein-positive patients with CIN2+ was >2-fold
higher than that of CIN2− patients, which was
statistically significant (P=0.000). However, this high specificity
was not observed in HC2-positive patients with high risk of
developing CC, although the sensitivity of the HC2 test was much
higher than E6/E7 protein-positive patients (96.6 vs. 71.3%).
According to the results of TCT, only HSIL+ cases were
statistically significant (P<0.0001).
 | Table I.Characteristics of the patients
according to histological diagnosis. |
Table I.
Characteristics of the patients
according to histological diagnosis.
| Characteristic | CIN2+ (n=348) | CIN2- (n=102) | P-value |
|---|
| Mean age ± SD,
years | 39.7±8.9 | 42.2±9.7 | 0.166 |
| HPV E6/E7 protein
detection, n (%) |
|
| <0.001 |
|
Positive | 248 (71.3) | 33 (32.4) |
|
|
Negative | 100 (28.7) | 69 (67.6) |
|
| HR-HPVHC2 test, n
(%) |
|
| 0.270 |
|
Positive | 336 (96.6) | 96 (94.1) |
|
|
Negative | 12 (3.4) | 6 (5.9) |
|
| TCT, n (%) |
|
| <0.001 |
|
HSIL | 126 (36.2) | 12 (11.8) |
|
|
LSIL | 108 (31.0) | 39 (38.2) |
|
|
ASC-H | 48 (13.8) | 6 (5.9) |
|
|
ASC-US | 45 (12.9) | 21 (20.6) |
|
|
Normal | 21 (6.0) | 24 (23.5) |
|
Comparison of diagnostic value of HPV
E6/E7 proteins with other tests about sensitivity and
specificity
For the diagnosis of CIN2+ cases in
Table II, the specificity of E6/E7
protein detection was markedly improved compared with that of the
HC2 test (67.6 vs. 5.9%), although the sensitivity was lower (71.3
vs. 96.6). Compared with that of TCT, the sensitivity of E6/E7
protein detection was much higher (71.3 vs. 36.2%), while the
specificity was lower (67.6 vs. 88.2%). According to previous
studies, the sensitivity of E6 protein detection by OncoE6 Cervical
Test was only 42.4%, which is lower than the 71.3% of HPV E6/E7
protein detection, but it exhibited 99.1% specificity for E6
protein detection (18). The
sensitivity of E6/E7 mRNA detection gradually increases when the
number of HR-HPV types increases (17,19,20). When
the number of HR-HPV types was >9, the sensitivity was >90%,
with a satisfactory specificity (19,20). In
addition, the sensitivity of HPV E6/E7 protein detection was higher
than that of E6/E7 mRNA detection (71.3 vs. 56.3%, respectively)
with regard to HPV types 16 and 18 (19).
 | Table II.Diagnostic value of HPV E6/E7 protein
detection, HC2 test and TCT on high-grade histological diagnosis
(positive cervical intraepithelial neoplasia2). |
Table II.
Diagnostic value of HPV E6/E7 protein
detection, HC2 test and TCT on high-grade histological diagnosis
(positive cervical intraepithelial neoplasia2).
| Indicator | HPV type | Triage | Cases, n |
Sensitivitya, % (95% CI) |
Specificityb, % (95% CI) | PPVc, % (95% CI) | NPVd, % (95% CI) |
|---|
| HPV E6/E7
proteins | 16 and 18 | Western
blotting | 450 | 71.3
(66.5–76.0) | 67.6
(58.4–76.9) | 88.3
(84.5–92.0) | 40.8
(33.3–48.3) |
| HPV DNA | 13 types of
HR-HPV | HC2 test | 450 | 96.6
(94.6–98.5) | 5.9 (1.2–10.5) | 77.8
(73.8–81.7) | 33.3
(9.2–57.5) |
| Morphocytology | – | TCT | 450 | 36.2
(31.1–41.3) | 88.2
(81.2–94.6) | 91.3
(86.5–96.1) | 28.8
(23.8–33.9) |
As shown in Table
III, the specificity of E6/E7 protein detection for patients
with CIN2+ plus ASC-US or LSIL was higher than that of
HPV DNA testing, suggesting that E6/E7 protein detection could be a
critical adjuvant of HPV DNA testing and TCT. According to previous
studies on HPV mRNA, the sensitivity and specificity of E6/E7 mRNA
detection for patients with CIN2+ plus ASC-US or LSIL or
ASC-US/LSIL were statistically similar for different types of HPV,
ranging from 66.0 to 86.8%, and from 52.2 to 82.0%, respectively
(21–23).
 | Table III.Diagnostic value of HPV E6/E7 protein
detection and HC2 test in CIN2+ plus LSIL or ASC-US patients. |
Table III.
Diagnostic value of HPV E6/E7 protein
detection and HC2 test in CIN2+ plus LSIL or ASC-US patients.
| A, CIN2+ plus
LSIL |
|---|
|
|---|
| Indicator | HPV type | Triage | Cases, n |
Sensitivitya, % (95% CI) |
Specificityb, % (95% CI) | PPVc, % (95% CI) | NPVd, % (95% CI) |
|---|
| HPV E6/E7
proteins | 16 and 18 | Western
blotting | 147 | 77.8
(69.8–85.8) | 56.4
(40.1–72.7) | 83.2
(75.8–90.6) | 47.8
(32.8–62.8) |
| HPV DNA | 13 types of
HR-HPV | HC2 test | 147 | 97.2
(94.1–100.0) | 7.7 (−1.1 to
16.4) | 74.5
(67.2–81.8) | 7.7 (−1.1 to
16.4) |
|
| B, CIN2+ plus
ASC-US |
|
| Indicator | HPV type | Triage | Cases, n |
Sensitivitya, % (95% CI) |
Specificityb, % (95% CI) | PPVc, % (95% CI) | NPVd, % (95% CI) |
|
| HPV E6/E7
proteins | 16 and 18 | Western
blotting | 66 | 78.4
(68.0–88.7) | 66.7
(47.7–85.7) | 83.3
(73.1–93.6) | 27.3
(16.2–38.3) |
| HPV DNA | 13 types of
HR-HPV | HC2 test | 66 | 93.5
(88.5–98.6) | 11.1 (−1.6 to
23.8) | 78.4
(70.6–86.2) | 33.3 (−5.1 to
71.8) |
Previous evidence indicated that E6/E7 protein
detection exhibited better sensitivity than TCT or E6/E7 mRNA
detection for the same HPV types, and better specificity than HPV
DNA testing by HC2. For CIN2+ plus ASC-US or LSIL cases,
the sensitivity and specificity of E6/E7 protein detection were
similar to those of E6/E7 mRNA detection for the same HPV types.
However, markedly different results were obtained in terms of PPV
and NPV (83.2 vs. 37.1% for PPV; and 47.8 vs. 88.9% for NPV,
respectively) (23). Therefore, E6/E7
protein detection exhibited a higher PPV than that of E6/E7 mRNA
detection. The sensitivity and specificity of E6/E7 protein
detection for CIN2+ patients with HSIL and positive HPV
DNA testing were 71.8 and 58.3%, respectively (Table IV), while E6/E7 mRNA testing
exhibited higher specificity and similar sensitivity compared with
those of E6/E7 protein detection. According to previous studies on
HPV mRNA detection, there may be no significant differences in
terms of sensitivity or specificity between the screening
indicators of E6/E7 mRNA and E6/E7 protein detection (21). However, the PPV in the E6/E7 protein
test was higher than the NPV, while the NPV in the E6/E7 mRNA test
was higher than the PPV.
 | Table IV.Diagnostic value of HPV E6/E7 protein
detection in CIN2+ patients with HSIL- and HPV DNA+. |
Table IV.
Diagnostic value of HPV E6/E7 protein
detection in CIN2+ patients with HSIL- and HPV DNA+.
| Patients | Indicator | HPV type | Triage | Cases, n |
Sensitivitya, % (95% CI) |
Specificityb, % (95% CI) | PPVc, % (95% CI) | NPVd, % (95% CI) |
|---|
| CIN2+
plus HSIL− and HPV DNA+ | HPV E6/E7
proteins | 16 and 18 | Western
blotting | 295 | 71.8
(65.7–77.9) | 58.3
(47.6–69.1) | 81.4
(75.8–87.0) | 45.0
(35.5–54.4) |
| CIN2+ plus LSIL and
HPV 16 DNA | E6/E7 mRNA | 16, 18, 31, 33 and
45 | Proofer | 254 | 67 (52–80) | 72 (65–78) | 36 (26–46) | 90 (85–94) |
| CIN2+
plus ASC-US and HPV 16 DNA | E6/E7 mRNA | 16, 18, 31, 33 and
45 | Proofer | 103 | 72 (51–88) | 74 (63–84) | 47 (31–64) | 89 (79–96) |
Discussion
Approximately 565,000 new cases of CC occur in the
world each year, and the incidence rate in developing countries is
3-fold higher than that in developed countries (24,25).
Approximately 50% of all CC cases in the world were recorded in
China and India (26). Nearly 181,500
people are diagnosed with CC each year in China, while >30,000
women succumbed to CC (27). In
Europe, ~38,000 CC cases are diagnosed each year, and >2/3 of
those would be expected to be cured and survive (28).
Usually, women with abnormal results of TCT or/and
HPV DNA detection should undergo subsequent pathological biopsy in
order to diagnose the existence and staging of CINs. Histology is
recognized as the gold standard for diagnosing the pathological
process of CC. CIN2+ is considered a precancerous lesion and, if
left untreated, this may progress to cervical cancer; therefore, if
a patient exhibits a CIN2+ lesion, this should be treated (17,29).
However, it is unknown which kind of lesions will finally lead to
infiltrative cancers. In developed countries, primary screening
based on TCT could prevent >80% of CC. Abnormal diseases are
often missed or misdiagnosed due to the limitations of testing
sensitivity and sampling techniques (28,30).
As a result, numerous studies have focused on
improving the techniques of CC screening. Chen et al
(31) evaluated the efficacy of the
Pap test combined with TCT in CC screening, and observed that the
combination had high sensitivity and specificity. Subsequently,
other reports revealed that the combined detection of HR-HPV by HC2
test and TCT may improve the sensitivity and specificity of CIN
diagnosis and the prediction of its postoperative recurrence
(32). In 2010, Ratnam et al
(17) assessed Proofer and HC2 tests
in a cross-sectional study, and noticed that Proofer is more
specific than HC2 in identifying women with CIN2+, but
has a lower sensitivity. The introduction of the above methods may
improve the accuracy of CC screening, and may avoid a large number
of unnecessary colposcopy and biopsy procedures, while effectively
predicting the development of tumor lesions.
Although the aforementioned methods are popular and
are recommended worldwide by guidelines for primary CC screening, a
few potential patients are still missed and could not be dug out as
detection of traditional activity showed lower sensitivity or
specificity. Previous studies on the diagnosis of HPV and the
occurrence of CC were usually conducted at the cellular, RNA or DNA
level, with few studies performed at the protein level. Therefore,
further evaluations are necessary for improved accurate
screening.
In 2011, Ratnam et al (33) demonstrated that the sensitivity and
specificity of the E6/E7 transcription-mediated amplification
method for the detection of CIN2+ were 100.0 and 88.4%,
respectively, in a study comprising 1,373 women. The Aptima HPV
test, which detected E6/E7 mRNA of 14 oncogenic types, was more
specific for detecting CIN2+ than the HC2 test. However,
in that study, Aptima and HC2 testing were performed upon routine
CC screening, subjects of which were confirmed CIN2+
already (33). Following infection by
HPV, HPV E6/E7 proteins would be overexpressed upon HPV invasion
into the host's cervical cells in the form of episomal HPV DNA or
upon viral integration into the host's genome (34). These events were closely and directly
associated with the development of cancer; thus, the major cause of
pre-CC lesions appears to be the functional expression of HR-HPV
E6/E7 proteins (34,35). Therefore, E6/E7 protein expression may
be directly associated with CC risk.
A pilot clinical study on the application of OncoE6
Cervical Test indicated that HPV 16/18/45 E6 protein detection had
higher specificity than HPV DNA testing for CIN3+
detection (36). In another clinical
trial, this new technology was demonstrated to have higher
specificity compared with that of HC2 testing [98.9%; 95%
confidence interval (CI)=98.6–99.2% vs. 86.8%; 95% CI=85.9–87.7%,
respectively], but lower sensitivity (67.3%; 95% CI=52.5–80.1% vs.
98.0%; 95% CI=89.1–99.9%, respectively) for CIN3+
detection only (37). Based on those
findings, the present study introduced a new type of biomarker,
E6/E7 protein detection, and evaluated this new methodology for the
diagnosis and progression determination of cervical pre-cancerous
lesions and CC.
In the present study, the combination of E6 and E7
protein testing could enhance the accuracy of the test; thus, the
differences between E6/E7 protein detection, HC2 test and TCT were
assessed in a Chinese population. According to the present results,
the application of E6/E7 protein detection in CC screening could
reduce the limitations of other recent technologies, such as the
Pap test and HC2 test, and the indicators (E6/E7 proteins) for both
cervical pre-cancer and progression to CC could be quantified with
the standard detection method established in the present study
(17,21). However, not all types of anti-E6/E7
monoclonal antibodies exhibit high specificity currently (8,38).
Therefore, further studies about the combination of E6/E7 mRNA and
protein testing may be introduced to improve the specificity and
sensitivity of CC screening. Further clinical studies with larger
samples and multi-center studies are required to evaluate the
efficiency of E6/E7 protein detection as a new indicator for the
screening and diagnosis of CC.
In conclusion, the present study provided evidence
that E6/E7 proteins may be potential new biomarkers with
satisfactory diagnostic values for HPV types 16 and 18. The
relative diagnostic value may be further improved by combination
with E6/E7 mRNA detection. Furthermore, the number of HPV types
being tested could be increased in the future.
Acknowledgements
The authors would like to thank the patients,
volunteers, clinicians and registry staff involved in the project,
as well as the Natural Science Foundation of China (grant no.
09ZR1435000) for providing financial support.
References
|
1
|
Di Bonito P, Grasso F, Mochi S, Accardi L,
Donà MG, Branca M, Costa S, Mariani L, Agarossi A, Ciotti M, et al:
Serum antibody response to human papillomavirus (HPV) infections
detected by a novel ELISA technique based on denatured recombinant
HPV16 L1, L2, E4, E6 and E7 proteins. Infect Agent Cancer. 1:62006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chelimo C, Wouldes TA, Cameron LD and
Elwood JM: Risk factors for and prevention of human
papillomaviruses (HPV), genital warts and cervical cancer. J
Infect. 66:207–217. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Muñoz N, Bosch FX, de Sanjosé S, Herrero
R, Castellsagué X, Shah KV, Snijders PJ and Meijer CJ:
International Agency for Research on Cancer Multicenter Cervical
Cancer Study Group: Epidemiologic classification of human
papillomavirus types associated with cervical cancer. N Engl J Med.
348:518–527. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
DeCaprio JA: Human papillomavirus type 16
E7 perturbs DREAM to promote cellular proliferation and mitotic
gene expression. Oncogene. 33:4036–4038. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Delury CP, Marsh EK, James CD, Boon SS,
Banks L, Knight GL and Roberts S: The role of protein kinase A
regulation of the E6 PDZ-binding domain during the
differentiation-dependent life cycle of human papillomavirus type
18. J Virol. 87:9463–9472. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ruttkay-Nedecky B, Jimenez Jimenez AM,
Nejdl L, Chudobova D, Gumulec J, Masarik M, Adam V and Kizek R:
Relevance of infection with human papillomavirus: The role of the
p53 tumor suppressor protein and E6/E7 zinc finger proteins
(Review). Int J Oncol. 43:1754–1762. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McGraw SL and Ferrante JM: Update on
prevention and screening of cervical cancer. World J Clin Oncol.
5:744–752. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zumbach K, Kisseljov F, Sacharova O,
Shaichaev G, Semjonova L, Pavlova L and Pawlita M: Antibodies
against oncoproteins E6 and E7 of human papillomavirus types 16 and
18 in cervical-carcinoma patients from Russia. Int J Cancer.
85:313–318. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chouhy D, Gil LB, Nocito AL, Wojdyla D,
Ornella L, Cittadini J, Gardiol D and Giri AA: Development and
evaluation of a colorimetric PCR system for the detection and
typing of human papillomaviruses. Int J Mol Med. 18:995–1003.
2006.PubMed/NCBI
|
|
10
|
Cheng J, Bian M, Cong X, Sun A, Li M, Ma
L, Chen Y and Liu J: Evaluation of a novel real-time fluorescent
polymerase chain reaction assay for high-risk human papilloma virus
DNA genotypes in cytological cervical screening. Biomed Rep.
1:280–284. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu X, Chen G, Qiu J, Lu J, Zhu W, Chen J,
Zhuo S and Yan J: Visualization of basement membranes in normal
breast and breast cancer tissues using multiphoton microscopy.
Oncol Lett. 11:3785–3789. 2016.PubMed/NCBI
|
|
12
|
Paavonen J, Jenkins D, Bosch FX, Naud P,
Salmerón J, Wheeler CM, Chow SN, Apter DL, Kitchener HC,
Castellsague X, et al: Efficacy of a prophylactic adjuvanted
bivalent L1 virus-like-particle vaccine against infection with
human papillomavirus types 16 and 18 in young women: An interim
analysis of a phase III double-blind, randomised controlled trial.
Lancet. 369:2161–2170. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pyne M, Law C, Hillyard D and Schlaberg R:
Testing and genotyping of high-risk human papillomavirus by the
cobas HPV Test and the Hybrid Capture 2 high-risk HPV DNA test
using cervical and vaginal samples. J Clin Microbiol. 52:1720–1723.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Munagala R, Kausar H, Munjal C and Gupta
RC: Withaferin A induces p53-dependent apoptosis by repression of
HPV oncogenes and upregulation of tumor suppressor proteins in
human cervical cancer cells. Carcinogenesis. 32:1697–1705. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen CL, Hsieh FC, Lieblein JC, Brown J,
Chan C, Wallace JA, Cheng G, Hall BM and Lin J: Stat3 activation in
human endometrial and cervical cancers. Br J Cancer. 96:591–599.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Perkins RB, Anderson BL, Gorin SS and
Schulkin JA: Challenges in cervical cancer prevention: A survey of
U.S. obstetrician-gynecologists. Am J Prev Med. 45:175–181. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ratnam S, Coutlee F, Fontaine D, Bentley
J, Escott N, Ghatage P, Gadag V, Holloway G, Bartellas E, Kum N, et
al: Clinical performance of the PreTect HPV-Proofer E6/E7 mRNA
assay in comparison with that of the Hybrid Capture 2 test for
identification of women at risk of cervical cancer. J Clin
Microbiol. 48:2779–2785. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao FH, Jeronimo J, Qiao YL, Schweizer J,
Chen W, Valdez M, Lu P, Zhang X, Kang LN, Bansil P, et al: An
evaluation of novel, lower-cost molecular screening tests for
humanpapillomavirus in rural China. Cancer Prev Res (Phila).
6:938–948. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hovland S, Arbyn M, Lie AK, Ryd W, Borge
B, Berle EJ, Skomedal H, Kadima TM, Kyembwa L, Billay EM, et al: A
comprehensive evaluation of the accuracy of cervical pre-cancer
detection methods in a high-risk area in East Congo. Br J Cancer.
102:957–965. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Waldstrøm M and Ornskov D: Clinical
performance of a human papillomavirus messenger RNA test (Aptima
HPV Assay) on residual material from archived 3-year-old PreservCyt
samples with low-grade squamous intraepithelial lesion. Arch Pathol
Lab Med. 135:1052–1056. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Benevolo M, Vocaturo A, Caraceni D, French
D, Rosini S, Zappacosta R, Terrenato I, Ciccocioppo L, Frega A and
Rossi Giorgi P: Sensitivity, specificity, and clinical value of
human papillomavirus (HPV) E6/E7 mRNA assay as a triage test for
cervical cytology and HPV DNA test. J Clin Microbiol. 49:2643–2650.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stoler MH, Wright TC Jr, Cuzick J, Dockter
J, Reid JL, Getman D and Giachetti C: APTIMA HPV assay performance
in women with atypical squamous cells of undetermined significance
cytology results. Am J Obstet Gynecol. 208:144.e1–8. 2013.
View Article : Google Scholar
|
|
23
|
Castro Perez S, Iñarrea Fernández A,
González Lamas MJ, Diez Sarán MT, Lama Cid A, Martín Alvarez MJ,
Mosquera Pato M, López-Miragaya I, Estévez N, Piñón Torres J and
Navarro Oña M: Human papillomavirus (HPV) E6/E7 mRNA as a triage
test after detection of HPV 16 and HPV 18 DNA. J Med Virol.
85:1063–1068. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Singh GK, Azuine RE and Siahpush M: Global
inequalities in cervical cancer incidence and mortality are linked
to deprivation, low socioeconomic status, and human development.
Int J MCH AIDS. 1:17–30. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Knaul FM, Bhadelia A, Gralow J,
Arreola-Ornelas H, Langer A and Frenk J: Meeting the emerging
challenge of breast and cervical cancer in low- and middle-income
countries. Int J Gynaecol Obstet. 119 Suppl 1:S85–S88. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Thun MJ, DeLancey JO, Center MM, Jemal A
and Ward EM: The global burden of cancer: Priorities for
prevention. Carcinogenesis. 31:100–110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lei T, Mao WM, Lei TH, Dai LQ, Fang L,
Chen WQ and Zhang SW: Incidence and mortality trend of cervical
cancer in 11 cancer registries of China. Chin J Cancer Res.
1:10–14. 2011. View Article : Google Scholar
|
|
28
|
Shepherd JH: Cervical cancer. Best Pract
Res Clin Obstet Gynaecol. 26:293–309. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sangoi AR, Rogers WM, Longacre TA, Montoya
JG, Baron EJ and Banaei N: Challenges and pitfalls of morphologic
identification of fungal infections in histologic, and cytologic
specimens: A ten-year retrospective review at a single institution.
Am J Clin Pathol. 131:364–375. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jemal A, Siegel R, Ward E, Murray T, Xu J,
Smigal C and Thun MJ: Cancer statistics, 2006. CA Cancer J Clin.
56:106–130. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen H, Shu HM, Chang ZL, Wang ZF, Yao HH,
Zhu HM, Lu TM, Ma QY and Yang BL: Efficacy of Pap test in
combination with ThinPrep cytological test in screening for
cervical cancer. Asian Pac J Cancer Prev. 13:1651–1655. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu XY, Zhao W, Li GJ, Jin XY and Li YZ:
Clinical significance of combined detection of thinprep cytology
test and high risk human papillomavirus hybrid capture 2 assay in
the screening and recurrence predication of cervical
intraepithelial neoplasia. Zhonghua Yi Xue Za Zhi. 92:2503–2505.
2012.(In Chinese). PubMed/NCBI
|
|
33
|
Ratnam S, Coutlee F, Fontaine D, Bentley
J, Escott N, Ghatage P, Gadag V, Holloway G, Bartellas E, Kum N, et
al: Aptima HPV E6/E7 mRNA test is as sensitive as Hybrid Capture 2
Assay but more specific at detecting cervical precancer and cancer.
J Clin Microbiol. 49:557–564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Peh WL, Middieton K, Christensen N,
Nicholls P, Egawa K, Sotlar K, Brandsma J, Percival A, Lewis J, Liu
WJ and Doorbar J: Life cycle heterogeneity in animal models of
human papillomavirus-associated disease. J Virol. 76:10401–10416.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bodily JM, Mehta KP, Cruz L, Meyers C and
Laimins LA: The E7 open reading frame acts in cis and in trans to
mediate differentiation-dependent activities in the human
papillomavirus type 16 life cycle. J Virol. 85:8852–8862. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schweizer J, Lu PS, Mahoney CW,
Berard-Bergery M, Ho M, Ramasamy V, Silver JE, Bisht A, Labiad Y,
Peck RB, et al: Feasibility study of a human papillomavirus E6
oncoprotein test for diagnosis of cervical precancer and cancer. J
Clin Microbiol. 48:4646–4648. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cuzick J, Bergeron C, von Knebel Doeberitz
M, Gravitt P, Jeronimo J, Lorincz AT, Meijer J L M C,
Sankaranarayanan R, Snijders J F P and Szarewski A: New
technologies and procedures for cervical cancer screening. Vaccine.
30 Suppl 5:F107–F116. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Achour M, Ben Younes R, Kochbati L, Kahla
S, Zeghal D, Maalej M, Zouari F and Oueslati R: Production of
recombinant proteins GST L1, E6 and E7 tag HPV 16 for antibody
detection of tunisian cervical cancer patients. African J
Biotechnol. 8:369–374. 2009.
|