Introduction
Colorectal cancer (CRC) is the third most commonly
diagnosed cancer in the world and its morbidity has increased in
recent years (1). Dysregulation of
mammalian target of rapamycin (mTOR) and mTOR signaling is
frequently observed in a variety of human cancers (2). mTOR exists in two functionally distinct
complexes: mTOR complex 1 (mTORC1), containing mTOR and
regulatory-associated protein of mTOR, and mTORC2, containing mTOR
and rapamycin-insensitive companion of mTOR (Rictor) (3). mTORC1 is sensitive to rapamycin and
responds to multiple stimuli, including energy status, growth
factors, amino acids and inflammation (4). mTORC2 affects cell morphology and actin
polymerization (5,6), and mainly promotes cell proliferation
and survival through phosphorylation of Akt and
serine/threonine-protein kinases (STK) (7,8). Rictor,
which is insensitive to rapamycin, forms mTORC2 by binding to
mammalian lethal with SEC13 protein 8, mammalian stress-activated
protein kinase interacting protein 1 and protein observed with
Rictor (9). Currently, only a limited
number of reports indicate that Rictor has certain biological
functions in malignant tumors. For example, it has been reported
that microRNA (miR)-152 acts as a tumor suppressor by targeting
Rictor in gynecological cancers (10). Although Rictor may be involved in
cancer progression, its expression in CRC remains unclear.
Thus, the present study evaluated the expression
levels of Rictor in CRC tissue (experimental group) vs.
paracarcinoma tissue (control group) using immunohistochemistry in
62 CRC paraffin-embedded tissue samples excised during operations.
The difference in expression was further verified at the cellular
level by comparing the differences in the expression level of
Rictor between CRC cells and human normal liver cells. The
association between the expression levels of Rictor protein and the
clinicopathological features of the two groups of patients was
compared using the χ2 test, while the association
between the expression of Rictor and the overall survival rate of
CRC patients was analyzed using the Kaplan-Meier survival analysis
method.
Materials and methods
Materials and chemicals
HCT116, SW480 and LoVo cells were obtained from the
American Type Culture Collection (Manassas, VA, USA), which
performed cell line authentication using DNA fingerprinting by
short tandem repeat analysis. The normal human hepatocyte cell line
HL-7702 was purchased from the Cell Bank of the Chinese Academy of
Sciences (Shanghai, China).
RPMI-1640 medium, trypsin, EDTA, fetal bovine serum
(FBS), penicillin and streptomycin were purchased from Gibco
(Thermo Fisher Scientific, Inc., Waltham, MA, USA). Monoclonal
anti-Rictor antibody (used for IHC and western blot analysis)
produced in mouse (SAB5300210) was purchased from Sigma-Aldrich;
Merck KGaA (Darmstadt, Germany). Alexa Fluor®
594-conjugated goat anti-mouse IgG secondary antibody (ZF-0513) was
purchased from ZSGB-BIO Technology Co., Ltd. (Beijing, China), and
used for IHC and western blot analysis. Anti-β-tubulin (MG7) mouse
monoclonal antibody was acquired from Beijing Ray Antibody Biotech
(RM2003; Beijing, China). Goat anti-mouse IgG was obtained from
Beijing Biosynthesis Biotechnology Co., Ltd. (cat. no. bs-0296G;
Beijing, China) and used for western blot analysis. ECL Western
Blotting kit was acquired from Biyuntian Biotech Co., Ltd.
(Shanghai, China).
Sample collection
A total of 62 CRC paraffin-embedded tissue samples
excised during operations were collected from Zhujiang Hospital
Affiliated to Nanfang Medical University (Guangzhou, China) during
June 2008 and August 2010. Patients included in the study did not
receive any chemical treatment or other anticancer therapies prior
to surgery. Patients excluded from the study were those who did not
have complete follow-up or clinicopathological data, and those who
did not provide informed consent for participation in the study, as
required by the Medical Ethics Committee Affiliated to Zhujiang
Hospital.
Immunohistochemistry
Paraffin was removed from the slides by incubation
in xylene three times (10 min each at room temperature), followed
by rehydration in a graded series of ethanol concentrations (100,
95, 85 and 75% ethanol, 10 min each at room temperature). Antigen
retrieval was performed in 100°C water with 0.01 M citrate buffer
for 30 min. The sections were incubated at 37°C with 0.3% hydrogen
peroxide for 30 min and blocked at room temperature with 5% bovine
serum albumin (Gibco; Thermo Fisher Scientific, Inc.) for 1 h.
Next, the sections were incubated with anti-Rictor antibody (1:800)
for 30 min at 37°C, followed by incubation with Alexa
Fluor® 594-conjugated goat anti-mouse IgG secondary
antibody (1:500) at room temperature for 1 h. The slides were then
rinsed with PBS-Tween-20 (three times, 5 min each), stained with
3,3′-diaminobenzidine (Thermo Fisher Scientific, Inc.), rinsed with
deionized water and counterstained with hematoxylin under a light
microscope at ×200 magnification. Scoring was conducted based on
the staining intensity and the percentage of positively stained
cells as follows: 0–5% scored 0; 6–35% scored 1; 36–70% scored 2;
and >70% scored 3. A final score of ≤1 was assigned to the
negative expression group, while a final score of ≥2 was assigned
to the positive expression group. Two senior pathologists
determined the scores independently.
Cell culture and western blotting
HCT116, SW480, LoVo and HL-7702 (a liver cancer cell
line was included to investigate CRC vs. paracarcinoma, as hepatic
metastases are common in patients with CRC) cells were cultured as
follows: Each cell line was seeded into 6-well plates at a density
of 2.0×105 cells/well in triplicate. Cells were grown in
3 ml growth medium (90% RPMI-1640 medium and 10% FBS) at 37°C, and
cell lysis was conducted at ~85% confluence. HCT116, SW480, LoVo
and HL-7702 cells were harvested and lysed following routine
methods as previously described (11). Proteins were quantified using the BCA
method and 30 µg proteins/lane were separated by 6% SDS-PAGE.
Following transfer to a polyvinylidene fluoride membrane, samples
were blocked with 5% FBS at room temperature for 1 h. Subsequently,
samples were incubated overnight at 4°C with anti-Rictor (1:1,000)
and anti-β-tubulin (1:1,000) primary antibodies. Finally, the
membrane was incubated with Alexa Fluor® 594-conjugated
goat anti-mouse IgG secondary antibody (1:100) and goat anti-mouse
IgG (1:100) for 30 min at room temperature. Proteins were
visualized using ECL Western Blotting kit (Biyuntian Biotech Co.,
Ltd.) and In Vivo Imaging System F (Kodak, Rochester, NY,
USA).
Statistical analysis
Statistical analyses were conducted using SPSS 20.0
software (IBM SPSS, Armonk, NY, USA). Fisher's exact or
χ2 tests were used for the comparison of the Rictor
expression rate between groups, while the correlation between
survival time and clinicopathological variables was analyzed using
the Kaplan-Meier method. For survival analysis, survival curves
were compared using the log-rank test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Immunohistochemistry
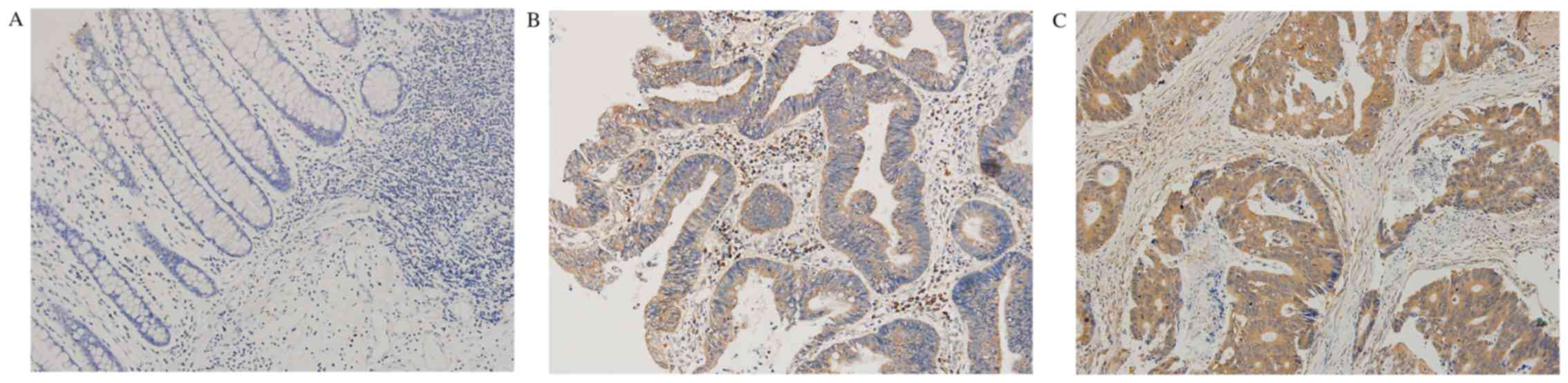
The immunohistochemical results indicated that
Rictor protein mainly existed in the cytoplasm (Fig. 1). The positive expression rate of
Rictor in CRC tissues was 58.1% (36/62), while it was 14.5% (9/62)
in normal tissues adjacent to carcinoma. The expression level of
Rictor protein in CRC tissues was significantly higher than that in
normal tissues adjacent to colorectal carcinoma (P<0.0001). The
results are shown in Table I.
 | Table I.Association between Rictor expression
and clinicopathological features of CRC patients. |
Table I.
Association between Rictor expression
and clinicopathological features of CRC patients.
| Pathological
results | n | Rictor-negative, n
(%) | Rictor-positive, n
(%) | χ2 | P-value |
|---|
| Tissue |
|
|
| 25.428 | <0.001 |
| CRC | 62 | 26 (41.9) | 36 (58.1) |
|
|
|
Normal | 62 | 53 (85.5) | 9 (14.5) |
|
|
| Sex |
|
|
| 0.147 | 0.701 |
| Male | 34 | 15 (44.1) | 19 (55.9) |
|
|
|
Female | 28 | 11 (39.3) | 17 (60.7) |
|
|
| Age, years |
|
|
| 0.045 | 0.831 |
|
>65 | 42 | 18 (42.9) | 24 (57.1) |
|
|
|
<65 | 20 | 8 (40.0) | 12 (60.0) |
|
|
| Differentiation
degree |
|
|
| 0.421 | 0.516 |
| High | 21 | 10 (47.6) | 11 (52.4) |
|
|
| Low | 41 | 16 (39.0) | 25 (61.0) |
|
|
| Tumor size, cm |
|
|
| 0.047 | 0.829 |
|
>3 | 32 | 13 (40.6) | 19 (59.4) |
|
|
|
<3 | 30 | 13 (43.3) | 17 (56.7) |
|
|
| Depth of
invasion |
|
|
| 0.633 | 0.426 |
| Muscular
layer | 25 | 12 (48.0) | 13 (52.0) |
|
|
| Serosal
layer | 37 | 14 (37.8) | 23 (62.2) |
|
|
| Lymphatic
metastasis |
|
|
| 14.600 | <0.001 |
|
With | 32 | 6 (18.8) | 26 (81.2) |
|
|
|
Without | 30 | 20 (66.7) | 10 (33.3) |
|
|
| Dukes stage |
|
|
| 14.090 | <0.001 |
|
A+B | 28 | 19 (67.9) | 9 (32.1) |
|
|
|
C+D | 34 | 7 (20.6) | 27 (79.4) |
|
|
To investigate the correlation between Rictor
expression and clinicopathological characteristics, 62 CRC samples
were collected to detect the expression of Rictor protein by
immunochemistry. The positive rate of Rictor protein expression and
the χ2 values are listed in Table I. Statistical correlation analysis
revealed that the expression level of Rictor protein in CRC tissues
was associated with Dukes stage (P=0.000174) and lymph node
metastasis (P=0.000133), but not with patients' age, sex, tumor
size, differentiation degree, depth of invasion or histological
type (P>0.05). The results are presented in Table I.
Western blotting
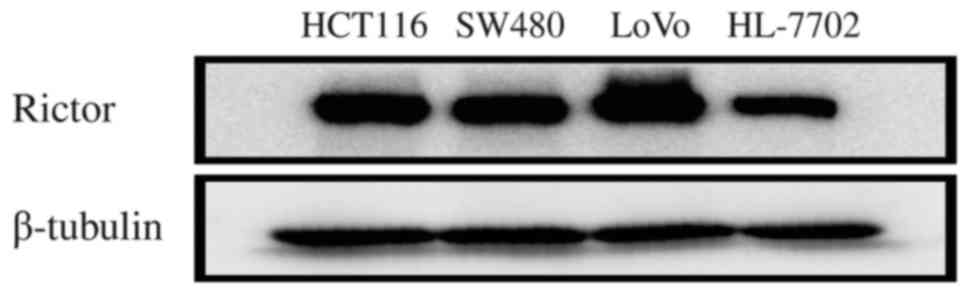
In order to identify the expression and significance
of Rictor protein in CRC at the cellular level, Rictor protein
expression was firstly detected in HCT116, SW480, LoVo and HL-7702
cells using western blotting. Rictor protein expression in these
CRC cell lines was higher than that in the normal liver cell line
HL-7702 (Fig. 2). Thus, these in
vitro results also suggested that Rictor overexpression existed
in CRC, which implies that Rictor may be involved in tumorigenesis
and cancer progression in CRC.
Rictor protein expression and survival
analysis
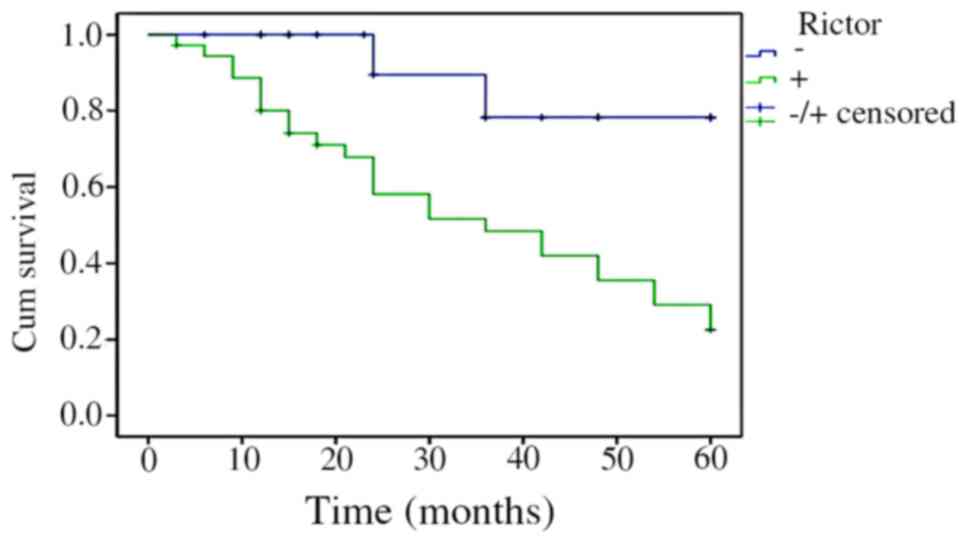
The Kaplan-Meier survival analysis results indicated
that the survival time of patients with CRC was correlated with
Rictor protein expression levels. Patients whose Rictor protein
expression was positive had a shorter survival time than those with
negative expression. Log-rank test revealed that the difference was
statistically significant (P=0.000415). The results are shown in
Fig. 3.
Discussion
The initiation and progression of CRC is a
multi-step process involving gradual changes. Numerous genes are
closely associated with the progression of CRC. Therefore,
identifying therapeutic targets for CRC and improving the 5-year
survival rate of patients with CRC has important theoretical and
practical significance. Rictor, as a member of mTORC2, is important
in the process of tumor proliferation, migration, invasion,
epithelial-mesenchymal transition (EMT) and poor prognosis
(12–16). For example, Akt activated by Rictor
phosphorylation induced the expression of c-Myc and cyclin E, and
this process could regulate the proliferation and cell cycle of
colon cancer cells. Blocking Rictor phosphorylation, which was able
to lessen the inhibition of the mTORC2-Akt signaling pathway, was
applied to stop tumor progression and cancer cell proliferation
(17). Bashir et al reported
that the Rictor gene was involved in cancer cell proliferation in
malignant glioma and had a high recurrence rate in hepatic
carcinoma (18). A previous study
indicated that integrin-linked kinase (ILK), a focal adhesion
adaptor, is an STK that regulates cell proliferation, survival and
EMT. The ILK-Rictor complex acts as a potential molecular target
for preventing/reversing fibrosis caused by EMT, cancer progression
and metastasis (13). Tsuruta et
al observed that Rictor, as a target gene of miR-152,
participated in regulating the proliferation of cancer cells in
endometrial carcinoma (10).
Therefore, it was hypothesized that Rictor protein may be involved
in the proliferation, migration and invasion of CRC.
Based on this theory, the expression level of Rictor
protein was analyzed in 62 CRC tissues. The results indicated that
Rictor expression in CRC tissues was markedly higher than that in
adjacent normal tissues. Western blotting results also confirmed
this finding at the cellular level, suggesting that overexpression
of Rictor protein existed in CRC, which may participate in the
occurrence and development of CRC. However, the specific biological
function of Rictor is still unknown. It has been reported recently
that the upregulation of Rictor is associated with an increased
rate of relapse in hepatocellular carcinoma. Through analysis of
the correlation between Rictor protein and clinicopathological
parameters, the present study further confirmed that Rictor protein
had a significant correlation with Dukes stage and lymph node
metastasis, which further verified our original experimental
hypothesis. Thus, it could be speculated that Rictor has the
potential to be one of the new targets of oncotherapy and may be
applied to clinical diagnosis, similarly to other tumor
markers.
In the present study, Kaplan-Meier survival analysis
was used to evaluate the association between the survival time of
patients with CRC and Rictor protein expression level. The results
revealed that positive expression of Rictor protein in patients
with CRC led to a shorter overall survival compared with that in
patients exhibiting negative expression, indicating that Rictor
protein can be used as a prognostic indicator of CRC. A previous
study also revealed that Rictor overexpression may result in
ovarian cancer patients becoming resistant to Taxol (17), which suggests that targeted control of
Rictor in cancer therapy may be used to improve drug sensitivity.
Several adenosine triphosphate-competitive and selective mTOR
inhibitors, which target simultaneously mTORC1 and mTORC2, have
been developed (19–21). A previous study demonstrated that the
anti-proliferative efficacy of these inhibitors is superior to that
of rapamycin (20). However, if these
inhibitors had larger toxicity in vivo than rapamycin, this
could be a major concern. Another study also suggested that
targeting only mTORC2 was sufficient to prevent colon cancer
progression (22).
Future studies may provide a rational answer about
whether drugs that specifically target Rictor could be used to
treat CRC or other malignant tumors. CRC has been treated with
rapamycin for a long period of time; however, the clinical results
are discouraging (23). One proposed
mechanism of resistance to rapamycin is the inability of this drug
to inhibit mTORC2 (24,25). It has been reported that Rictor
contributed to cisplatin resistance in human ovarian cancer cells
(26). This supports the hypothesis
that mTORC2 compensates for the loss of mTORC1 activity upon
rapamycin treatment, thereby leading to rapamycin resistance.
Knockdown of Rictor was reported to induce G1 arrest in the breast
cancer cell line MCF7 and in the prostate cancer cell line PC3
(27). In addition, inhibition of the
Akt-mTOR signaling pathway was shown to induce G2-M arrest and
autophagy in the breast cancer cell line MDA-MB-231 (28). Considering the present results, it can
be proposed that Rictor may be key to improve the therapeutic
regimen for treating CRC patients.
In summary, the present study demonstrated that
Rictor expression may be associated with CRC initiation and
progression, and also confirmed that Rictor expression is
associated with Dukes stage and lymph node metastasis.
Additionally, it was further verified that Rictor had a correlation
with the prognosis of patients with CRC. Thus, Rictor could play a
crucial role in directing treatment and prognosis evaluation in CRC
patients.
Acknowledgements
The authors thank Professor Zhao Liang (Department
of Pathology, Southern Medical University, Guangzhou, China) for
providing useful guiding ideas and discussions.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chiarini F, Evangelisti C, McCubrey JA and
Martelli AM: Current treatment strategies for inhibiting mTOR in
cancer. Trends Pharmacol Sci. 36:124–135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Iwaya T, Yokobori T, Nishida N, Kogo R,
Sudo T, Tanaka F, Shibata K, Sawada G, Takahashi Y, Ishibashi M, et
al: Downregulation of miR-144 is associated with colorectal cancer
progression via activation of mTOR signaling pathway.
Carcinogenesis. 33:2391–2397. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee DF, Kuo HP, Chen CT, Hsu JM, Chou CK,
Wei Y, Sun HL, Li LY, Ping B, Huang WC, et al: IKK beta suppression
of TSC1 links inflammation and tumor angiogenesis via the mTOR
pathway. Cell. 130:440–455. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sarbassov DD, Ali SM, Kim DH, Guertin DA,
Latek RR, Erdjument-Bromage H, Tempst P and Sabatini DM: Rictor, a
novel binding partner of mTOR, defines a rapamycin-insensitive and
raptor-independent pathway that regulates the cytoskeleton. Curr
Biol. 14:1296–1302. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jacinto E, Loewith R, Schmidt A, Lin S,
Rüegg MA, Hall A and Hall MN: Mammalian TOR complex 2 controls the
actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol.
6:1122–1128. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sarbassov DD, Guertin DA, Ali SM and
Sabatini DM: Phosphorylation and regulation of Akt/PKB by the
rictor-mTOR complex. Science. 307:1098–1101. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jacinto E, Facchinetti V, Liu D, Soto N,
Wei S, Jung SY, Huang Q, Qin J and Su B: SIN1/MIP1 maintains
rictor-mTOR complex integrity and regulates Akt phosphorylation and
substrate specificity. Cell. 127:125–137. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pearce LR, Huang X, Boudeau J, Pawłowski
R, Wullschleger S, Deak M, Ibrahim AF, Gourlay R, Magnuson MA and
Alessi DR: Identification of Protor as a novel Rictor-binding
component of mTOR complex-2. Biochem J. 405:513–522. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tsuruta T, Kozaki K, Uesugi A, Furuta M,
Hirasawa A, Imoto I, Susumu N, Aoki D and Inazawa J: miR-152 is a
tumor suppressor microRNA that is silenced by DNA hypermethylation
in endometrial cancer. Cancer Res. 71:6450–6462. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McDonald PC, Oloumi A, Mills J, Dobreva I,
Maidan M, Gray V, Wederell ED, Bally MB, Foster LJ and Dedhar S:
Rictor and integrin-linked kinase interact and regulate Akt
phosphorylation and cancer cell survival. Cancer Res. 68:1618–1624.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Agarwal NK, Chen CH, Cho H, Boulbès DR,
Spooner E and Sarbassov DD: Rictor regulates cell migration by
suppressing RhoGDI2. Oncogene. 32:2521–2526. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Serrano I, McDonald PC, Lock FE and Dedhar
S: Role of the integrin-linked kinase (ILK)/Rictor complex in
TGFβ-1-induced epithelial-mesenchymal transition (EMT). Oncogene.
32:50–60. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Verreault M, Weppler SA, Stegeman A,
Warburton C, Strutt D, Masin D and Bally MB: Combined RNAi-mediated
suppression of Rictor and EGFR resulted in complete tumor
regression in an orthotopic glioblastoma tumor model. PLoS One.
8:e595972013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Oneyama C, Kito Y, Asai R, Ikeda J,
Yoshida T, Okuzaki D, Kokuda R, Kakumoto K, Takayama K, Inoue S, et
al: miR-424/503-mediated Rictor upregulation promotes tumor
progression. PLoS One. 8:e803002013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang F, Zhang X, Li M, Chen P, Zhang B,
Guo H, Cao W, Wei X, Cao X, Hao X and Zhang N: mTOR complex
component Rictor interacts with PKCzeta and regulates cancer cell
metastasis. Cancer Res. 70:9360–9370. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen CH, Shaikenov T, Peterson TR,
Aimbetov R, Bissenbaev AK, Lee SW, Wu J, Lin HK and dos Sarbassov
D: ER stress inhibits mTORC2 and Akt signaling through
GSK-3β-mediated phosphorylation of rictor. Sci Signal. 4:ra102011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bashir T, Cloninger C, Artinian N,
Anderson L, Bernath A, Holmes B, Benavides-Serrato A, Sabha N,
Nishimura RN, Guha A and Gera J: Conditional astroglial Rictor
overexpression induces malignant glioma in mice. PLoS One.
7:e477412012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Feldman ME, Apsel B, Uotila A, Loewith R,
Knight ZA, Ruggero D and Shokat KM: Active-site inhibitors of mTOR
target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol.
7:e382009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu K, Toral-Barza L, Shi C, Zhang WG,
Lucas J, Shor B, Kim J, Verheijen J, Curran K, Malwitz DJ, et al:
Biochemical, cellular, and in vivo activity of novel
ATP-competitive and selective inhibitors of the mammalian target of
rapamycin. Cancer Res. 69:6232–6240. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thoreen CC, Kang SA, Chang JW, Liu Q,
Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM and Gray NS: An
ATP-competitive mammalian target of rapamycin inhibitor reveals
rapamycin-resistant functions of mTORC1. J Biol Chem.
284:8023–8032. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Roulin D, Cerantola Y, Dormond-Meuwly A,
Demartines N and Dormond O: Targeting mTORC2 inhibits colon cancer
cell proliferation in vitro and tumor formation in vivo. Mol
Cancer. 9:572010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Faivre S, Kroemer G and Raymond E: Current
development of mTOR inhibitors as anticancer agents. Nat Rev Drug
Discov. 5:671–688. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guertin DA and Sabatini DM: Defining the
role of mTOR in cancer. Cancer Cell. 12:9–22. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gulhati P, Cai Q, Li J, Liu J, Rychahou
PG, Qiu S, Lee EY, Silva SR, Bowen KA, Gao T and Evers BM: Targeted
inhibition of mammalian target of rapamycin signaling inhibits
tumorigenesis of colorectal cancer. Clin Cancer Res. 15:7207–7216.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Im-aram A, Farrand L, Bae SM, Song G, Song
YS, Han JY and Tsang BK: The mTORC2 component rictor contributes to
cisplatin resistance in human ovarian cancer cells. PLoS One.
8:e754552013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hietakangas V and Cohen SM: TOR complex 2
is needed for cell cycle progression and anchorage-independent
growth of MCF7 and PC3 tumor cells. BMC Cancer. 8:2822008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kuo PL, Hsu YL and Cho CY: Plumbagin
induces G2-M arrest and autophagy by inhibiting the AKT/mammalian
target of rapamycin pathway in breast cancer cells. Mol Cancer
Ther. 5:3209–3221. 2006. View Article : Google Scholar : PubMed/NCBI
|