Introduction
Gastric cancer (GC) is the fifth most frequently
diagnosed cancer, and a leading cause of cancer-associated
mortality, with ~1,000,000 new cases reported and resulting in
~723,000 mortalities globally in 2012 (1). In China, GC is the third most common
type of cancer and a leading cause of cancer-related mortality;
with ~420,000 new cases and 290,000 mortalities reported in 2011
(2). Despite progress being made in
the diagnosis and treatment of GC, the long-term survival rate
remains relatively low, with 70–75% of patients with GC ultimately
succumbing to the disease (3). As a
consequence, there is an urgent requirement to identify novel
prognostic factors in patients with GC (4). Furthermore, identifying molecular
subgroups associated with GC may lead to identifying patients who
may potentially benefit from targeted drug therapies (5). The pathogenesis of gastric
carcinogenesis remains incompletely characterized and there are
only a limited number of reliable molecular biomarkers, which are
currently targeted in clinical practice. Therefore, the challenge
remains to improve our understanding of the molecular mechanisms
underlying GC, in order to identify biomarkers for early diagnosis,
prognosis prediction and potential therapeutic targets.
The heparan sulfate 6-O-sulfotransferase
(HS6ST) family comprises three isoforms (HS6ST1, 2,
and 3), whose major function consists of attaching a sulfate
to heparan sulfate proteoglycans (HSPGs) (6). HS6ST2, a member of the HS6ST family, has
an alternatively spliced form named HS6ST-2S with 40 amino acids
deleted. Despite the differences in sequences, the two enzymes
catalyze the transfer of sulfate groups from adenosine
3′-phosphate, 5′-phosphosulphate (PAPS) to the 6-O position of the
glucosamine residues in HSPGs (7). By
way of this mechanism, HSPGs subsequently participate in diverse
biological functions including blood clotting, cell recognition,
adhesion, proliferation, and differentiation by interactions with
diverse cytokines (8,9). Previous studies have revealed that
HS6ST2 is associated with the progression of malignant tumors, and
is upregulated in various tumor types including thyroid (10,11),
colorectal (12), pancreatic
(13), ovarian (14), breast cancer (15), and chondrosarcomas (16). However, to the best of our knowledge,
the involvement of HS6ST2 in GC has not yet been elucidated. In the
present study, we evaluated the expression of HS6ST2 in GC and
non-tumor tissues, and evaluated the association with
clinicopathological features and prognostic values, to determine
whether HS6ST2 represents a novel biomarker for patients with
GC.
Materials and methods
Human tissue specimens
A total of 46 paired fresh tumor and adjacent
non-tumor tissue samples were collected from patients who underwent
curative surgery for gastric cancer at Zhejiang Provincial People's
Hospital, (Zhejiang, China), between January 2011 and December 2012
and January 2006 and December 2008. A total of 110 fresh tissues
were formalin-fixed, paraffin-embedded or immediately snap frozen
in liquid nitrogen (−196°C) following resection and stored at −80°C
until analysis. A total of 10 paired samples were profiled on the
GeneChip® Human Genome U133 Plus 2.0 Array platform
(Affymetrix; Thermo Fisher Scientific, Inc.) and 36 paired samples
were used for reverse transcription quantitative polymerase chain
reaction analysis (RT-qPCR). Patient ages ranged from 30–81 years
(median age, 59 years), of which 80 were males and 30 were females.
Tumor location, tumor size, differentiation status, invasion depth,
lymph node metastasis, distant metastasis and tumor-node metastasis
(TNM) stage were also recorded. The pathological diagnosis was
confirmed independently by two pathologists, according to the 7th
edition of the American Joint Committee on Cancer staging manual
(17). The last follow-up was
December 2015. In addition, 60 non-tumor gastric tissue specimens
were acquired by endoscopy from patients without tumors and used as
controls.
The present study was approved by the Ethics
Committee of Zhejiang Provincial People's Hospital. Written
informed consent was obtained from all patients enrolled in the
present study, for the use of all resultant specimens. None of the
patients enrolled in the present study had received
chemoradiotherapy treatment prior to surgery.
Microarray analysis
Total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and examined
using an Agilent Bioanalyzer 2100 (Agilent Technologies, Inc.,
Santa Clara, CA, USA) according to the manufacturer's protocol.
Total RNA was amplified, labeled, and purified to obtain biotin
labeled cRNA using a GeneChip® 3′IVT Express kit
(Affymetrix; Thermo Fisher Scientific Inc.) Array hybridization and
washes were performed using GeneChip® Hybridization,
Wash and Stain kit (Affymetrix; Thermo Fisher Scientific Inc.), and
a Hybridization Oven 645 and a Fluidics Station 450 (Affymetrix;
Inc). Slides were scanned using GeneChip® Scanner 3000
(Affymetrix; Thermo Fisher Scientific Inc.) and Command Console
Software 3.1 (Affymetrix; Thermo Fisher Scientific Inc.) with
default settings. Raw data were normalized by MAS 5.0 algorithm in
Gene Spring Software 11.0 (Agilent Technologies, Inc.).
Differentially expressed genes in GC compared with adjacent
non-tumor tissues were identified through SAM (significance
analysis of microarray). Genes were regarded as differentially
expressed when the N (normal) vs. T (tumor) signal log ratio values
were ≤0.5 or ≥2.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR) analysis
The expression of HS6ST2 mRNA was evaluated
by RT-qPCR analysis. Total RNA was isolated using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and reverse-transcribed to cDNA using a PrimeScript™ RT
Reagent kit (Takara Biotechnology Co., Ltd., Dalian, China),
according to the manufacturer's protocol.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was
selected as an internal control. qPCR was performed in a 20 µl
total reaction volume using SYBR® Premix Ex Taq™
II kit (Takara Biotechnology Co., Ltd.) on the Mx3000P qPCR System
(Stratagene; Agilent Technologies, Inc.). The primer sequences were
as follows: HS6ST2 forward, 5′-GAAGCAGAACTCAGGCAAGG-3′ and
reverse, 5′-CCAATGAAGGAAGCAGGATGT-3′; GAPDH forward,
5′-TGAAGGTCGGAGTCAACGG-3′ and reverse, 5′-CTGGAAGATGGTGATGGGATT-3′.
The PCR reaction was run as follows: 95°C for 4 min as an initial
denaturation step, amplification for 40 cycles with denaturation at
95°C for 10 sec, annealing at 57°C for 30 sec, and extension at
72°C for 30 sec. Melting curve analysis was performed at the
termination of the PCR cycle. Each qPCR was performed in
triplicate, from which the mean value was calculated. The
expression level of HS6ST2 mRNA was expressed as
2−∆∆Cq in which DCq = Cq (HS6ST2)-Cq (GAPDH) (18).
Immunohistochemistry (IHC)
analysis
Paraffin-embedded tissues (n=110), which were
formalin fixed in 10% (v/v) formalin for 24 h at room temperature
and cut into 4 µm sections for IHC. Sections were deparaffinized
and rehydrated through a descending series of alcohols. Tissue
sections were placed in a 10 mM sodium citrate buffer (pH, 6.0)
(120°C for 3 min) to perform antigen retrieval and washed by PBS.
An SP-9000 Detection kit (OriGene Technologies, Inc., Beijing,
China) was used for performing IHC. Endogenous peroxidase activity
was blocked by 3% (v/v) hydrogen peroxide for 15 min at room
temperature. Non-specific binding was blocked by incubation with
10% (v/v) goat serum (OriGene Technologies, Inc., Beijing, China)
or 20 min at room temperature. The sections were incubated
overnight at 4°C with a primary antibody against HS6ST2 (1:200;
cat. no. ab122220; Abcam, Cambridge, UK). The sections were then
incubated with a biotin-labeled secondary antibody (ready-to-use
kit; cat. no. SP9000; goat anti-mouse IgG; OriGene Technologies,
Inc., Beijing, China) for 15 min at room temperature and incubated
with horseradish peroxidase-labeled streptavidin (OriGene
Technologies, Inc.) for 20 min at room temperature. Slides were
stained with 3,3′-diaminobenzidine (DAB; OriGene Technologies,
Inc.) for 5 min at room temperature and counterstained with
hematoxylin. For the negative control, immunohistochemical staining
was performed by omitting the primary anti-HS6ST2 antibody.
All slides were independently examined by two
pathologists (Department of Pathology, Zhejiang Provincial People's
Hospital Zhejiang, China) blinded to the individual patient data,
using an optical microscope (Nikon Corporation, Tokyo, Japan) to
examine five random fields of view at magnification, ×200 and ×400.
The immunoreactivity of HS6ST2 was evaluated according to a
combined scoring system based on the staining intensity and
percentage of positive stained cells. Intensity scores from 0 to 3
were defined as follows: 0, no staining; 1+, weak staining; 2+,
moderate staining and 3+, strong staining. The extent of staining
was scored from 0 to 4 as follows: 0, <5% positive cells; 1+,
5–25% positive cells; 2+, 26–50% positive cells; 3+, 51–75%
positive cells; and 4+, 76–100% positive cells. Individual patient
scores were obtained by multiplying the intensity and extent score.
Scores from 0 to 3 were regarded as negative staining, while scores
from 4 to 12 were deemed positive.
Gene expression omnibus dataset and
survival analysis
A gene expression dataset composed of 876 patients
with gastric cancer was obtained as previously described (19). In brief, the key words ‘gastric’,
‘cancer’, ‘GPL570’ and ‘GPL96’ were searched for in the GEO dataset
(http://www.ncbi.nlm.nih.gov/geo/)
(20). Publications containing only
raw gene expression files, survival information, and a minimum of
30 patients were used for subsequent analyses. Raw. CEL files were
MAS 5.0 normalized using R (http://www.r-project.org). A second scaling
normalization was performed to reduce batch effects (21). In the present study, the expression
level of HS6ST2 was evaluated in 629 GC samples from the
merged dataset. All percentiles between lower and upper quartiles
of HS6ST2 were computed and the best performing threshold
was used as cutoff (cutoff =31) (22). GraphPad Prism® version 5.0,
GraphPad Software Inc., La Jolla, CA, USA) was employed for
Kaplan-Meier analysis.
Statistical analysis
Statistical analysis was performed using SPSS
(version 19.0; IBM Corp., Armork, NY, USA). The mRNA expression of
HS6ST2 in paired adjacent non-tumor and tumor samples was
evaluated using the Wilcoxon signed-rank test. The association
between the expression of HS6ST2 protein and clinicopathological
data was performed using a χ2 test. The survival
analyses were evaluated by Kaplan-Meier survival curves and the
differences between the two groups were compared by a log-rank
test. The survival data was also evaluated by univariate and
multivariate Cox regression analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
Evaluation of HS6ST2 mRNA expression
by microarray analysis
In the present study, differentially expressed genes
between 10 pairs of adjacent non-tumor and gastric cancer tissues
were analyzed by microarray, and the raw data was uploaded to GEO
(accession no. GSE79973). The results from the present study
indicated that the expression of HS6ST2 was significantly
upregulated in GC tissues compared with adjacent non-tumor tissues
(>12-fold; false discovery rate <0.01; Table I). As a result, HS6ST2 was
selected as a potential biomarker, and its involvement in GC was
further investigated.
 | Table I.Selected genes significantly
upregulated in gastric cancer. |
Table I.
Selected genes significantly
upregulated in gastric cancer.
| Probe set ID | False discovery
rate (FDR) | Fold-change (T vs.
N) | Gene symbol | Entrez Gene ID | Chromosomal
location | UniGene ID |
|---|
| 227140_at | 0.000326 | 27.848961 | INHBA | 3624 | chr7p15-p13 | Hs.583348 |
| 203820_s_at | 0.001551 | 16.163251 | IGF2BP3 | 10643 | chr7p11 | Hs.700696 |
| 204051_s_at | 0.001501 | 15.832003 | SFRP4 | 6424 | chr7p14.1 | Hs.658169 |
| 212353_at | 0.000653 | 13.410469 | SULF1 | 23213 | chr8q13.1 | Hs.409602 |
| 230030_at | 0.009619 | 12.400589 | HS6ST2 | 90161 | chrXq26.2 | Hs.385956 |
| 226237_at | 0.000653 | 11.848787 | COL8A1 | 1295 | chr3q12.3 | Hs.654548 |
| 209875_s_at | 0.001303 | 10.858137 | SPP1 | 6696 | chr4q22.1 | Hs.313 |
| 223475_at | 0.002232 | 9.8014127 |
CRISPLD1 | 83690 | chr8q21.11 | Hs.436542 |
| 219087_at | 0.001643 | 8.9984428 | ASPN | 54829 | chr9q22 | Hs.435655 |
| 213905_x_at | 0.002647 | 8.8140516 | BGN | 633 | chrXq28 | Hs.821 |
| 225681_at | 0.000595 | 8.4565955 | CTHRC1 | 115908 | chr8q22.3 | Hs.405614 |
| 202363_at | 0.003312 | 8.0317287 | SPOCK1 | 6695 | chr5q31 | Hs.596136 |
| 225664_at | 0.001047 | 7.6351305 | COL12A1 | 1303 | chr6q12-q13 | Hs.101302 |
| 227566_at | 0.000806 | 7.4371010 | NTM | 50863 | chr11q25 | Hs.504352 |
HS6ST2 mRNA expression in GC and
adjacent non-tumor tissues
In the present study, the microarray data was
further validated by RT-qPCR analysis of the expression of
HS6ST2 mRNA in 36 paired GC and adjacent non-tumor
specimens. The expression of HS6ST2 mRNA was significantly
increased in tumor tissue compared with that of adjacent non-tumor
tissue in 29 (80.6%) cases. The mean expression level of
HS6ST2 mRNA in GC was significantly higher than that
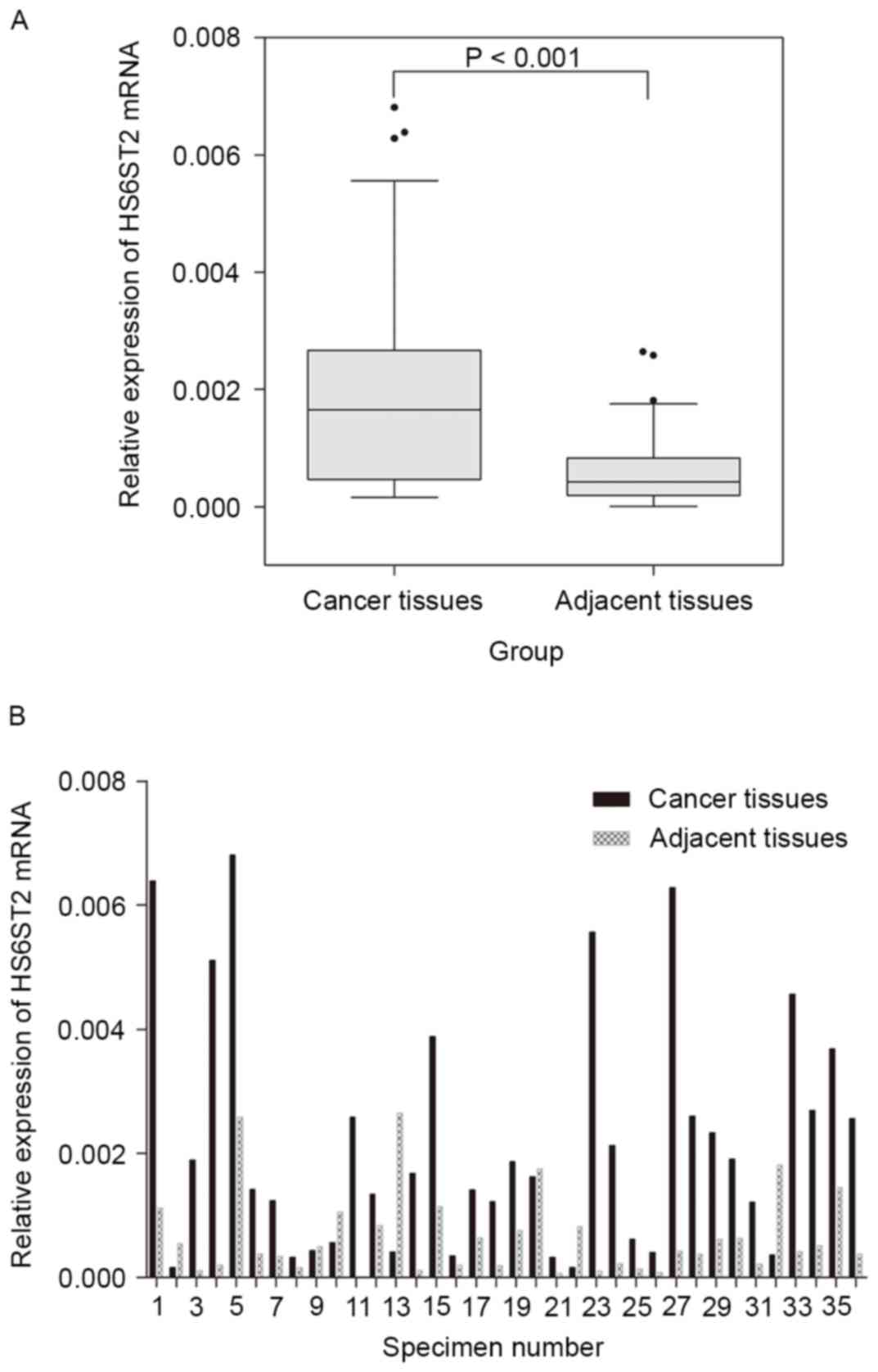
observed in non-tumor tissues (P<0.001; Fig. 1A). The relative HS6ST2 mRNA
expression level of each specimen was presented in Fig. 1B.
HS6ST2 protein levels in GC and
non-tumor tissues
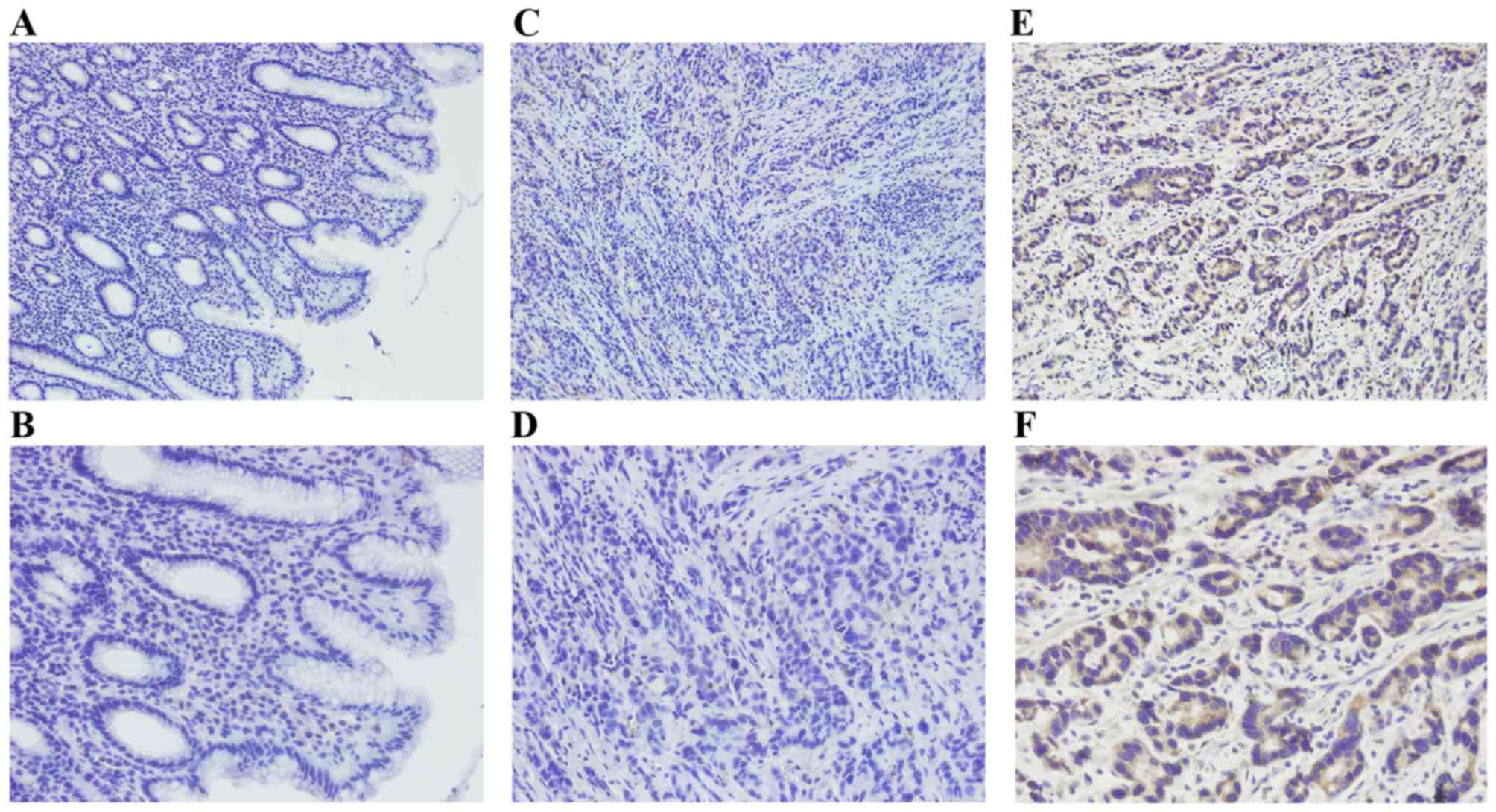
In the present study, HS6ST2 protein levels were
examined in 110 GC tissues and 60 non-tumor gastric tissues by IHC.
Positive immunostaining for HS6ST2 protein was high in GC (70.9%;
78/110) compared with that observed in non-tumor gastric mucosa
(18.3%, 11/60; P<0.001). HS6ST2 staining was detected by a brown
stain in the cytoplasm of cells (Fig.
2).
Associations between HS6ST2 expression
and clinicopathological features
The analysis of the association between HS6ST2
protein expression and clinicopathological parameters, within the
cohort were then evaluated using a χ2 test. A positive
association was observed between the expression of HS6ST2 protein
with invasion depth (P=0.028), distant metastasis (P=0.011), and
tumor-node metastasis stage (P=0.039). However, associations were
not observed between HS6ST2 expression and patient sex (P=0.732),
age (P=0.473), tumor size (P=0.661), tumor location (P=0.517),
lymph node metastasis (P=0.788) or the degree of pathological
differentiation (P=0.732; Table
II).
 | Table II.Associations between HS6ST2
expression and clinicopathological data. |
Table II.
Associations between HS6ST2
expression and clinicopathological data.
|
|
| HS6ST2
expression |
|
|---|
|
|
|
|
|
|---|
| Parameter | Cases (n) | Positive | Negative | P-value |
|---|
| Sex |
|
|
| 0.732 |
|
Male | 80 | 56 | 24 |
|
|
Female | 30 | 22 | 8 |
|
| Age, years |
|
|
| 0.473 |
|
≥60 | 54 | 40 | 14 |
|
|
<60 | 56 | 38 | 18 |
|
| Tumor size, mm |
|
|
| 0.661 |
|
≥50 | 62 | 45 | 17 |
|
|
<50 | 48 | 33 | 15 |
|
| Tumor location |
|
|
| 0.517 |
|
Fundus | 26 | 20 | 6 |
|
|
Body | 38 | 28 | 10 |
|
|
Antrum | 46 | 30 | 16 |
|
|
Differentiation |
|
|
| 0.732 |
| Well or
moderate | 44 | 32 | 12 |
|
|
Poor | 66 | 46 | 20 |
|
| Local invasion |
|
|
| 0.028a |
|
T1-T2 | 26 | 14 | 12 |
|
|
T3-T4 | 84 | 64 | 20 |
|
| Node
metastasis |
|
|
| 0.788 |
|
Yes | 81 | 58 | 23 |
|
| No | 29 | 20 | 9 |
|
| Distant
metastasis |
|
|
| 0.011a |
|
Yes | 24 | 22 | 2 |
|
| No | 86 | 56 | 30 |
|
| TNM stage |
|
|
| 0.039a |
| I | 13 | 7 | 6 |
|
| II | 22 | 13 | 9 |
|
|
III | 51 | 36 | 15 |
|
| IV | 24 | 22 | 2 |
|
Overall survival analysis
Overall survival analysis based on IHC and evaluated
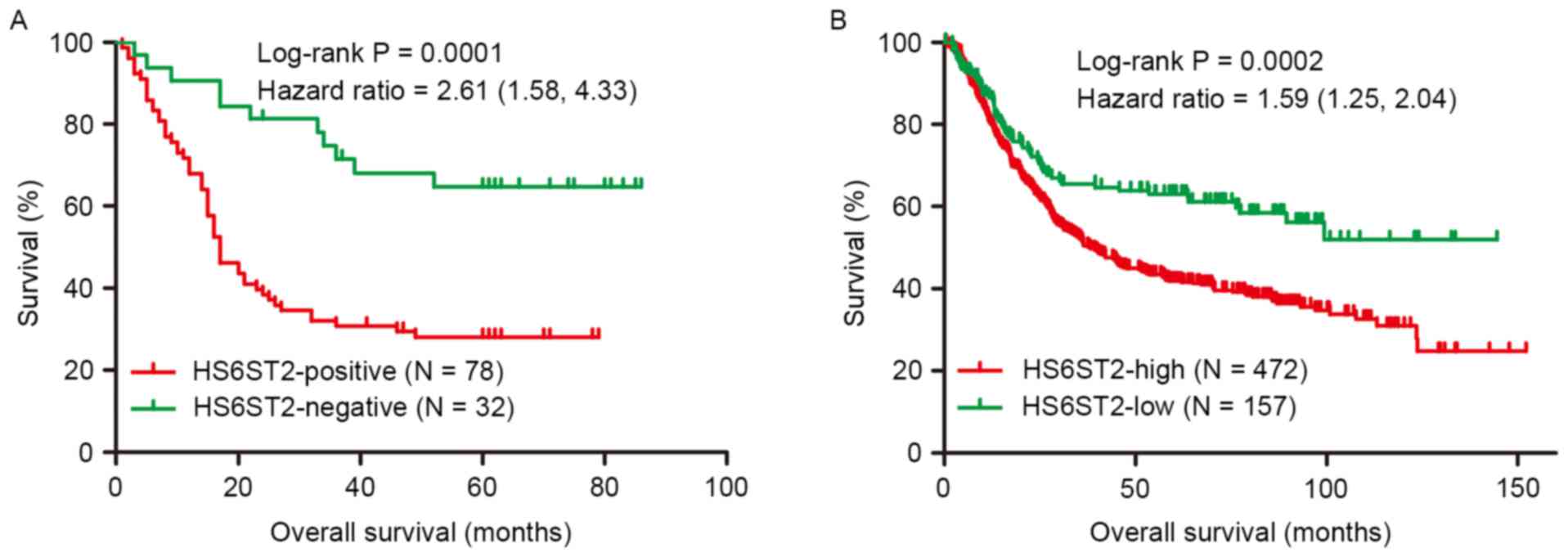
by Kaplan-Meier curves, revealed that the 5-year survival rate of
the HS6ST2-positive (28%) group was significantly reduced compared
with the HS6ST2-negative group (64.7%; Fig. 3A). Cox regression analysis indicated
that the independent negative prognostic factors for GC were HS6ST2
protein expression (P=0.038) and local invasion (P=0.031; Table III). These results were validated
using a non-overlapping cohort of 629 patients. In the validation
cohort, patients with low expression of HS6ST2 demonstrated
increased overall survival, compared with the HS6ST2-high group
(P=0.0002). Furthermore, the 5-year survival rate in HS6ST2-high
group was 41.9% compared with 63.1% in HS6ST2-low group (Fig. 3B).
 | Table III.Multivariate Cox regression survival
analysis of clinicopathological data and HS6ST2 expression in
patients with gastric cancer. |
Table III.
Multivariate Cox regression survival
analysis of clinicopathological data and HS6ST2 expression in
patients with gastric cancer.
| Variable | Cases (n) | Hazard ratio | 95% confidence
interval | P-value |
|---|
| Tumor location,
fundus/body/antrum | 26/38/46 | 0.748 | 0.551–1.104 | 0.062 |
| Local invasion,
T3-T4/T1-T2 | 84/26 | 2.938 | 1.106–7.805 | 0.031a |
| Node metastasis,
yes/no | 81/29 | 1.201 | 0.507–2.843 | 0.677 |
| Distant metastasis,
yes/no | 24/86 | 2.215 | 0.764–6.422 | 0.143 |
| TNM stage,
IV/III/II/I | 24/51/22/13 | 1.452 | 0.671–3.139 | 0.344 |
| HS6ST2 expression,
positive/negative | 78/32 | 2.042 | 1.040–4.012 | 0.038a |
Discussion
The treatment of gastric cancer currently involves
combinatorial therapies, including surgery, chemotherapy and
radiotherapy; however, long-term survival rates remain relatively
low (3,23). TNM classification based on the
clinicopathological characteristics of tumors; including local
invasive depth, lymph node involvement, and distant metastasis is
the current gold standard for prognosis prediction and available
treatment options for GC (24,25).
However, even when symptoms of GC are detected at the early stages,
patients are still susceptible to a reduced life span (25).
In previous decades, molecular profiling has
redefined our understanding of multiple types of human cancer.
Molecular biomarkers are not only useful for diagnosis but may
represent potential targets with prognostic significance in
multiple types of cancer (26).
However, there are a limited number of biomarkers identified, which
are specific to GC. The application of microarray technology has
proved beneficial in the investigation of the differential gene
expression profile of GC and the identification of genes
specifically expressed in GC (27).
Thus, the microarray-based approach in the present study was used
to screen for genes aberrantly expressed in GC, and to select novel
molecular biomarkers, which may prove to have prognostic
significance in GC. In the present study, microarray was used to
analyze differentially-expressed genes in 10 pairs of adjacent
non-tumor and gastric cancer tissues, and the result showed that
the expression of HS6ST2 was significantly upregulated in GC
tissues compared with adjacent non-tumor tissues. The results
indicated that HS6ST2 may perform an important role in GC,
and the biological mechanism of HS6ST2 requires further
exploration.
Heparan sulfate 6-O-sulfotransferase 2 (HS6ST2) is a
golgi-resident enzyme that catalyzes the formation of 6-O-sulfation
of heparan sulfate (HS) in proteoglycans (HSPGs), which regulate
numerous developmental processes (6,28).
Following the 6-O-sulfation of HS, HSPGs participate in the
regulation of numerous signaling pathways by binding and activating
cytokines. Growth factors including epidermal growth factor (EGF),
fibroblast growth factor (FGF) and vascular endothelial growth
factor (VEGF) are dependent on the 6-O-sulfation of HS to assemble
tri-molecular signaling complexes (HS-growth factor-receptor) for
signal transduction (29–32). Previous emerging evidence has
indicated that HS6ST2 is a critical factor involved in regulating
processes of angiogenesis and epithelial-mesenchymal transition
(EMT) during carcinogenesis. Through the regulation of HS
6-O-sulfation, HS6ST2 influences angiogenic processes by inducing
heparin-binding (HB)-EGF receptor signaling in ovarian cancer
(32). EMT is an important mechanism
which induces tumor-related epithelial cells to acquire mesenchymal
features; including increased motility and reduced cell-cell
contact in the early stage of tumorigenesis (33). Song et al (13) revealed that the activation of HS6ST2,
in pancreatic cancer (PC), participated in EMT and angiogenesis in
the progression of this disease. The potential mechanism by which
HS6ST2 potentiates PC carcinogenesis is mainly attributed to the
activation of the notch-signaling pathway, which mediates EMT and
angiogenesis (13). The
notch-signaling pathway is involved in the processes of tumor cell
proliferation, invasion, and the establishment of a mesenchymal
phenotype (33,34). In thyroid carcinomas, HS6ST2
was identified as a target gene of twist family bhlh transcription
factor 1, a critical regulator of EMT (10,35).
Inhibition of H6ST2 in tumor cells impairs cell migration,
invasion, tubule formation, and may reverse EMT (10,13,32).
Notably, previous reports revealed that the specific inhibition of
HS6ST2 by a high molecular weight Escherichia coli
K5-derived heparin-like polysaccharide (K5-NSOS), prevented tumor
progression in a mouse model of breast cancer metastasis (15). These results indicated that K5-NSOS
may be a potential anticancer agent in cancer therapy. In addition,
a clinical study revealed that the overexpression of HS6ST2 was
associated with colorectal cancer (CRC); and may serve as a
predictor for poor prognosis in patients with CRC (12). However, the function of HS6ST2 in GC
remains largely unexplored. To the best of our knowledge, the
present study is the first investigation into the expression of
HS6ST2 in gastric cancer and its clinical significance in the
development and progression of this disease.
Gene chip analysis revealed that HS6ST2 was
upregulated (>12-fold) in tumor tissues as compared with
adjacent non-tumor gastric mucosa. In addition, analysis by RT-qPCR
and IHC, confirmed that HS6ST2 mRNA and protein expression
levels were significantly higher in tumor tissues. Furthermore,
HS6ST2 expression was revealed to be positively associated with TNM
stage, invasion depth, and distant metastasis in patients with GC.
The prognosis analysis based on IHC and the merged dataset
demonstrated that patients with high HS6ST2 expression were
associated with a poor prognosis of GC in comparison to those
patients with a low expression. Furthermore, multivariate analysis
demonstrated that the expression status of HS6ST2 was an
independent prognostic factor in patients with GC. It was
identified that HS6ST2 is involved in GC as a molecular biomarker,
which provides evidence supporting an association between the
biological activity of HS6ST2 and GC prognosis. The present study
indicates that HS6ST2 may serve as a potential marker for GC and
may be useful for the development of effective treatments against
GC. In the present study, the results revealed that HS6ST2 was
upregulated in GC tissues and may represent a useful prognostic
marker. However, the function of HS6ST2 in these processes is
largely unclear, and further studies (such as cell biological
experiments in vitro and animal experiments in vivo)
are needed to characterize the molecular mechanisms by which HS6ST2
participates in the carcinogenesis of GC.
In summary, HS6ST2 was significantly overexpressed
in GC tissues, and upregulation of HS6ST2 was associated with
aggressive histopathological features and poor prognosis in
patients with GC. Taken together, these results indicate that
HS6ST2 may be a potential therapeutic target or a valuable
prognostic biomarker for patients with GC.
Acknowledgements
The present study was supported by Zhejiang
Provincial Natural Science Foundation of China (grant no.
LZ12H16004).
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zhang S, Zhao P, Zeng H,
Zou X and He J: Annual report on status of cancer in China, 2010.
Chin J Cancer Res. 26:48–58. 2014.PubMed/NCBI
|
|
3
|
Allemani C, Weir HK, Carreira H, Harewood
R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A,
et al: Global surveillance of cancer survival 1995–2009: Analysis
of individual data for 25,676,887 patients from 279
population-based registries in 67 countries (CONCORD-2). Lancet.
385:977–1010. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lin LL, Huang HC and Juan HF: Revealing
the molecular mechanism of gastric cancer marker annexin A4 in
cancer cell proliferation using exon arrays. PLoS One.
7:e446152012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kasaian K and Jones SJ: A new frontier in
personalized cancer therapy: Mapping molecular changes. Future
Oncol. 7:873–894. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Habuchi H, Tanaka M, Habuchi O, Yoshida K,
Suzuki H, Ban K and Kimata K: The occurrence of three isoforms of
heparan sulfate 6-O-sulfotransferase having different specificities
for hexuronic acid adjacent to the targeted N-sulfoglucosamine. J
Biol Chem. 275:2859–2868. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Habuchi H, Miyake G, Nogami K, Kuroiwa A,
Matsuda Y, Kusche-Gullberg M, Habuchi O, Tanaka M and Kimata K:
Biosynthesis of heparan sulphate with diverse structures and
functions: Two alternatively spliced forms of human heparan
sulphate 6-O-sulphotransferase-2 having different expression
patterns and properties. Biochem J. 371:131–142. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nagai N, Habuchi H, Esko JD and Kimata K:
Stem domains of heparan sulfate 6-O-sulfotransferase are required
for Golgi localization, oligomer formation and enzyme activity. J
Cell Sci. 117:3331–3341. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sasisekharan R, Shriver Z, Venkataraman G
and Narayanasami U: Roles of heparan-sulphate glycosaminoglycans in
cancer. Nat Rev Cancer. 2:521–528. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Di Maro G, Orlandella FM, Bencivenga TC,
Salerno P, Ugolini C, Basolo F, Maestro R and Salvatore G:
Identification of targets of Twist1 transcription factor in thyroid
cancer cells. J Clin Endocrinol Metab. 99:E1617–E1626. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schulten HJ, Al-Mansouri Z, Baghallab I,
Bagatian N, Subhi O, Karim S, Al-Aradati H, Al-Mutawa A, Johary A,
Meccawy AA, et al: Comparison of microarray expression profiles
between follicular variant of papillary thyroid carcinomas and
follicular adenomas of the thyroid. BMC Genomics 16 Suppl.
1:S72015. View Article : Google Scholar
|
|
12
|
Hatabe S, Kimura H, Arao T, Kato H,
Hayashi H, Nagai T, Matsumoto K, DE Velasco M, Fujita Y, Yamanouchi
G, et al: Overexpression of heparan sulfate 6-sulfotransferase-2 in
colorectal cancer. Mol Clin Oncol. 1:845–850. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song K, Li Q, Peng YB, Li J, Ding K, Chen
LJ, Shao CH, Zhang LJ and Li P: Silencing of hHS6ST2 inhibits
progression of pancreatic cancer through inhibition of Notch
signalling. Biochem J. 436:271–282. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Backen AC, Cole CL, Lau SC, Clamp AR,
McVey R, Gallagher JT and Jayson GC: Heparan sulphate synthetic and
editing enzymes in ovarian cancer. Br J Cancer. 96:1544–1548. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pollari S, Käkönen RS, Mohammad KS,
Rissanen JP, Halleen JM, Wärri A, Nissinen L, Pihlavisto M,
Marjamäki A, Perälä M, et al: Heparin-like polysaccharides reduce
osteolytic bone destruction and tumor growth in a mouse model of
breast cancer bone metastasis. Mol Cancer Res. 10:597–604. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Waaijer CJ, de Andrea CE, Hamilton A, van
Oosterwijk JG, Stringer SE and Bovée JV: Cartilage tumour
progression is characterized by an increased expression of heparan
sulphate 6O-sulphation-modifying enzymes. Virchows Arch.
461:475–481. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Györffy B and Schäfer R: Meta-analysis of
gene expression profiles related to relapse-free survival in 1,079
breast cancer patients. Breast Cancer Res Treat. 118:433–441. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fekete T, Rásó E, Pete I, Tegze B, Liko I,
Munkácsy G, Sipos N, Rigó J Jr and Györffy B: Meta-analysis of gene
expression profiles associated with histological classification and
survival in 829 ovarian cancer samples. Int J Cancer. 131:95–105.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mihaly Z, Kormos M, Lánczky A, Dank M,
Budczies J, Szász MA and Győrffy B: A meta-analysis of gene
expression-based biomarkers predicting outcome after tamoxifen
treatment in breast cancer. Breast Cancer Res Treat. 140:219–232.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Orditura M, Galizia G, Sforza V,
Gambardella V, Fabozzi A, Laterza MM, Andreozzi F, Ventriglia J,
Savastano B, Mabilia A, et al: Treatment of gastric cancer. World J
Gastroenterol. 20:1635–1649. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Washington K: 7th edition of the AJCC
cancer staging manual: Stomach. Ann Surg Oncol. 17:3077–3079. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sawada T, Yashiro M, Sentani K, Oue N,
Yasui W, Miyazaki K, Kai K, Fushida S, Fujimura T, Ohira M, et al:
New molecular staging with G-factor supplements TNM classification
in gastric cancer: A multicenter collaborative research by the
Japan Society for Gastroenterological Carcinogenesis G-project
committee. Gastric Cancer. 18:119–128. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Harris TJ and McCormick F: The molecular
pathology of cancer. Nat Rev Clin Oncol. 7:251–265. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
D'Angelo G, Di Rienzo T and Ojetti V:
Microarray analysis in gastric cancer: A review. World J
Gastroenterol. 20:11972–11976. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bernfield M, Götte M, Park PW, Reizes O,
Fitzgerald ML, Lincecum J and Zako M: Functions of cell surface
heparan sulfate proteoglycans. Annu Rev Biochem. 68:729–777. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Robinson CJ, Harmer NJ, Goodger SJ,
Blundell TL and Gallagher JT: Cooperative dimerization of
fibroblast growth factor 1 (FGF1) upon a single heparin saccharide
may drive the formation of 2:2:1 FGF1.FGFR2c.heparin ternary
complexes. J Biol Chem. 280:42274–42282. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Robinson CJ, Mulloy B, Gallagher JT and
Stringer SE: VEGF165-binding sites within heparan sulfate encompass
two highly sulfated domains and can be liberated by K5 lyase. J
Biol Chem. 281:1731–1740. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Esko JD and Lindahl U: Molecular diversity
of heparan sulfate. J Clin Invest. 108:169–173. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cole CL, Rushton G, Jayson GC and
Avizienyte E: Ovarian cancer cell heparan sulfate
6-O-sulfotransferases regulate an angiogenic program induced by
heparin-binding epidermal growth factor (EGF)-like growth
factor/EGF receptor signaling. J Biol Chem. 289:10488–10501. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jie Zou, Peng Li, Fei Lu, Na Liu, Jianjian
Dai, Jingjing Ye, Xun Qu, Xiulian Sun, Daoxin Ma, Jino Park and
Chunyan Ji: Notch1 is required for hypoxia-induced proliferation,
invasion and chemoresistance of T-cell acute lymphoblastic leukemia
cells. J Hematol Oncol. 6:32013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang J, Mani SA, Donaher JL, Ramaswamy S,
Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A and
Weinberg RA: Twist, a master regulator of morphogenesis, plays an
essential role in tumor metastasis. Cell. 117:927–939. 2004.
View Article : Google Scholar : PubMed/NCBI
|