Introduction
Gastric cancer is responsible for 10% of total
mortalities worldwide every year, and the rate of the disease is
particularly high in in males and developing countries (1). While the precise underlying
tumorigenesis mechanism of gastric cancer has not been fully
understood, a number of risk factors have been reported, including
a low intake of vegetables, an excess intake of salty food and
smoking (2). Another major factor
that is associated with the development of gastric cancer is
chronic inflammation, which is caused by Helicobacter pylori (H.
pylori) infection (3). In
addition to H, pylori, recent studies have also revealed
various intrinsic mediators that are involved in
inflammation-associated carcinogenesis (4).
Caudal-type homeobox 2 (CDX2) is a caudal-related
homeobox transcription factor, which is involved in intestinal
differentiation in normal and in aberrant locations. The authors of
the present study and others have previously reported that CDX2 may
exert a suppressive role in progression and carcinogenesis of
gastric carcinoma (5,6). Consistent with these results, four
reports have demonstrated that negative CDX2 expression was
associated with poor survival in gastric cancer patients (7–10).
However, other studies reveled that CDX2, which is ectopically
observed in intestinal metaplasia in the stomach, may increase the
risk of gastric cancer development (11,12).
Therefore, further studies will be required to elucidate the
functional role of CDX2 in inflammation-associated gastric
carcinogenesis and investigate the uses of prognostic
biomarkers.
MicroRNAs (miRNAs/miRs) are short noncoding RNAs
that specifically regulate the translation of target genes
(13). Through this function, miRNAs
affect a variety of cellular functions, including proliferation,
differentiation, and apoptosis and have a causal role in
tumorigenesis including inflammation-associated carcinogenesis.
Signal transducer and activator of transcription 3 (STAT3) is a
downstream of interleukin (IL)-6 and form an inflammatory positive
feedback loop with nuclear factor κB, Lin28B, miR-181b and miR-21
(14). Because CDX2 is one of the
targets of miR-181b, the regulation of CDX2 may be involved in the
IL-6/STAT3 signaling pathway via miR-181b (15). Furthermore, the IL-6/STAT3 signaling
pathway also forms a feedback loop with miR-34a that is induced by
p53 (16).
The present study collected 210 patients with
gastric cancer and focused exclusively on inflammation-associated
carcinogenesis. Based on a previous report by the present authors
(5), the present study first
confirmed that negative CDX2 expression was associated with
cancer-specific survival in patients with gastric cancer.
Subsequently, in order to elucidate the functional role of CDX2
suppression in inflammation-associated carcinogenesis, the
interactions between the IL-6/STAT3 signaling pathway, miR-181b,
miR-34a, and p53 expression were investigated experimentally.
Materials and methods
Tissue samples
Primary gastric adenocarcinoma specimens were
procured from patients undergoing surgical resections. In a total
of 210 cases, 78 cases were recruited from Ohta Nishinouchi
Hospital (Fukushima, Japan) from January 2001 to December 2003. A
total of 132 cases were recruited from Fukushima Medical University
(Fukushima, Japan) from January 1991 to December 2004. The cases
from Fukushima Medical University were included in a previous study
by the authors (5). All patients were
Japanese, and no patients received chemotherapy or radiotherapy
prior to surgery. Data including age, gender, tumor-node-metastasis
(TNM) stage (17,18) and pathological diagnosis, (lymphatic
and venous invasion), were retrospectively collected. As with the
previous study by the authors (5),
the cases were divided into two types, differentiated and
undifferentiated. The differentiated type was defined as patients
with well and moderately differentiated tubular or papillary
adenocarcinomas, and the undifferentiated type included patients
with poorly differentiated adenocarcinomas and signet-ring cell
carcinomas. The average overall 5-year survival rate was 68.8%. The
present study was approved by the Ethics Committee of the Fukushima
Medical University. Written informed consent was obtained from all
patients.
Immunohistochemistry (IHC)
HC was performed as previously described (5). Briefly, histological sections
(thickness, 4 µm) were fixed in 10% buffered formalin using a
polymer peroxidase method (Envision+/HRP; Dako; Agilent
Technologies, Inc., Santa Clara, CA, USA) (19). Following deparaffinization with xylene
and rehydration using a descending alcohol series, the tissue
sections were treated with 0.3% hydrogen peroxide in methanol for
30 min at room temperature to block endogenous peroxidase activity.
Following rinsing in PBS, the sections were incubated with
anti-CDX2 antibody (1:1,000; cat. no. MU392A-UC; clone CDX2-88,
Biogenex, San Ramon, CA, USA) and anti-p53 (1:1,000; cat. no.
M7000101-2; DO-7, Dako; Agilent Technologies, Inc.) at 4°C
overnight. An additional wash in PBS was followed by treatment with
ready-to-use peroxidase-labeled polymer conjugated to goat
anti-rabbit immunoglobulins (cat. no. SM801; ENvision + kit; Dako;
Agilent Technologies, Inc.) as the secondary antibody for 30 min at
room temperature. The staining was visualized using
diaminobenzidine at room temperature for 5 min, followed by
counterstaining with hematoxylin at room temperature for 30
sec.
Expression of these proteins was evaluated using
optical microscopy (BX43; Olympus Corporation, Tokyo, Japan) and
was positive when the nucleus of the cancerous tissue was stained.
The staining of each tissue sample was evaluated at ×40 or ×400
magnification by two investigators, M.S. and K.S. (Fukushima
Medical University School of Medicine, Fukushima, Japan), who were
blinded to the sample name and the clinical outcomes. The rate of
positive stained cancer cells was evaluated in three randomly
selected areas (size, 200×200 µm) from the tumor tissue samples.
When the average positive tumor rate was >10%, the tumor was
defined as being positively stained.
Cell culture and transfection
Human gastric carcinoma cell lines AGS, KATOIII,
MKN1, MKN45, MKN74, NUGC3, NUGC4 and Okajima used in the present
study were originally obtained from the American Type Culture
Collection (Manassas, VA, USA). All cells were maintained according
to recommended protocols (20) and
media with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.).
The monolayer cells were maintained in a 37°C incubator with 5%
CO2, observed regularly under a light microscope
(magnification, ×40) and subcultured when they reached 80–90%
confluency. KATOIII and MKN74 cells were transfected with
pre-miR-181b (ID, PM12442) and miR Mimic negative control#1
(Ambion; Thermo Fisher Scientific, Inc., Waltham, MA, USA,) using
Lipofectamine (RNAiMAX; Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. One day prior to
transfection, a gastric cancer cell line was seeded at
5×105 cells per well on a 6-cm plate. Transfection with
a final concentration of 40 nM pre-miR-181b and control was
performed when the cell density was 30–50% on the plates, and the
plates were then incubated for 48 h at 37°C. Total RNA was
extracted 48 h following transfection using TRIZOL (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. Recombinant human IL-6 (Peprotech, Rocky hill, NJ,
USA) was used at 40 ng/ml for transfection of KATOIII and MKN74
cells.
Reverse transcription polymerase chain
reaction (RT-PCR)
Total RNA was isolated from cultured cell lines
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions and was quantified by
NanoDrop (Thermo Fisher Scientific, Inc.). Subsequently, 5 µg total
RNA was used for cDNA synthesis using the SuperScript III First
Strand cDNA Synthesis kit (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Conventional PCR
was performed using Takara ExTaq DNA polymerase (Takara Bio, Inc.,
Otsu, Japan). CDX2 and β-actin mRNA transcripts were amplified
using the following primers: CDX2 forward,
5′-CGGCTGGAGCTGGAGAAGG-3′ and reverse, 5′-TCAGCCTGGAATTGCTCTGC-3′;
β-actin forward, 5′-GCTCGTCGTCGACAACGGCTC-3′ and reverse,
5′-CAAACATGATCTGGGTCATCTTCTC-3′. The amplification was performed at
94°C for 5 min with 30 cycles (25 cycles for β-actin) of 94°C for 1
min, 60°C for 30 sec (55°C for β-actin) and 72°C for 1 min.
Quantitative reverse transcription
polymerase chain reaction (RT-qPCR)
RT-qPCR for mRNA and mature microRNA was performed
using TaqMan MicroRNA assays (Applied Biosystems; Thermo Fisher
Scientific, Inc.) according to manufacturer's instructions with the
7900 HT Fast Real-Time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.) at 94°C for initial denaturation for 5
min, followed by 35 cycles at 94°C for 1 min, 55–60°C for 45 sec
and 72°C for 45 sec (21,22). All assays were performed in triplicate
and investigators were blinded to the experiment. The TaqMan probes
[hsa-miR-181b (ID, 001098), hsa-miR-34a (ID, 00425), CDX2 (ID,
Hs01078080_m1), STAT3 (ID, Hs01047580_m1), RNU66 (ID 001002) and
GAPDH (ID, Hs99999905_ml)] were purchased from Applied Biosystems
(Thermo Fisher Scientific, Inc.). Relative miRNA or mRNA expression
was determined using RNU66 or GAPDH, which were used as a
normalization controls, respectively, using the 2−ΔΔCq
method, according to the supplier's protocol (Thermo Fisher
Scientific, Inc.) (23).
MicroRNA prediction
A publically available comprehensive database,
miRWalk (zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk) was
used to identify the miRNAs with the potential to directly suppress
CDX2 expression (24). This database
provides information on miRNA predicted as well as validated miRNA
binding site information on miRNAs for human.
Statistical analysis
The patients were divided into high or low
expression groups based on the intensity of immunostaining for CDX2
and p53, according to previously described criteria (5). Univariate and multivariate Cox
regression was used to evaluate the associations between clinical
factors and cancer-specific survival and overall survival in JMP 10
(www.jmp.com; SAS Institute, NC, USA). For all
analyses, age was treated as a categorical variable as >65 or
<65 years. Histology was categorized as undifferentiated vs.
differentiated. TNM staging (17,18) was
categorized as stage I vs. stage II/III. For multivariable Cox
regression models, all variables were included with cancer-specific
survival. Kaplan-Meier analysis was performed using Graphpad Prism
(v.5.0; GraphPad Software, Inc., La Jolla, CA, USA). Quantitation
of relative expression of microRNA was calculated with RQ manager
1.2 (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Differences in mRNA and microRNA expression between control cells
and treated cells were analyzed by unpaired t-test (GraphPad
Software, Inc.). All data are expressed as the mean ± standard
deviation. P<0.05 was considered to indicate a statistically
significant difference.
Results
CDX2 expression is associated with
clinicopathological factors
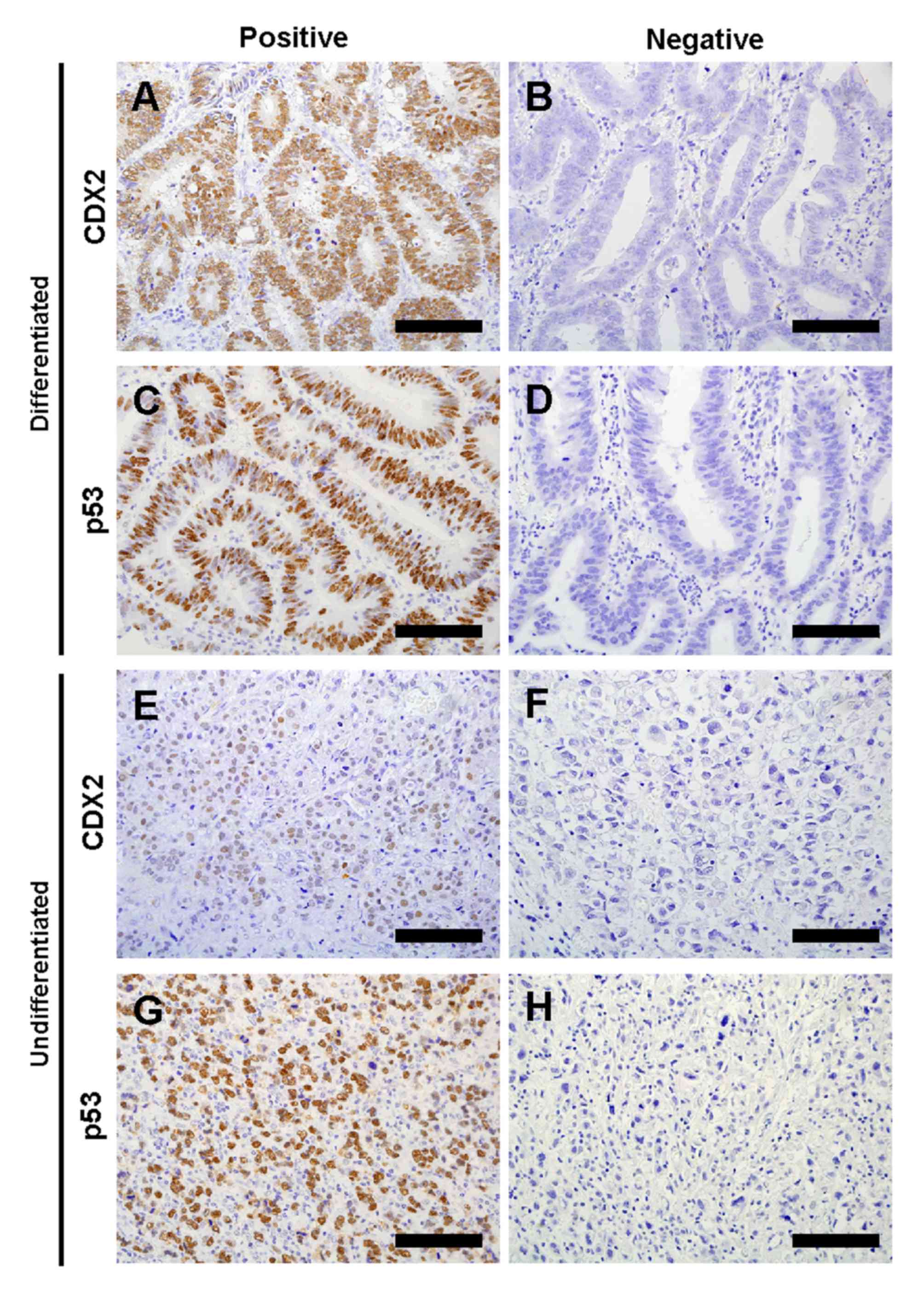
The authors of the present study evaluated CDX2 and
p53 expression in 210 gastric cancer tumor samples by IHC (Fig. 1). Characteristics of these patients
subjected to CDX2 expression are summarized in Table I. CDX2 expression was positive in 101
cases (48.1%), whereas negative in 109 cases (51.9%). The
percentage of negative CDX2 expression was significantly higher in
females compared with males (P=0.006) and in undifferentiated
histological type compared with differentiated type (P=0.002). In
addition, there was a statistically significant inverse association
between CDX2 expression and depth of invasion (P<0.001) or stage
(P<0.001), respectively.
 | Table I.Characteristics of patients with
gastric cancer in the study. |
Table I.
Characteristics of patients with
gastric cancer in the study.
|
|
| CDX2 expression |
|
|---|
|
|
|
|
|
|---|
| Parameter | Total, n | Positive, n (%) | Negative, n (%) | P-valuea |
|---|
| n | 210 | 101 | 109 |
|
| Age, year |
|
|
| 0.111 |
| Mean
(range) | 64.0 (29–87) | 65.5 (33–87) | 62.6 (29–87) |
|
| Gender |
|
|
| 0.006 |
| Male | 149 | 81 (80) | 68 (62) |
|
|
Female | 61 | 20 (20) | 41 (38) |
|
| Histological
type |
|
|
| 0.002 |
|
Differentiated | 112 | 65 (64) | 47 (43) |
|
|
Undifferentiated | 98 | 36 (36) | 62 (57) |
|
| Depth of
invasion |
|
|
| <0.001 |
| T1 | 79 | 48 (48) | 31 (28) |
|
| T2 | 84 | 43 (43) | 41 (38) |
|
| T3 | 46 | 9 (9) | 37 (34) |
|
| T4 | 1 | 1 (1) | 0 |
|
| Lymphatic
invasion |
|
|
| 0.857 |
|
Present | 173 | 84 (83) | 89 (82) |
|
|
Absent | 37 | 17 (17) | 20 (18) |
|
| Venous
invasion |
|
|
| 0.065 |
|
Present | 152 | 67 (66) | 85 (78) |
|
|
Absent | 58 | 34 (34) | 24 (22) |
|
| LN metastasis |
|
|
| 0.383 |
|
Positive | 138 | 63 (62) | 75 (69) |
|
|
Negative | 72 | 38 (38) | 34 (31) |
|
| TNM stage |
|
|
| <0.001 |
| I | 111 | 68 (67) | 43 (39) |
|
| II | 40 | 16 (16) | 24 (22) |
|
|
III | 59 | 17 (17) | 42 (39) |
|
| p53 expression |
|
|
| 0.097 |
|
Positive | 104 | 56 (55) | 48 (44) |
|
|
Negative | 106 | 44 (45) | 62 (56) |
|
CDX2 is associated with poor prognosis
in gastric cancer
In order to investigate whether CDX2 is a
potential prognostic biomarker, Cox regression analysis was
performed. Cancer-specific survival and overall survival times were
used as an endpoint for the present cohort. Negative expression of
CDX2 was significantly associated with poor cancer-specific
survival (hazard ratio [HR]=2.60, 95% confidence interval
[CI]=1.37–5.2; Table II) but not
overall survival (HR=1.47, 95% CI=0.92–2.40; Table III). Multivariate Cox regression
analysis demonstrated that CDX2 expression was associated with poor
cancer-specific survival in the present cohort independent of
staging (HR=2.10, 95% CI=1.05–4.41; Table II).
 | Table II.Univariate and multivariate Cox
regression analysis of CDX2 expression and other parameters for
cancer-specific survival. |
Table II.
Univariate and multivariate Cox
regression analysis of CDX2 expression and other parameters for
cancer-specific survival.
| A, Univariate
analysis |
|---|
|
|---|
| Variable | HR (95% CI) | P-value |
|---|
| CDX2
expression |
|
|
|
Negative vs. positive | 2.60
(1.37–5.27) | 0.003 |
| Age, years |
|
|
| >65
vs. ≤65 | 1.48
(0.81–2.75) | 0.206 |
| Gender |
|
|
| Male
vs. female | 0.95
(0.51–1.89) | 0.888 |
| Histological
type |
|
|
|
Undifferentiated vs.
differentiated | 1.07
(0.59–1.96) | 0.814 |
|
Stage |
|
|
| III vs.
I & II | 6.50
(3.52–12.50) | <0.001 |
|
| B, Multivariate
analysis |
|
| Variable | HR (95% CI) | P-value |
|
| CDX2
expression |
|
|
|
Negative vs. positive | 2.10
(1.05–4.41) | 0.035 |
| Age, years |
|
|
| >65
vs. ≤65 | 2.11
(1.14–4.00) | 0.018 |
| Gender |
|
|
| Male
vs. female | 0.99
(0.50–2.02) | 0.967 |
| Histological
type |
|
|
|
Undifferentiated vs.
differentiated | 0.69
(0.36–1.32) | 0.266 |
| Stage |
|
|
| III vs.
I and II | 7.10
(3.72–14.08) | <0.001 |
 | Table III.Univariate analysis of CDX2
expression and parameters for overall survival. |
Table III.
Univariate analysis of CDX2
expression and parameters for overall survival.
| Variable | HR (95% CI) | P-value |
|---|
| CDX2
expression |
|
|
|
Negative vs. positive | 1.47
(0.92–2.40) | 0.107 |
| Age, year |
|
|
| >65
vs. ≤65 | 1.91
(1.18–3.13) | 0.008 |
| Gender |
|
|
| Male
vs. female | 1.43
(0.84–2.59) | 0.191 |
| Histological
type |
|
|
|
Undifferentiated vs.
differentiated | 0.92
(0.57–1.47) | 0.731 |
| Stage |
|
|
| III vs.
I and II | 3.20
(2.00–5.12) | <0.001 |
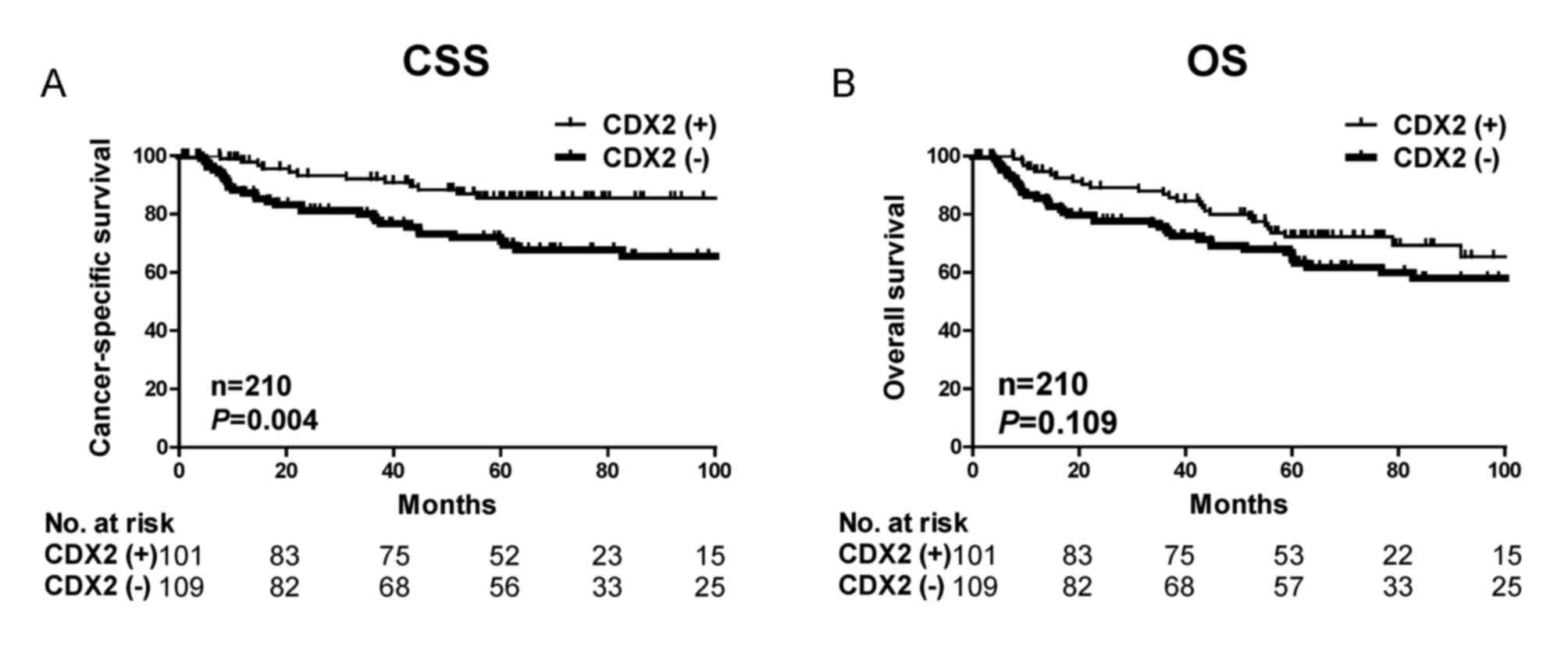
Kaplan-Meier analysis further demonstrated that
patients with decreased CDX2 expression had significantly poorer
cancer-specific survival outcome (P=0.004) but not overall survival
(P=0.109) in the present cohort (Fig.
2). The significant associations between CDX2 expression and
cancer-specific survival suggest that CDX2 expression may be a
useful prognostic biomarker for gastric cancer.
CDX2 expression is dependent on p53
expression and histological differentiation
The rate of positive p53 expression of was less
frequent in gastric cancer tissues with negative CDX2 expression
compared with tissues with positive CDX2 expression (P=0.097;
Table I). When the authors focused on
CDX2-negative gastric cancer tissues, it was identified that cases
with p53 positive expression were significantly less frequent in
undifferentiated type compared with differentiated type (P=0.021;
Table IV). These results indicated
that the TP53 gene was less frequently mutated in
undifferentiated gastric cancer with negative CDX2 expression
compared with differentiated gastric cancer.
 | Table IV.Evaluation of IHC for CDX2 and p53 in
differentiated and undifferentiated gastric cancer. |
Table IV.
Evaluation of IHC for CDX2 and p53 in
differentiated and undifferentiated gastric cancer.
| A, Negative CDX2
expression |
|---|
|
|---|
| Expression | Differentiated |
Undifferentiated |
P-valuea |
|---|
| Positive p53
expression | 27 | 21 | 0.021 |
| Negative p53
expression | 21 | 41 |
|
|
| B, Positive CDX2
expression |
|
| Expression | Differentiated |
Undifferentiated |
P-valuea |
|
| Positive p53
expression | 37 | 19 | 0.678 |
| Negative p53
expression | 27 | 17 |
|
CDX2 is suppressed during
inflammation-associated carcinogenesis
Recent studies have revealed that gene expression
can be specifically controlled by miRNAs. Therefore, the authors of
the present study have investigated to find out which miRNA has the
potential to directly suppress CDX2 expression. Based on the
published data (15) and miRwalk
(24), which consists of 8
established miRNA-target prediction programs, the authors of the
present study selected miR-181b, which is part of the IL-6/STAT3
signaling pathway. To examine whether miR-181b may directly
suppress CDX2 expression, the authors examined the level of CDX2
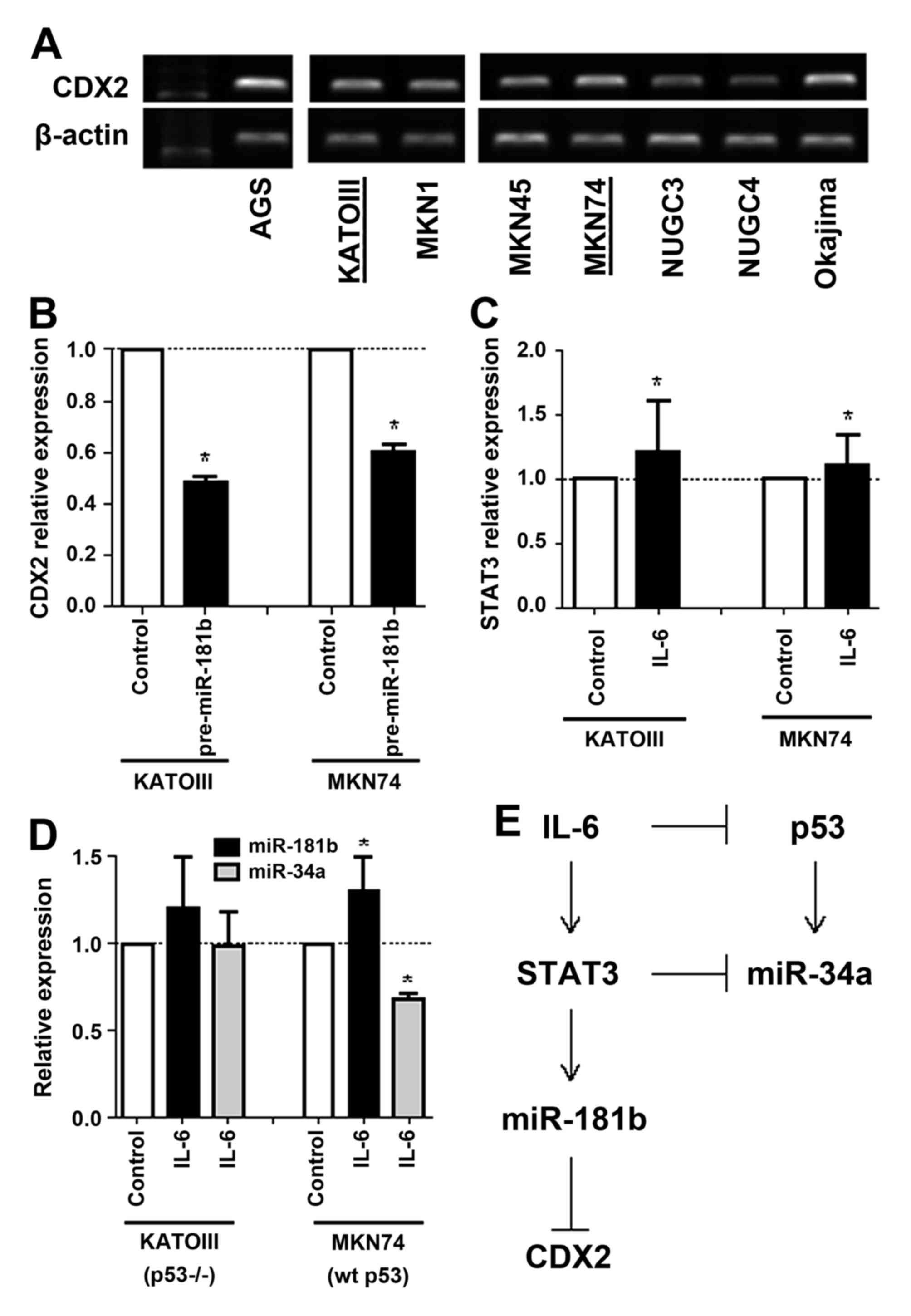
expression in gastric cancer cell lines and selected KATOIII
(p53-null) and MKN74 (p53-wild type) cells for further experiments
(Fig. 3A). Overexpression of mature
miR-181b by pre-miR-181b transfection suppressed CDX2 mRNA
expression in KATOIII and MKN74 gastric cancer cell lines (Fig. 3B). Compared with the control cells,
CDX2 was 0.5- and 0.6-fold lower in both cell lines that
overexpress miR-181b, respectively, suggesting that miR-181b may
directly suppress CDX2 expression in vitro.
Next, overexpression of IL-6 by transfection of
recombinant IL-6 induced STAT3 mRNA expression in KATOIII and MKN74
cancer cell lines (Fig. 3C). However,
while induction of miR-181b and suppression of miR-34b expression
by overexpression of IL-6 were observed in p53 wild-type MKN74
cells, those were not observed in p53 deleted KATOIII cells
(Fig. 3D). These results suggest that
activation of the IL-6/STAT3 signaling pathway suppresses CDX2
expression by inducing miR-181b and also may suppress p53 and
miR-34a expression in tumor with wild-type p53 (Fig. 3E).
Discussion
In the present study, the authors have demonstrated
that the association of decreased CDX2 expression with poorer
gastric cancer prognosis was significant. According to a previous
meta-analysis of CDX2 expression in gastric cancer, which analyzed
a combination of 4 different cohorts containing 475 patients, CDX2
was identified as a prognostic factor in gastric cancer (25). Although whether CDX2 may be associated
with patient outcome remains controversial, to the best our
knowledge the present study is the largest study to date with 210
cases, which provides additional evidence that CDX2 is an important
prognostic biomarker for gastric cancer.
A previous report demonstrated that nuclear staining
of CDX2 in IHC examination was a robust biomarker for predicting
prognosis in gastric cancer, whereas cytoplasmic staining of CDX2
was not (5). Although the clinical
significance of nuclear and cytoplasmic staining has not fully
understood, previous studies by the authors (5) and the present study also used nuclear
staining of CDX2 for analysis and confirmed positive association
with patient outcome but not cytoplasmic staining (data not shown).
Consistent with previous reports (5,25), in the
present study decreased CDX2 expression was associated with a
higher proportion of cases with poorly differentiated histological
type. The authors also performed IHC analysis of p53 and revealed
that there was a significant low frequency of positive p53
expression in gastric cancer tissues that are negative for CDX2
expression and undifferentiated. This result suggests that gastric
cancer with negative CDX2 expression occurs when there is a low
frequency of TP53 mutation or high frequency of p53
inactivation.
STAT3 is a downstream effector of IL-6 and form an
inflammatory positive feedback loop with miR-181b (14). The miR-181 family exhibits
cancer-specific expression patterns, and miR-181b is upregulated in
various malignant neoplasms, including breast, pancreas, prostate,
esophagus and gastric cancer (15,26–29). In
gastric cancer cell lines, miR-181b significantly increased cell
proliferation, migration and invasion (30). The present study revealed that
miR-181b directly regulates CDX2 expression in vitro and
thus that miR-181b has a central role in inflammation-associated
carcinogenesis. It was also found that miR-34a is suppressed by
activation of the IL-6/STAT3 signaling pathway. Since miR-34a is
induced by activation of p53, suppression of miR-34a may frequently
occur in tumor with wild-type p53. Notably, a previous report
demonstrated that the IL-6/STAT3 signaling pathway was also able to
downregulate p53 expression (31).
Considering the IHC results in the present study, the activation of
the IL-6/STAT3 signaling pathway may contribute to gastric
carcinogenesis via suppression of CDX2 and inactivation of p53,
resulting in a poor prognostic outcome.
The present study has several limitations. While the
present study has attempted to elucidate a functional relationship
between inflammation and gastric carcinogenesis in vitro,
further studies to investigate the interactions between IL-6/STAT3
and p53 signaling pathways are required. The present study has only
suggested that CDX2 expression is associated with p53 inactivation.
Additionally, it was not possible to collect patient history of
H. pylori infection. H. pylori infection is a sign
for chronic inflammation status, which is the main risk factor for
gastric cancer development. Therefore, this risk factor was not
examined. Previously, it was shown that STAT3 can be activated by
H. pylori infection (32),
therefore further studies that take this factor into account are
required.
Acknowledgements
The present study was supported by JSPS KAKENHI
(grant no. 24791439).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Sauer AM, Chen MS Jr,
Kagawa-Singer M, Jemal A and Siegel RL: Cancer statistics for Asian
Americans, Native Hawaiians, and Pacific Islanders, 2016:
Converging incidence in males and females. CA Cancer J Clin.
66:182–202. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Polk DB and Peek RM Jr: Helicobacter
pylori: Gastric cancer and beyond. Nat Rev Cancer. 10:403–414.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schetter AJ, Heegaard NH and Harris CC:
Inflammation and cancer: Interweaving microRNA, free radical,
cytokine and p53 pathways. Carcinogenesis. 31:37–49. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Okayama H, Kumamoto K, Saitou K, Hayase S,
Kofunato Y, Sato Y, Miyamoto K, Nakamura I, Ohki S, Sekikawa K and
Takenoshita S: CD44v6, MMP-7 and nuclear Cdx2 are significant
biomarkers for prediction of lymph node metastasis in primary
gastric cancer. Oncol Rep. 22:745–755. 2009.PubMed/NCBI
|
|
6
|
Liu Q, Teh M, Ito K, Shah N, Ito Y and
Yeoh KG: CDX2 expression is progressively decreased in human
gastric intestinal metaplasia, dysplasia and cancer. Mod Pathol.
20:1286–1297. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bai Z, Ye Y, Chen D, Shen D, Xu F, Cui Z
and Wang S: Homeoprotein Cdx2 and nuclear PTEN expression profiles
are related to gastric cancer prognosis. APMIS. 115:1383–1390.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fan Z, Li J, Dong B and Huang X:
Expression of Cdx2 and hepatocyte antigen in gastric carcinoma:
Correlation with histologic type and implications for prognosis.
Clin Cancer Res. 11:6162–6170. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ge J and Chen Z, Wu S, Yuan W, Hu B and
Chen Z: A clinicopathological study on the expression of
cadherin-17 and caudal-related homeobox transcription factor (CDX2)
in human gastric carcinoma. Clin Oncol (R Coll Radiol). 20:275–283.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang X, Tsukamoto T, Mizoshita T, Ban H,
Suzuki H, Toyoda T and Tatematsu M: Expression of osteopontin and
CDX2: Indications of phenotypes and prognosis in advanced gastric
cancer. Oncol Rep. 21:609–613. 2009.PubMed/NCBI
|
|
11
|
Silberg DG, Sullivan J, Kang E, Swain GP,
Moffett J, Sund NJ, Sackett SD and Kaestner KH: Cdx2 ectopic
expression induces gastric intestinal metaplasia in transgenic
mice. Gastroenterology. 122:689–696. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mutoh H, Sakurai S, Satoh K, Tamada K,
Kita H, Osawa H, Tomiyama T, Sato Y, Yamamoto H, Isoda N, et al:
Development of gastric carcinoma from intestinal metaplasia in
Cdx2-transgenic mice. Cancer Res. 64:7740–7747. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: Contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk
ML and Struhl K: STAT3 activation of miR-21 and miR-181b-1 via PTEN
and CYLD are part of the epigenetic switch linking inflammation to
cancer. Mol Cell. 39:493–506. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ji J, Yamashita T, Budhu A, Forgues M, Jia
HL, Li C, Deng C, Wauthier E, Reid LM, Ye QH, et al: Identification
of microRNA-181 by genome-wide screening as a critical player in
EpCAM-positive hepatic cancer stem cells. Hepatology. 50:472–480.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rokavec M, Öner MG, Li H, Jackstadt R,
Jiang L, Lodygin D, Kaller M, Horst D, Ziegler PK, Schwitalla S, et
al: IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated
colorectal cancer invasion and metastasis. J Clin Invest.
124:1853–1867. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sobin LH and Compton CC: TNM seventh
edition: What's new, what's changed: Communication from the
international union against cancer and the american joint committee
on cancer. Cancer. 116:5336–5339. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sobin LH, Gospodarowicz MK and Wittekind
CH: International Union against Cancer(UICC): TNM Classification of
Malignant Tumors 7th edition. Oxford, UK: Wiley-Blackwell; 2009
|
|
19
|
Tachibana K, Saito M, Imai JI, Ito E,
Yanagisawa Y, Honma R, Saito K, Ando J, Momma T, Ohki S, et al:
Clinicopathological examination of dipeptidase 1 expression in
colorectal cancer. Biomed Rep. 6:423–428. 2017.PubMed/NCBI
|
|
20
|
Bai Y, Akiyama Y, Nagasaki H, Yagi OK,
Kikuchi Y, Saito N, Takeshita K, Iwai T and Yuasa Y: Distinct
expression of CDX2 and GATA4/5, development-related genes, in human
gastric cancer cell lines. Mol Carcinog. 28:184–188. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Saito M, Schetter AJ, Mollerup S, Kohno T,
Skaug V, Bowman ED, Mathé EA, Takenoshita S, Yokota J, Haugen A and
Harris CC: The association of microRNA expression with prognosis
and progression in early-stage, non-small cell lung adenocarcinoma:
A retrospective analysis of three cohorts. Clin Cancer Res.
17:1875–1882. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Saito M, Shiraishi K, Matsumoto K,
Schetter AJ, Ogata-Kawata H, Tsuchiya N, Kunitoh H, Nokihara H,
Watanabe S, Tsuta K, et al: A three-microRNA signature predicts
responses to platinum-based doublet chemotherapy in patients with
lung adenocarcinoma. Clin Cancer Res. 20:4784–4793. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang XT, Wei WY, Kong FB, Lian C, Luo W,
Xiao Q and Xie YB: Prognostic significance of Cdx2
immunohistochemical expression in gastric cancer: A meta-analysis
of published literatures. J Exp Clin Cancer Res. 31:982012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang J, Sai K, Chen FR and Chen ZP:
miR-181b modulates glioma cell sensitivity to temozolomide by
targeting MEK1. Cancer Chemother Pharmacol. 72:147–158. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang B, Hsu SH, Majumder S, Kutay H, Huang
W, Jacob ST and Ghoshal K: TGFbeta-mediated upregulation of hepatic
miR-181b promotes hepatocarcinogenesis by targeting TIMP3.
Oncogene. 29:1787–1797. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao Y, Schetter AJ, Yang GB, Nguyen G,
Mathé EA, Li P, Cai H, Yu L, Liu F, Hang D, et al: microRNA and
inflammatory gene expression as prognostic marker for overall
survival in esophageal squamous cell carcinoma. Int J Cancer.
132:2901–2909. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guo JX, Tao QS, Lou PR, Chen XC, Chen J
and Yuan GB: miR-181b as a potential molecular target for
anticancer therapy of gastric neoplasms. Asian Pac J Cancer Prev.
13:2263–2267. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brighenti E, Calabrese C, Liguori G,
Giannone FA, Trerè D, Montanaro L and Derenzini M: Interleukin 6
downregulates p53 expression and activity by stimulating ribosome
biogenesis: A new pathway connecting inflammation to cancer.
Oncogene. 33:4396–4406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Giraud AS, Menheniott TR and Judd LM:
Targeting STAT3 in gastric cancer. Expert Opin Ther Targets.
16:889–901. 2012. View Article : Google Scholar : PubMed/NCBI
|