Introduction
Lung cancer, the most common cause of
cancer-associated mortality in women and men worldwide (1,2), has a
high invasive and metastatic potential, which leads to drug
resistance and treatment failure (3).
A total of 85% of all cases of lung cancers are of the non-small
cell lung cancer (NSCLC) histotype, of which lung adenocarcinoma is
the most frequent type (accounting for 40%) (4). Although advances in treatment have been
made in recent years, patients with lung adenocarcinoma continue to
have poor prognoses when the disease is detected at an advanced
clinical stage (5). Owing to its
growth site in the periphery of the lung, adenocarcinoma readily
invades the pleural cavity (6).
Visceral pleural invasion (VPI) has been regarded as a pathological
descriptor and a poor independent prognostic factor for the past
several decades in NSCLC (7–9). In addition to the current
Tumor-Node-Metastasis (TNM) classification recommendations
(10), in which T1 tumors with VPI
are upgraded to T2a, T2a tumors with VPI should be classified as
T2b (11). Evidence indicates that
patients suffering with VPI had a poorer prognosis, even when
adjusting for tumor size (12–14). In
addition, VPI is also associated with extensive N2 involvement
(9) and lymph node metastasis
(9,15,16), which
reflects the fact that malignant tumor cells can move to the
cervical venous circulation through the mediastinal lymphatic
vessels, leading to a greater incidence of mortality (8). VPI has long been recognized as an
important clinical factor by thoracic surgeons, medical oncologists
and radiation oncologists when treating patients with NSCLC.
Although a number of studies have focused on VPI in NSCLC (7,8,14), few have assessed the role of VPI in
lung adenocarcinomas, let alone explored the molecular
mechanism.
The epidermal growth factor receptor (EGFR), one of
the members of the ErbB receptor family, is activated by the
binding of its extracellular domain to specific ligands, including
EGF and transforming growth factor-α (TGFα), which causes
trans-autophosphorylation of EGFR and activation of the
intracellular tyrosine kinase activity ultimately leading to cell
proliferation, survival, invasion, and metastasis (17,18).
Approximately 10% of Western and 40% of Asian patients with lung
cancer harbor somatic EGFR mutations (19), which have a notable role in the
pathogenesis of lung cancer (20),
particularly in lung adenocarcinomas. Tsai et al (21) found that lung adenocarcinomas
harboring the EGFR-L858R mutation tended to invade the adjacent
pleural cavity and be involved in the formation of malignant
pleural effusion (MPE). Shi et al (22) analyzed the associations between EGFR
mutations and clinicopathological factors in lung adenosquamous
cell carcinoma cases and found that the incidence of positive
pleural invasion was higher in patients positive for the EGFR-L858R
mutation than that in those negative for the mutation. In addition,
NSCLC patients with EGFR mutations are sensitive to EGFR tyrosine
kinase inhibitors (TKIs) such as gefitinib (23). Of the different types of EGFR
mutations, exon 19 deletions and missense mutations (L858R) in exon
21 are the two predominant types, and have been found to be the
most powerful biologic predictors of EGFR TKI sensitivity (24). Although studies investigating clinical
and histological characteristics of patients harboring EGFR
mutations have been published in recent years (19–22), the
prognostic value of EGFR mutations in patients with peripheral lung
adenocarcinomas exhibiting VPI is uncertain.
MicroRNAs (miRNAs/miRs), a class of small
non-coding, highly conserved single-stranded RNAs, can serve as
oncogenes or tumor suppressors when the expression is dysregulated
in malignancies (25). For example,
miR-483-5p promotes the epithelial-mesenchymal transition (EMT),
which is frequently accompanied by the invasive, metastatic
transformation of lung adenocarcinoma cells (26). miR-543 inhibits the proliferation and
metastasis of colorectal cancer (CRC) cells, which indicates that
it may serve as a favorable diagnostic and biomarker for CRC
metastasis (27). However, the
potential to use molecules similar to these for cancer diagnosis
has not yet been systematically explored. A previous study revealed
that microRNA expression could be used to distinguish between
tumors of different developmental origin, and between tumors and
normal tissue, and reflect the state of cellular differentiation
(28). miRNAs may serve as useful
clinical biomarkers for the screening of high-risk populations and
detection of solid tumors in the early stages of cancer
progression, and may also be novel targets for cancer therapy
(29,30). Valeri et al demonstrated that
use of anti-miR-135b in CRC mouse models has substantial
therapeutic potential, which indicated that miRNAs are a promising
therapeutic target (30). Previous
findings demonstrated that miR-135b was upregulated in many types
of cancer, including colorectal cancer (31), hepatocellular carcinoma (32), head and neck squamous cell carcinoma
(33) and NSCLC (3). Additionally, miR-135b participates in
the development and progression of cancer by promoting the
proliferation, invasion and metastasis of tumor cells, (34). Although Lin et al (3) demonstrated that miR-135b can promote
lung cancer cell invasion in vitro and metastasis in
vivo, the role of miR-135b in lung adenocarcinomas was not
clear. In addition, the role of miR-135b in the formation of VPI
has not yet been reported.
Despite the general appreciation of the role of VPI
in lung cancer dissemination, little is known about the molecular
characteristics and gene expression profile that may attract cancer
cells to the pleural space (35). In
order to study the association between the miR-135b level, EGFR
mutations and VPI in peripheral lung adenocarcinoma, the present
study examined the expression level of miR-135b and EGFR mutations
using the reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) and DNA sequencing, respectively. The results of
the present study demonstrated that miR-135b was significantly
upregulated in lung adenocarcinoma compared with adjacent normal
tissue and positively associated EGFR mutations in peripheral lung
adenocarcinoma. Furthermore, it was identified that lung
adenocarcinomas with EGFR mutations and miR-135b overexpression
were more likely to invade visceral pleura. The present study
highlights the significance of miR-135b and EGFR mutations with
VPI; therefore, the present study provides potential drug targets
in peripheral lung adenocarcinoma therapy.
Materials and methods
Specimens
A total of 65 paraffin-embedded peripheral lung
adenocarcinoma tissue and corresponding pericarcinomatous tissue
that had been surgically excised between January 2012 and October
2014 from the Affiliated Zhoushan Hospital of Wenzhou Medical
University (Zhejiang, China) were used in this study. No patient
received radiotherapy, chemotherapy or any other antineoplastic
therapy prior to surgery. The cases were divided into those of
adenocarcinoma in situ/minimally invasive adenocarcinoma
(AIS/MIA) and invasive adenocarcinoma (IAC); there were 40 cases
(61.5%) of AIS/MIA and 25 cases (38.5%) of IAC. In total, 21
patients were male (32.3%) and 44 were female (67.7%). The median
age was 56 years (range, 31–83 years) (Table I). At least three qualified and
certified pathologists conducted strict identification of the
tissues, the classification of each sample was undetermined until
at least two-thirds of the recognized experts reached a consensus.
Staging and pathological classification standard reference to the
World Health Organization classification of lung cancer, 2011
(36). The present study was approved
by the Ethics Committee of the Affiliated Zhoushan Hospital of
Wenzhou Medical University.
 | Table I.Patient characteristics and
demographic data. |
Table I.
Patient characteristics and
demographic data.
|
Characteristics | Patients, n
(%) |
|---|
| Sex |
|
|
Male | 21 (32.3) |
|
Female | 44 (67.7) |
| Age, years |
|
|
≥60 | 25 (38.5) |
|
<60 | 40 (61.5) |
| Smoking status |
|
|
Yes | 14 (21.5) |
| No | 51 (78.5) |
| TNM clinical
stage |
|
| 0 | 26 (40.0) |
| IA | 22 (33.8) |
|
IB-IV | 17 (21.2) |
| Tumor size, cm |
|
| ≥3 | 60 (92.3) |
|
>3 | 5 (7.7) |
| Site |
|
|
Left | 28 (43.1) |
|
Right | 37 (56.9) |
| CEA, µg/ml |
|
| ≥5 | 8 (12.3) |
|
<5 | 57 (87.7) |
| VPI |
|
|
Yes | 17 (26.2) |
| No | 48 (73.8) |
| Histological
types |
|
|
AIS/MIA | 40 (61.5) |
|
IAC | 25 (38.5) |
Total RNA extraction
Total RNA, including miRNA, was extracted from 65
paraffin-embedded samples of tumor tissue and corresponding
pericarcinomatous tissue using RecoverAll™ Total Nucleic Acid
Isolation kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA)
and the concentration and purity of total RNA was measured using
the Q-3000 micro-ultraviolet spectrophotometer (Quawell Technology,
Inc., San Jose, CA, USA). Total RNA was extracted and stored at
−80°C until use.
Taqman-based RT-qPCR
The isolated total RNA was reverse transcribed using
TaqMan MicroRNA Reverse Transcription kit (Life Technologies;
Thermo Fisher Scientific, Inc.) for mature microRNA-135b and U6
internal control by MyCycler Thermal Cycler (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). Reaction conditions were: 16°C for 30
min, 42°C for 30 min, and 85°C for 5 min. PCR was then performed on
cDNA using TaqMan 2X Universal PCR Master Mix with no AmpErase
Uracil-N-Glycosylase and TaqMan Human MiRNA Assay kits (cat nos.
miR-135b, 002261; U6, 001973; Applied Biosystems; Thermo Fisher
Scientific, Inc.) on an Applied Biosystems 7500 real-time PCR
system (Thermocycling conditions were: 95°C for 10 min, followed by
40 cycles of 95°C for 15 sec and 60°C for 1 min). All qPCR
reactions were performed in triplicate. The quantity of miR-135b in
each peripheral lung adenocarcinoma tissue was calculated using the
2−ΔΔCq method (37) by SDS
2.0.1 software (Applied Biosystems; Thermo Fisher Scientific,
Inc.), ΔΔCq=ΔCq (cancer group)-ΔCq(adjacent cancer group), where
ΔCq=Cq(miR-135b)-Cq(U6).
Genomic DNA extraction
Genomic DNA was isolated from paraffin-embedded lung
adenocarcinoma tissue using the DNA FFRPE Tissue kit (Qiagen, Inc.,
Valencia, CA, USA), according to the manufacturer's instructions.
The concentration and purity of total DNA was measured using a
Q-3000 micro-ultraviolet spectrophotometer (Quawell Technology,
Inc.). DNA was authenticated by 1.2% agarose gel
electrophoresis.
PCR and product purification
PCR was used to analyze exons 19 and 21 of the EGFR
gene. The primers used were: exon 19 forward,
5′-CCCCAGCAATATCAGCCTTA-3′ and reverse,
5′-TGTGGGAGATGAGCAGGGTCT-3′; and exon 21 forward,
5′-GAATTCGGATGCAGAGCTTC-3′ and reverse, 5′-TCATTCACTGTCCCAGCAAG-3′.
The reaction system was determined as follows: 10X Buffer 5 µl, 5X
Q-Solution 10 µl, 100 mmol dNTP 0.1 µl (Applied Biosystems; Thermo
Fisher Scientific, Inc.), 2.5 units Hot-Star Taq DNA polymerase
(Qiagen, Inc.), 50 ng template DNA 5 µl, ddH2O up to 50
µl. PCR thermocycling conditions: 95°C for 15 min; 94°C for 15 sec,
60°C for 30 sec, 72°C for 1 min, 10 cycles; 94°C for 15 sec; 56°C
for 30 sec, 72°C for 1 min, 25 cycles; 72°C for 10 min. PCR
products was authenticated by 1.5% agarose gel electrophoresis and
purified using the EasyPure PCR Purification kit (Beijing Transgen
Biotech Co., Ltd., Beijing, China), according to the manufacturer's
instructions.
Gene sequencing for EGFR mutation
status
Sequencing reaction system: 1 µl BigDye, 0.5 ml
BigDye seqBuffer, 1 M primer, 1 µl PCR products that have been
purified, 1.5 µl ddH2O (96°C for 1 min; 96°C for 10 sec, 50°C for 5
sec, 60°C for 4 min, followed by 25 cycles) (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The PCR products of sequencing
reaction were purified using the ethanol and EDTA precipitation
method (19), and dissolved with
Hi-Di Formamide, followed by 95°C for 4 min, ice water for 4 min,
then separated by electrophoresis with ABI3100XL genetic analyzer.
The original sequencing results were analyzed using ABI Sequencing
Analysis V5.4 software (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The occurrence of EGFR mutations were confirmed
by comparative analysis between the results on ABI SeqScap software
version 5 (Applied Biosystems; Thermo Fisher Scientific, Inc.) and
the reference EGFR gene sequence (19), in combination with manual
checking.
Statistical analysis
Statistical analyses were performed using SPSS
v.21.0 (IBM Corp., Armonk, New York) and GraphPad Prism 5 (GraphPad
Software, Inc., La Jolla, CA, USA). Paired Student's t-test was
used to analyze the difference in the expression of miR-135b
between 65 pairs of peripheral lung adenocarcinomas and adjacent
non-cancerous tissue. Mann-Whitney U-test was used to compare the
expression level of miR-135b in different pathological
classification of lung adenocarcinomas. Pearson's χ2 test and
Fisher's exact test were used to evaluate the association between
miR-135b expression and clinicopathological characteristics in
peripheral lung adenocarcinomas, the association between EGFR
mutation status and clinicopathological characteristics, the
association between miR-135b overexpression and EGFR mutations, and
the association between EGFR mutations with miR-135b overexpression
and VPI. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-135b is markedly upregulated in
invasive lung adenocarcinoma compared to AIS/MIA
miR-135b expression was analyzed by TaqMan RT-qPCR,
relative to the endogenous control U6 snRNA, in 65 paired lung
adenocarcinomas and adjacent non-cancerous lung tissue specimens.
Compared with the adjacent tissue, miR-135b expression was
downregulated in 15 lung adenocarcinomas tissues (15/65, 23.1%) and
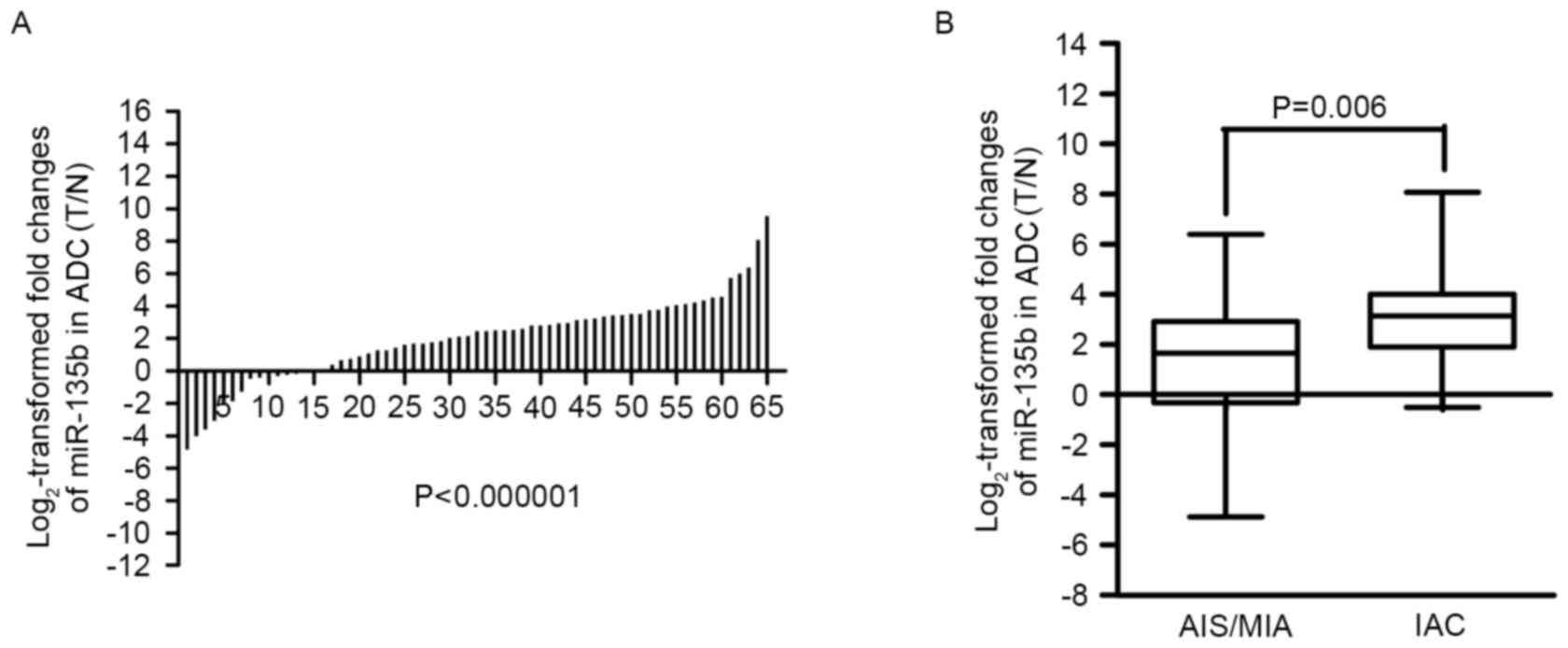
upregulated in 50 (50/65, 76.9%). The results revealed that
miR-135b is significantly upregulated in lung adenocarcinomas,
compared with the adjacent non-cancerous tissue (P=7.1248×10–8)
(Fig. 1A), indicating that miR-135b
may promote the development of lung adenocarcinomas.
To evaluate the biological functions of miR-135b in
peripheral lung adenocarcinomas, the expression of miR-135b was
analyzed in AIS/MIA and IAC. The present study revealed that
miR-135b was expressed at a higher level in IAC (P=0.006,
Mann-Whitney U-test) (Fig. 1B) than
AIS/MIA, which further indicated that miR-135b overexpression was
associated with the invasiveness of lung adenocarcinomas.
Additionally, Pearson's χ2 test and Fisher's exact
test were used to evaluate the association between miR-135b
expression and clinicopathological characteristics in peripheral
lung adenocarcinomas. There was no significant statistical
difference between miR-135b expression and the following
clinicopathological characteristics: gender, tumor size, smoking,
tumor site, TNM clinical stage, carcinoembryonic antigen (CEA)
levels and VPI. By contrast, higher levels of miR-135b were found
in patients with peripheral lung adenocarcinoma of advanced age
(≥60 years of age) (P=0.008) (Table
II).
 | Table II.Statistical analysis between miR-135b
expression and clinicopathological characteristics. |
Table II.
Statistical analysis between miR-135b
expression and clinicopathological characteristics.
|
| miR-135b Expression
in ADC (T/N) |
|
|---|
|
|
|
|
|---|
| Variable | Overexpressing | Not
overexpressing |
P-valuea |
|---|
| Sex |
|
| 0.368 |
|
Male | 17 | 4 |
|
|
Female | 31 | 13 |
|
| Age, years |
|
| 0.008 |
|
≥60 | 23 | 2 |
|
|
<60 | 25 | 15 |
|
| Tumor size, cm |
|
| 0.297 |
| ≥3 | 6 | 0 |
|
|
<3 | 42 | 17 |
|
| Smoking |
|
| 0.425 |
|
Yes | 12 | 2 |
|
| No | 36 | 15 |
|
| Site |
|
| 0.572 |
|
Left | 22 | 6 |
|
|
Right | 26 | 11 |
|
| TNM clinical
stage |
|
| 0.058 |
|
0-IA | 32 | 16 |
|
|
IB-IV | 16 | 1 |
|
| CEA, µg/ml |
|
| 0.171 |
| ≥5 | 8 | 0 |
|
|
<5 | 40 | 17 |
|
| VPI |
|
| 0.058 |
|
Yes | 16 | 1 |
|
| No | 32 | 16 |
|
EGFR mutation rate in invasive lung
adenocarcinoma was higher than that in AIS/MIA
Of the 65 samples of peripheral lung adenocarcinoma
in the present study, 23 were positive for EGFR mutations (35.4%),
with 20% (13 cases) and 15.4% (10 cases) positive for mutations in
exons 19 and 21, respectively. The in-frame deletions in exon 19
include 9 incidences of the ΔE746-A750 mutation, 1 incidence of the
ΔL747-A750 mutation and 3 incidences of the ΔL747-S752 mutation,
whereas the point mutations in exon 21 were 9 incidences of L858R
and one incidence of L861Q.
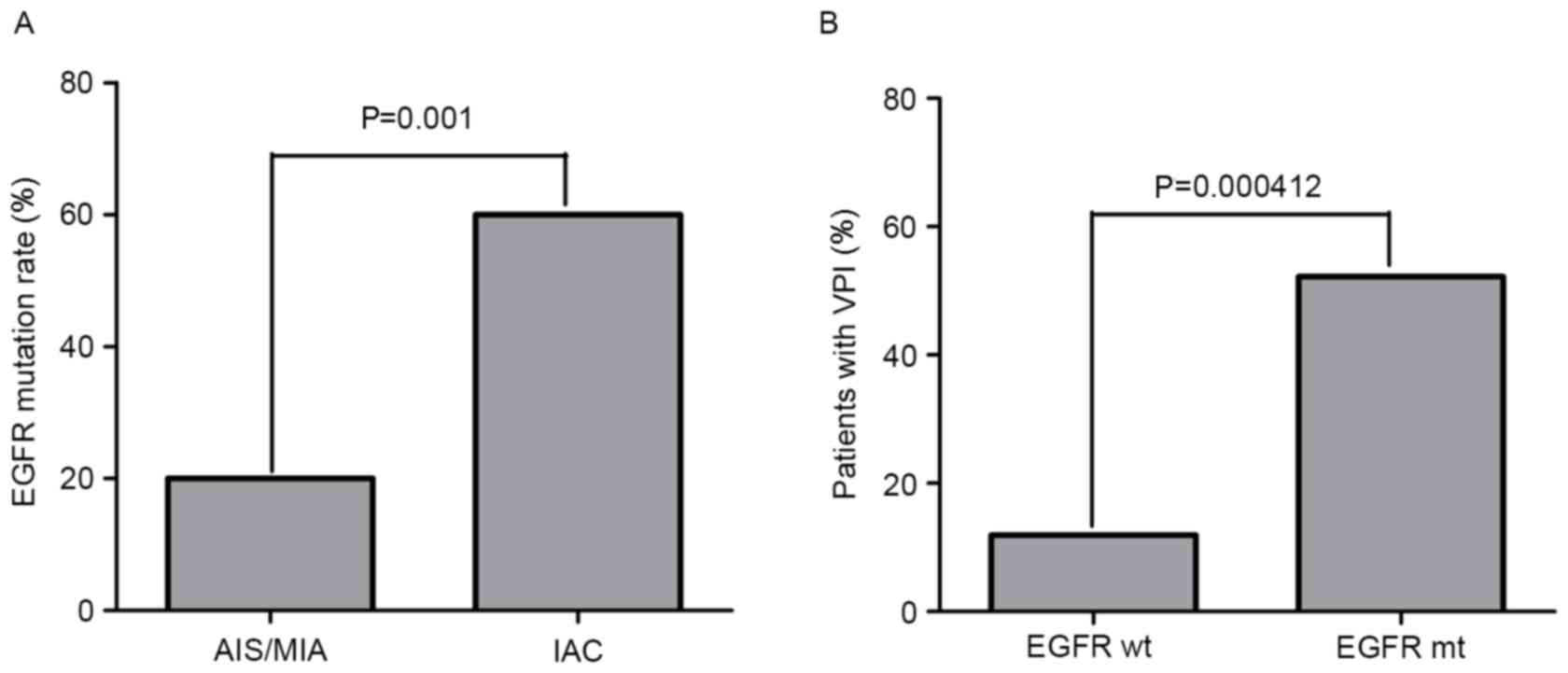
The EGFR mutation rate in IAC was 60%, which was
significantly higher than that in AIS/MIA (20%) (P=0.001) (Fig. 2A), demonstrating that EGFR mutation
was positively associated with the invasion of lung
adenocarcinoma.
Additionally, Pearson's χ2 test and Fisher's exact
test were used to evaluate the association between EGFR mutation
status and clinicopathological characteristics in peripheral lung
adenocarcinomas. Higher EGFR mutation rates were observed in
peripheral lung adenocarcinoma patients with characteristics
including advanced age (P=0.035), large tumor (P=0.033), advanced
TNM stage (P=0.001), high level of CEA (P=0.011) and visceral
pleural invasion (P=0.001) (Table
III). Of note is that patients harboring EGFR mutations had a
higher incidence of VPI (52.2%) than those with wild-type EGFR
(11.9%) (P=0.000412; Fig. 2B).
 | Table III.Statistical analysis between EGFR
mutation status and clinicopathological characteristics in
peripheral lung adenocarcinomas. |
Table III.
Statistical analysis between EGFR
mutation status and clinicopathological characteristics in
peripheral lung adenocarcinomas.
|
| Mutation status of
EGFR in ADC (T/N) |
|
|---|
|
|
|
|
|---|
| Variable | Mutated | Wild type | P-value |
|---|
| Sex |
|
| 0.154 |
|
Male | 10 | 11 |
|
|
Female | 13 | 31 |
|
| Age, years |
|
| 0.027 |
|
≥60 | 13 | 12 |
|
|
<60 | 10 | 30 |
|
| Tumor size, cm |
|
| 0.033 |
| ≥3 | 5 | 1 |
|
|
<3 | 18 | 41 |
|
| Smoking |
|
| 0.108 |
|
Yes | 8 | 6 |
|
| No | 15 | 36 |
|
| Site |
|
| 0.105 |
|
Left | 13 | 15 |
|
|
Right | 10 | 27 |
|
| TNM stage |
|
| <0.001 |
|
0-IA | 11 | 37 |
|
|
IB-IV | 12 | 5 |
|
| CEA, µg/ml |
|
| 0.011 |
| ≥5 | 6 | 1 |
|
|
<5 | 17 | 41 |
|
| VPI |
|
| <0.001 |
|
Yes | 12 | 5 |
|
| No | 11 | 37 |
|
Expression level of miR-135b in EGFR
mutation group is higher than that in EGFR wild-type group
According to the aforementioned results, it appears
that high expression levels of miR-135b and EGFR mutations were
positively associated with the invasion of peripheral lung
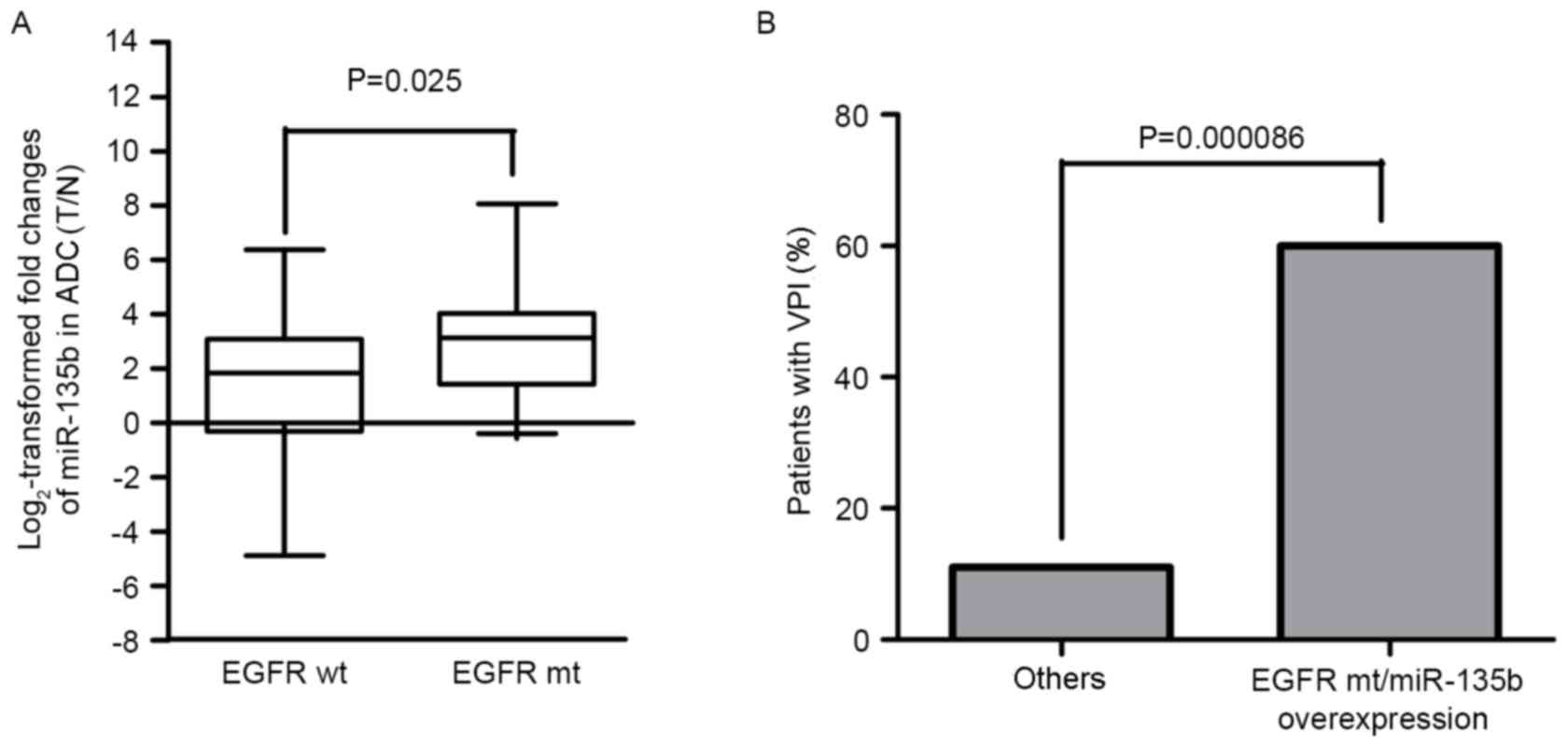
adenocarcinoma. To investigate whether miR-135b was associated with
mutations to EGFR, the 65 cases were divided into the EGFR mutation
and EGFR wild-type groups. The expression levels of miR-135b in the
EGFR mutation group were significantly higher than that in EGFR
wild-type group by χ2 test (P=0.020; Table IV) and Mann-Whitney U-test (P=0.025;
Fig. 3A), which revealed that
miR-135b expression was positively associated with mutations to
EGFR. Thus, we can also consider that the peripheral lung
adenocarcinomas patients harboring EGFR mutations usually have
higher expression levels of miR-135b.
 | Table IV.Patients harboring EGFR mutations
have higher expression levels of miR-135b than those with wild-type
EGFR. |
Table IV.
Patients harboring EGFR mutations
have higher expression levels of miR-135b than those with wild-type
EGFR.
|
| miR-135b Expression
in ADC (T/N) |
|
|---|
|
|
|
|
|---|
| Variable | Overexpressing |
Underexpressing |
P-valuea |
|---|
| Mutant EGFR | 21 | 2 | 0.020 |
| Wild-type EGFR | 27 | 15 |
|
Peripheral lung adenocarcinomas
harboring EGFR mutations with miRNA-135b overexpression are more
likely to invade visceral pleura
As a poor prognostic indicator in lung cancer, VPI
can be caused by invasion of lung cancer. As aforementioned, the
expression level of miR-135b was positively associated with
mutations to EGFR; the two had a synergistic role in the invasion
of peripheral lung adenocarcinoma. Thus, the association between
miR-135b expression, EGFR mutation status and VPI was analyzed.
Analysis using Pearson's χ2 and Fisher's exact tests
revealed that peripheral lung adenocarcinomas harboring EGFR
mutations with miR-135b overexpression (60%) were more likely to
invade the visceral pleura (Fig. 3B)
compared with other groups (11%) (P=0.000086; Table V). Table
VI shows the association between clinicopathological
characteristics and VPI. Statistical analysis revealed the
following clinicopathological factors as significant: Sex, age,
tumor size, smoking, TNM stage, serum CEA level, EGFR mutation
status, which demonstrated that VPI is a poor prognostic predictor
for patients with lung adenocarcinomas In addition, patients
harboring EGFR mutations with miR-135b overexpression have higher
incidence in the elderly (P=0.000119), lager tumor size (P=0.019),
advanced TNM classification (P=0.000086) and VPI (P=0.000086)
(Table VII).
 | Table V.Peripheral lung adenocarcinomas
harboring EGFR mutations with miR-135b overexpression are more
likely to invade visceral pleura. |
Table V.
Peripheral lung adenocarcinomas
harboring EGFR mutations with miR-135b overexpression are more
likely to invade visceral pleura.
| Variable | Total | VPI | No VPI |
P-valuea |
|---|
| miR-135b
overexpression, mutated EGFR | 21 | 12 | 9 | <0.001 |
| No miR-135b
overexpression, mutated EGFR | 2 | 0 | 2 | 0.970 |
| miR-135b
overexpression, wild-type EGFR | 27 | 4 | 23 | 0.080 |
| No miR-135b
overexpression, wild-type EGFR | 15 | 1 | 14 | 0.105 |
 | Table VI.The association between
clinicopathological characteristics and VPI. |
Table VI.
The association between
clinicopathological characteristics and VPI.
| Variable | VPI | No VPI | P-value |
|---|
| Sex |
|
| 0.034 |
|
Male | 9 | 12 |
|
|
Female | 8 | 36 |
|
| Age, years |
|
| 0.002 |
|
≥60 | 12 | 13 |
|
|
<60 | 5 | 35 |
|
| Tumor size, cm |
|
| <0.001 |
|
≥3cm | 6 | 0 |
|
|
<3cm | 11 | 48 |
|
| Smoking |
|
| 0.008 |
|
Yes | 8 | 6 |
|
| No | 9 | 42 |
|
| Site |
|
| 0.127 |
|
Left | 10 | 18 |
|
|
Right | 7 | 30 |
|
| TNM stage |
|
| <0.001 |
|
0-IA | 0 | 48 |
|
|
IB-IV | 17 | 0 |
|
| CEA (µg/ml) |
|
| <0.001 |
| ≥5 | 7 | 0 |
|
|
<5 | 10 | 38 |
|
| EGFR mutation
status |
|
| <0.001 |
|
Mutated | 12 | 11 |
|
| Wild
type | 5 | 37 |
|
| miR-135b |
|
| 0.058 |
|
Overexpressed | 16 | 32 |
|
| Not
overexpressed | 1 | 16 |
|
 | Table VII.Association between
clinicopathological characteristics and patients harboring EGFR
mutations with miR-135b overexpression. |
Table VII.
Association between
clinicopathological characteristics and patients harboring EGFR
mutations with miR-135b overexpression.
|
| miR-135b expression
status |
|
|---|
|
|
|
|
|---|
| Variable | Overexpressed | Not
overexpressed | P-value |
|---|
| Sex |
|
|
|
|
Male | 10 | 11 | 0.068 |
|
Female | 11 | 33 |
|
| Age, years |
|
|
|
|
≥60 | 12 | 13 | 0.032 |
|
<60 | 9 | 31 |
|
| Tumor size, cm |
|
|
|
| ≥3 | 5 | 1 | 0.019 |
|
<3 | 16 | 43 |
|
| Smoking status |
|
|
|
|
Yes | 8 | 6 | 0.055 |
| No | 13 | 38 |
|
| Site |
|
|
|
|
Left | 12 | 16 | 0.114 |
|
Right | 9 | 28 |
|
| TNM stage |
|
|
|
|
0-IA | 9 | 39 | <0.001 |
|
IB-IV | 12 | 5 |
|
| CEA, µg/ml |
|
|
|
| ≥5 | 7 | 5 | 0.055 |
|
<5 | 16 | 42 |
|
| VPI |
|
|
|
|
Yes | 12 | 5 | <0.001 |
| No | 9 | 39 |
|
Discussion
The latest classification of lung adenocarcinoma was
raised by the International Association for the Study of Lung
Cancer/American Thoracic Society/European Respiratory Society in
2011, which suggested three adenocarcinoma subtypes: AIS, MIA, and
IAC (38). It is hypothesized that
the development of lung adenocarcinomas may be a stepwise process
triggered by the sequential accumulation of genetic and epigenetic
changes from atypical adenomatous hyperplasia (AAH) to AIS, and
ultimately to IAC (36). A previous
study indicated that the overall survival (OS) rate of AIS was 100%
and patients with MIA had ~100% OS rate if the disease was
completely resected (39); however,
lepidic-predominant ADC, acinar-predominant ADC, papillary
predominant ADC, micropapillary predominant ADC, solid with mucin
production-predominant ADC and invasive mucinous ADC exhibited 93,
67, 74, 62, 58 and 76% OS rate if the disease was completely
resected, respectively (40). Owing
to the highly similar patient outcomes, AIS and MIA are combined
into a single category (36). On the
other hand, Tsuta et al (40)
suggested that AIS and MIA should be reclassified as tumor in
situ. Samples were therefore divided into two groups: AIS/MIA
and IAC. Peripheral lung adenocarcinoma, which arises in the
peripheral lung parenchyma, represents the dominant histological
subtype of all lung cancers (41) and
frequently leads to aggressive local invasion and metastatic
phenotypes (42,43). In this retrospective study, 65
peripheral lung adenocarcinoma samples were collected and tested
for EGFR mutation status and miR-135b expression level. Mutations
to EGFR and miR-135b overexpression were positively associated with
the invasion of lung adenocarcinomas, which may supplement the
molecular mechanism of invasiveness.
Lung cancer arises through the progressive
accumulation of mutations in oncogenes and tumor-suppressor genes
(42). Mutation of EGFR, the most
common targetable driver mutation in advanced lung adenocarcinoma
with the mutation rate varying between 43.5 and 76.6% in Asian
cohorts and between 9.6 and 29.8% in non-Asian cohorts, has
attracted increasing attention (44).
Numerous studies have detailed the close association between lung
cancer invasion and EGFR mutation status (45–47).
Kadota et al (46) found that
lepidic predominant adenocarcinoma tumors were statistically more
likely to have activating EGFR mutations compared with other
subtypes, with no EGFR mutations observed in AIS specimens and low
mutation levels in MIA specimens. Soh et al (47) also found the number of EGFR mutations
was increased in 39.5 and 50% of non-invasive and invasive lesions,
respectively. Yoshida et al found that EGFR mutations in AAH
and bronchioloalveolar carcinoma (BAC) were less frequent than
those detected in invasive adenocarcinoma (48), which indicated that EGFR mutations
might have a role in the multi-stage pathogenesis of lung
adenocarcinoma. The present study observed the same phenomenon:
Patients with invasive lesions had higher EGFR mutation rates than
those without such lesions. However, EGFR mutations were not
significantly associated with sex or smoking status in the present
study, which is inconsistent with previous findings that EGFR
mutations were more prevalent in women and non-smokers (19). Another study revealed that EGFR
mutations facilitated the migration of cancer cells into the
pleural cavity (49) and had largely
focused on advanced-stage lung adenocarcinoma (50), while the present study found that EGFR
mutations were more prevalent in the elderly (≥60 years of age),
and those with larger tumors (≥3 cm), aggravated tumor TNM
classification (IB-IV), higher CEA levels (≥5 µg/ml) and VPI, which
demonstrated the positive association between EGFR mutations and
lung adenocarcinoma progression. The number of patients analyzed in
the present study was small, and EGFR mutation rates may fluctuate
owing to variations in geographic patient location and methods of
examination. Further studies are therefore required to confirm the
aforementioned results.
miR-135b has long been regarded as an oncogenic
miRNA in different cancer types (3,31–33). In the present study, the expression
level of miR-135b was significantly higher in lung adenocarcinoma
samples than that in corresponding non-cancerous tissue. Just as
previous research has shown that miR-135b can promote lung cancer
cell invasion in vitro (3),
the present study found miR-135b was markedly upregulated in
invasive lung adenocarcinoma compared to AIS/MIA, which may further
demonstrate that miR-135b can promote the development of lung
cancer by enhancing the invasiveness of tumor cells in vitro
and in vivo. Thus, miR-135b may be a novel biomarker for
lung cancer progression (30). In
addition, miR-135b overexpression was pervasive in the elderly
according to our study and patients with VPI tended to have higher
levels of miR-135b (P=0.058).
Different oncogenic pathways can converge to affect
the same miRNA, for example, miR-135b overexpression is associated
with gene mutations in many pathways. Valeri et al (30) revealed that miR-135b overexpression is
associated with mutations in specific colorectal cancer (CRC)
pathways [mutations to adenomatous polyposis coli,
phosphatidylinositol 3-kinase (PI3K), and SRC proto-oncogene,
non-receptor tyrosine kinase]. PI3K/protein kinase B, rat sarcoma
viral oncogene homolog (RAS)/mitogen-activated protein kinase and
signal transducer and activator of transcription 3 (STAT3) are
three major downstream pathways activated by EGFR phosphorylation,
which eventually results in tumor cell proliferation, migration and
metastasis, and evasion from apoptosis. A previous study also
revealed that EGFR signaling is required for tumor maintenance in
human lung adenocarcinoma-expressing EGFR mutants (51). EGFR mutations are hypothesized to
promote the expression of miR-135b through the activation of STAT3;
thus, miR-135b may be a biomarker and driver of resistance to TKI
therapy (52–55). The present study found that miR-135b
overexpression is significantly associated with EGFR mutation
status, generally lung adenocarcinoma patients harboring EGFR
mutations have higher miR-135b expression levels. As miRNAs often
act as downstream effectors of protein kinases or driver genes
mutated in cancer (56), miR-135b may
mediate its carcinogenic role by participating in these signaling
pathways. Targeting miR-135b may therefore represent a strategy to
increase the specificity and overcome drug resistance to
EGFR-mutation-targeted therapies.
In the present study, VPI was observed in 26.2%
(17/65) of the surgically resected NSCLC specimens, which nearly
matched the 26.8% reported by Kimihiro et al (55). VPI has a higher incidence in patients
with large tumors, high CEA levels, advanced TNM classification,
being elderly (≥60 years of age), EGFR mutations and men in the
present study, which conformed with previous studies that found
that VPI was observed significantly more frequently in tumors with
factors indicative of tumor aggressiveness, including poorer
histologic grade (57), larger tumor
size (8) and high serum CEA level
(57). In general, VPI in lung
adenocarcinoma patients indicates an invasive and aggressive
biology. Although many studies have reported the existence of an
association between clinicopathological characteristics and VPI in
NSCLC (34,58,59),
little is known regarding the genetic changes or signaling pathways
involved in the development of VPI in lung adenocarcinoma. Numerous
studies have shown that the incidence of EGFR mutations in patients
with MPE is significantly higher than in those without (49,60);
however, few studies have indicated the association between VPI and
EGFR mutations. The KRAS gene, a member of the RAS family, is a
downstream factor of the EGFR signaling pathway (47), and Raparia et al (61) found that peripheral lung
adenocarcinomas harboring KRAS mutations are more likely to invade
the visceral pleura, suggesting that there may be an association
between EGFR signaling pathway and visceral pleura involvement. In
the present study, VPI was positively associated with mutations to
EGFR. Furthermore, miR-135b is significantly associated with EGFR
mutation status and may be a downstream effector in EGFR signaling
pathway; therefore, we hypothesized that EGFR mutations and
miR-135b overexpression can promote the invasion of lung
adenocarcinoma through EGFR signaling pathway. Finally, lung
adenocarcinoma patients harboring EGFR mutations were more likely
to exhibit invasion of the visceral pleura when miR-135b was
overexpressed, which further indicated the potential signaling
pathway of EGFR mutations/miR-135b may promote the formation of VPI
and miR-135b may be a good biomarker of the tumor progression.
Additionally, targeting miR-135b may minimize drug resistance to
EGFR mutations and increase therapeutic potential.
Acknowledgements
The present study was supported by grants from the
Scientific Research Foundation of Ministry of Public Health of
China/Major Science and Technology Program of Medicine and Health
of Zhejiang Province of China (no. WKJ2014-2-021), the National
Natural Science Foundation of China (no. 81502106), the Natural
Science Foundation of Zhejiang Province (no. LY15H160047) the
Medical Bureau of Zhejiang Province (nos. 2015ZDA032 and
2016341029), the Science and Technology Bureau of Zhoushan (no.
2015C31029) and the Science Technology Department of Zhejiang
Province (no. 2016C37008).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Meoni G, Cecere FL, Lucherini E and Di
Costanzo F: Medical treatment of advanced non-small cell lung
cancer in elderly patients: A review of the role of chemotherapy
and targeted agents. J Geriatr Oncol. 4:282–290. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lin CW, Chang YL, Chang YC, Lin JC, Chen
CC, Pan SH, Wu CT, Chen HY, Yang SC, Hong TM and Yang PC:
MicroRNA-135b promotes lung cancer metastasis by regulating
multiple targets in the hippo pathway and LZTS1. Nat Commun.
4:18772013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Travis WD, Brambilla E and Riely GJ: New
pathologic classification of lung cancer: Relevance for clinical
practice and clinical trials. J Clin Oncol. 31:992–1001. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siegelin MD and Borczuk AC: Epidermal
growth factor receptor mutations in lung adenocarcinoma. Lab
Invest. 94:129–137. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee YC and Light RW: Management of
malignant pleural effusions. Respirology. 9:148–156. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rami-Porta R, Bolejack V, Crowley J, Ball
D, Kim J, Lyons G, Rice T, Suzuki K, Thomas CF Jr, Travis WD, et
al: The IASLC lung cancer staging project: Proposals for the
revisions of the t descriptors in the forthcoming eighth edition of
the tnm classification for lung cancer. J Thorac Oncol.
10:990–1003. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ou SH, Zell JA, Ziogas A and Anton-Culver
H: Prognostic significance of the non-size-based AJCC T2
descriptors: Visceral pleura invasion, hilar atelectasis, or
obstructive pneumonitis in stage IB non-small cell lung cancer is
dependent on tumor size. Chest. 133:662–669. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Manac'h D, Riquet M, Medioni J, Le
Pimpec-Barthes F, Dujon A and Danel C: Visceral pleura invasion by
non-small cell lung cancer: An underrated bad prognostic factor.
Ann Thorac Surg. 71:1088–1093. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rami-Porta R, Bolejack V, Crowley J, Ball
D, Kim J, Lyons G, Rice T, Suzuki K, Thomas CF Jr, Travis WD, et
al: The IASLC lung cancer staging project: Proposals for the
revisions of the T descriptors in the forthcoming eighth edition of
the tnm classification for lung cancer. J Thorac Oncol.
10:990–1003. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kawase A, Yoshida J, Miyaoka E, Asamura H,
Fujii Y, Nakanishi Y, Eguchi K, Mori M, Sawabata N, Okumura M, et
al: Visceral pleural invasion classification in non-small-cell lung
cancer in the 7th edition of the tumor, node, metastasis
classification for lung cancer: Validation analysis based on a
large-scale nationwide database. J Thorac Oncol. 8:606–611. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yoshida J, Nagai K, Asamura H, Goya T,
Koshiishi Y, Sohara Y, Eguchi K, Mori M, Nakanishi Y, Tsuchiya R,
et al: Visceral pleura invasion impact on non-small cell lung
cancer patient survival: Its implications for the forthcoming TNM
staging based on a large-scale nation-wide database. J Thorac
Oncol. 4:959–963. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kudo Y, Saji H, Shimada Y, Nomura M,
Matsubayashi J, Nagao T, Kakihana M, Usuda J, Kajiwara N, Ohira T
and Ikeda N: Impact of visceral pleural invasion on the survival of
patients with non-small cell lung cancer. Lung cancer. 78:153–160.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fibla JJ, Cassivi SD, Brunelli A, Decker
PA, Allen MS, Darling GE, Landreneau RJ and Putnam JB:
Re-evaluation of the prognostic value of visceral pleura invasion
in Stage IB non-small cell lung cancer using the prospective
multicenter ACOSOG Z0030 trial data set. Lung cancer. 78:259–262.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takizawa T, Terashima M, Koike T, Watanabe
T, Kurita Y, Yokoyama A and Honma K: Lymph node metastasis in small
peripheral adenocarcinoma of the lung. J Thorac Cardiovasc Surg.
116:276–280. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kang JH, Kim KD and Chung KY: Prognostic
value of visceral pleura invasion in non-small cell lung cancer.
Eur J Cardiothorac Surg. 23:865–869. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hirsch FR, Scagliotti GV, Langer CJ,
Varella-Garcia M and Franklin WA: Epidermal growth factor family of
receptors in preneoplasia and lung cancer: Perspectives for
targeted therapies. Lung cancer. 1 41 Suppl:S29–S42. 2003.
View Article : Google Scholar
|
|
18
|
Kazandjian D, Blumenthal GM, Yuan W, He K,
Keegan P and Pazdur R: FDA approval of gefitinib for the treatment
of patients with metastatic EGFR mutation-positive non-small cell
lung cancer. Clin Cancer Res. 22:1307–1312. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dacic S, Shuai Y, Yousem S, Ohori P and
Nikiforova M: Clinicopathological predictors of EGFR/KRAS
mutational status in primary lung adenocarcinomas. Mod Pathol.
23:159–168. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ikeda K, Nomori H, Ohba Y, Shibata H, Mori
T, Honda Y, Iyama K and Kobayashi T: Epidermal growth factor
receptor mutations in multicentric lung adenocarcinomas and
atypical adenomatous hyperplasias. J Thorac Oncol. 3:467–471. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tsai MF, Chang TH, Wu SG, Yang HY, Hsu YC,
Yang PC and Shih JY: EGFR-L858R mutant enhances lung adenocarcinoma
cell invasive ability and promotes malignant pleural effusion
formation through activation of the CXCL12-CXCR4 pathway. Sci Rep.
5:135742015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shi X, Wu H, Lu J, Duan H, Liu X and Liang
Z: Screening for major driver oncogene alterations in adenosquamous
lung carcinoma using PCR coupled with next-generation and Sanger
sequencing methods. Sci Rep. 6:222972016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mok TS, Wu YL, Thongprasert S, Yang CH,
Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et
al: Gefitinib or carboplatin-paclitaxel in pulmonary
adenocarcinoma. N Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang Y, Shi C, Sun H, Yin W, Zhou X, Zhang
L and Jiang G: Elderly male smokers with right lung tumors are
viable candidates for KRAS mutation screening. Sci Rep.
6:185662016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gasparini P, Cascione L, Landi L, Carasi
S, Lovat F, Tibaldi C, Alì G, D'Incecco A, Minuti G, Chella A, et
al: microRNA classifiers are powerful diagnostic/prognostic tools
in ALK-, EGFR-, and KRAS-driven lung cancers. Proc Natl Acad Sci
USA. 112:pp. 14924–14929. 2015, View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Song Q, Xu Y, Yang C, Chen Z, Jia C, Chen
J, Zhang Y, Lai P, Fan X, Zhou X, et al: miR-483-5p promotes
invasion and metastasis of lung adenocarcinoma by targeting RhoGDI1
and ALCAM. Cancer Res. 74:3031–3042. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fan C, Lin Y, Mao Y, Huang Z, Liu AY, Ma
H, Yu D, Maitikabili A, Xiao H, Zhang C, et al: MicroRNA-543
suppresses colorectal cancer growth and metastasis by targeting
KRAS, MTA1 and HMGA2. Oncotarget. 7:21825–21839. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang WB, Chen PH, Hsu T I, Fu TF, Su WC,
Liaw H, Chang WC and Hung JJ: Sp1-mediated microRNA-182 expression
regulates lung cancer progression. Oncotarget. 5:740–753. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Valeri N, Braconi C, Gasparini P, Murgia
C, Lampis A, Paulus-Hock V, Hart JR, Ueno L, Grivennikov SI, Lovat
F, et al: MicroRNA-135b promotes cancer progression by acting as a
downstream effector of oncogenic pathways in colon cancer. Cancer
cell. 25:469–483. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nagel R, le Sage C, Diosdado B, van der
Waal M, Vrielink Oude JA, Bolijn A, Meijer GA and Agami R:
Regulation of the adenomatous polyposis coli gene by the miR-135
family in colorectal cancer. Cancer Res. 68:5795–5802. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Y, Xu D, Bao C, Zhang Y, Chen D, Zhao
F, Ding J, Liang L, Wang Q, Liu L, et al: MicroRNA-135b, a HSF1
target, promotes tumor invasion and metastasis by regulating RECK
and EVI5 in hepatocellular carcinoma. Oncotarget. 6:2421–2433.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang L, Sun ZJ, Bian Y and Kulkarni AB:
MicroRNA-135b acts as a tumor promoter by targeting the
hypoxia-inducible factor pathway in genetically defined mouse model
of head and neck squamous cell carcinoma. Cancer Lett. 331:230–238.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu W, Wang Z, Yang P, Yang J, Liang J,
Chen Y, Wang H, Wei G, Ye S and Zhou Y: MicroRNA-135b regulates
metastasis suppressor 1 expression and promotes migration and
invasion in colorectal cancer. Mol Cell Biochem. 388:249–259. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Agalioti T, Giannou AD and Stathopoulos
GT: Pleural involvement in lung cancer. J Thorac Dis. 7:1021–1030.
2015.PubMed/NCBI
|
|
36
|
Travis WD, Brambilla E, Noguchi M,
Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ,
Van Schil PE, et al: International association for the study of
lung cancer/american thoracic society/european respiratory society
international multidisciplinary classification of lung
adenocarcinoma. J Thorac Oncol. 6:244–285. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kadota K, Villena-Vargas J, Yoshizawa A,
Motoi N, Sima CS, Riely GJ, Rusch VW, Adusumilli PS and Travis WD:
Prognostic significance of adenocarcinoma in situ, minimally
invasive adenocarcinoma, and nonmucinous lepidic predominant
invasive adenocarcinoma of the lung in patients with stage I
disease. Am J Surg Pathol. 38:448–460. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yoshizawa A, Motoi N, Riely GJ, Sima CS,
Gerald WL, Kris MG, Park BJ, Rusch VW and Travis WD: Impact of
proposed IASLC/ATS/ERS classification of lung adenocarcinoma:
Prognostic subgroups and implications for further revision of
staging based on analysis of 514 stage I cases. Mod Pathol.
24:653–664. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tsuta K, Kawago M, Inoue E, Yoshida A,
Takahashi F, Sakurai H, Watanabe S, Takeuchi M, Furuta K, Asamura H
and Tsuda H: The utility of the proposed IASLC/ATS/ERS lung
adenocarcinoma subtypes for disease prognosis and correlation of
driver gene alterations. Lung cancer. 81:371–376. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cagle PT, Allen TC, Dacic S, Beasley MB,
Borczuk AC, Chirieac LR, Laucirica R, Ro JY and Kerr KM: Revolution
in lung cancer: New challenges for the surgical pathologist. Arch
Pathol Lab Med. 135:110–116. 2011.PubMed/NCBI
|
|
42
|
Eto T, Suzuki H, Honda A and Nagashima Y:
The changes of the stromal elastotic framework in the growth of
peripheral lung adenocarcinomas. Cancer. 77:646–656. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nakanishi H, Matsumoto S, Iwakawa R, Kohno
T, Suzuki K, Tsuta K, Matsuno Y, Noguchi M, Shimizu E and Yokota J:
Whole genome comparison of allelic imbalance between noninvasive
and invasive small-sized lung adenocarcinomas. Cancer Res.
69:1615–1623. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Clay TD, Russell PA, Do H, Sundararajan V,
Conron M, Wright GM, Dobrovic A, Moore MM and McLachlan SA:
Associations between the IASLC/ATS/ERS lung adenocarcinoma
classification and EGFR and KRAS mutations. Pathology. 48:17–24.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yatabe Y, Takahashi T and Mitsudomi T:
Epidermal growth factor receptor gene amplification is acquired in
association with tumor progression of EGFR-mutated lung cancer.
Cancer Res. 68:2106–2111. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kadota K, Yeh YC, D'Angelo SP, Moreira AL,
Kuk D, Sima CS, Riely GJ, Arcila ME, Kris MG, Rusch VW, et al:
Associations between mutations and histologic patterns of mucin in
lung adenocarcinoma: Invasive mucinous pattern and extracellular
mucin are associated with KRAS mutation. Am J Surg Pathol.
38:1118–1127. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Soh J, Toyooka S, Ichihara S, Asano H,
Kobayashi N, Suehisa H, Otani H, Yamamoto H, Ichimura K, Kiura K,
et al: Sequential molecular changes during multistage pathogenesis
of small peripheral adenocarcinomas of the lung. J Thorac Oncol.
3:340–347. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yoshida Y, Shibata T, Kokubu A, Tsuta K,
Matsuno Y, Kanai Y, Asamura H, Tsuchiya R and Hirohashi S:
Mutations of the epidermal growth factor receptor gene in atypical
adenomatous hyperplasia and bronchioloalveolar carcinoma of the
lung. Lung cancer. 50:1–8. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Han HS, Eom DW, Kim JH, Kim KH, Shin HM,
An JY, Lee KM, Choe KH, Lee KH, Kim ST, et al: EGFR mutation status
in primary lung adenocarcinomas and corresponding metastatic
lesions: Discordance in pleural metastases. Clin Lung Cancer.
12:380–386. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Mok T, Wu YL and Zhang L: A small step
towards personalized medicine for non-small cell lung cancer.
Discov Med. 8:227–231. 2009.PubMed/NCBI
|
|
51
|
Mitsudomi T and Yatabe Y: Mutations of the
epidermal growth factor receptor gene and related genes as
determinants of epidermal growth factor receptor tyrosine kinase
inhibitors sensitivity in lung cancer. Cancer Sci. 98:1817–1824.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Alvarez JV, Greulich H, Sellers WR,
Meyerson M and Frank DA: Signal transducer and activator of
transcription 3 is required for the oncogenic effects of
non-small-cell lung cancer-associated mutations of the epidermal
growth factor receptor. Cancer Res. 66:3162–3168. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Matsuyama H, Suzuki HI, Nishimori H,
Noguchi M, Yao T, Komatsu N, Mano H, Sugimoto K and Miyazono K:
miR-135b mediates NPM-ALK-driven oncogenicity and renders
IL-17-producing immunophenotype to anaplastic large cell lymphoma.
Blood. 118:6881–6892. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Takata S, Takigawa N, Segawa Y, Kubo T,
Ohashi K, Kozuki T, Teramoto N, Yamashita M, Toyooka S, Tanimoto M
and Kiura K: STAT3 expression in activating EGFR-driven
adenocarcinoma of the lung. Lung Cancer. 75:24–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Khatri R and Subramanian S: MicroRNA-135b
and its circuitry networks as potential therapeutic targets in
colon cancer. Front Oncol. 3:2682013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Shimizu K, Yoshida J, Nagai K, Nishimura
M, Ishii G, Morishita Y and Nishiwaki Y: Visceral pleural invasion
is an invasive and aggressive indicator of non-small cell lung
cancer. J Thorac Cardiovasc Surg. 130:160–165. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Riquet M, Badoual C, Le Pimpec Barthes F,
Lhote FM, Souilamas R, Hubsch JP and Danel C: Visceral pleura
invasion and pleural lavage tumor cytology by lung cancer: A
prospective appraisal. Ann Thorac Surg. 75:353–355. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Butnor KJ and Cooper K: Visceral pleural
invasion in lung cancer: Recognizing histologic parameters that
impact staging and prognosis. Adv Anat Pathol. 12:1–6. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zou J, Bella AE, Chen Z, Han X, Su C, Lei
Y and Luo H: Frequency of EGFR mutations in lung adenocarcinoma
with malignant pleural effusion: Implication of cancer biological
behaviour regulated by EGFR mutation. J Int Med Res. 42:1110–1117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Raparia K, Villa C, Raj R and Cagle PT:
Peripheral lung adenocarcinomas with KRAS mutations are more likely
to invade visceral pleura. Arch Pathol Lab Med. 139:189–193. 2015.
View Article : Google Scholar : PubMed/NCBI
|