Introduction
Liver cirrhosis is defined as the histological
development of regenerative nodules surrounded by fibrous
connective tissue in response to chronic liver disease (1). Generally, liver cirrhosis is
asymptomatic and unsuspected. Liver biopsy is considered as the
gold standard for diagnosis of cirrhosis, followed by assessment of
risk progression according to sequential histological grading of
inflammation and staging of fibrosis. However, biopsy is prone to
considerable sampling variability in all liver diseases (2–5).
Considering the limitations of liver biopsy, developing a reliable
non-invasive and convenient method for liver cirrhosis has become
an urgent requirement for early diagnosis (6–8).
MicroRNAs (miRNAs/miRs) are a class of
evolutionarily conserved small (18–24 nucleotides) single-stranded
non-coding RNAs, which regulate target gene expression at the
post-transcriptional level by the degradation of target mRNA or the
suppression of mRNA translation subsequent to its specific binding
to target mRNA (9,10). In addition, miRNAs are highly stable
in serum and plasma, providing the possibility of evaluating
circulatory miRNA as a biomarker (11,12).
Dysregulated miRNA expression has been reported in nearly all types
of human cancer, including hepatocellular carcinoma (HCC) (13–15).
Several miRNAs were identified as potential biomarkers for liver
injury (16–20). A previous study also identified
frequent and extensive dysregulation of miRNA in liver adenoma and
cirrhosis, and found that different liver cancer stages may affect
the dysregulation of miRNA expression (21).
Expression microarray and next-generation sequencing
may reveal more differentially regulated miRNAs. However, due to
the heterogeneity of the disease, variations of sample sources and
diversity of analysis methods, the results of these studies are
inconsistent (13). In the present
study, the miRNA and mRNA expression profiles for liver cirrhosis
tissues from the Gene Expression Omnibus (GEO) database were
integrated. In addition, differentially expressed miRNA target
genes were predicted, and a miRNA-target regulatory network was
constructed. Certain key miRNAs were established as biomarkers for
diagnosis, prognosis and prediction of therapeutic response.
Materials and methods
Gene expression profiles
The miRNA and mRNA expression profiles for liver
cirrhosis tissues were obtained from the public repository of GEO
database (http://www.ncbi.nlm.nih.gov/geo) (22). The following key words were used:
‘Liver cirrhosis’ and ‘Homo sapiens’. Previous studies that
compared gene expression profiling between liver cirrhosis and
normal tissues or cultured cells were included in the present
study. The type of study was defined as ‘expression profiling by
array’ or ‘non-coding RNA profiling by array’.
Screening of differentially expressed
miRNAs and mRNAs
Raw data were preprocessed via background correction
and normalization. The Limma package in R (23) was used to identify the differentially
expressed miRNAs and mRNAs between liver cirrhosis tissues and
controls with an unpaired t-test, then a P-value was calculated.
P-values from multiple studies were combined using Fisher's exact
test, and the random effects model was used to combine effect size
from multiple studies. The miRNAs and mRNAs with a false discovery
rate of <0.01 were regarded as differently expressed miRNAs and
mRNAs.
Identification of regulatory targets
for differently expressed miRNA
As the miRNAs recognize their regulatory targets
through base pairing, computational methods have been invaluable
for identifying these targets. The target genes for human miRNAs
were downloaded from miRTarBase (http://mirtarbase.mbc.nctu.edu.tw), and the
transcriptional targets of the identified miRNAs in liver cirrhosis
tissues were predicted. In the present study, the differentially
expressed target genes whose expression was inversely correlated
with that of miRNAs were subjected to subsequent study. The
miRNA-target gene interaction network in liver cirrhosis tissues
was then constructed, and the network was visualized using
Cytoscape (http://www.cytoscape.org) (24).
Functional enrichment analysis of
differentially expressed target genes
The Database for Annotation, Visualization and
Integrated Discovery (25) is the
most common tool to analyze the functional enrichment of genes. To
gain insights into the biological functions of miRNA target genes,
Gene Ontology (GO) (26)
classification and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway (27) enrichment analysis
were performed. P<0.05 was selected as the cutoff criterion.
Clinical specimens
The blood samples were obtained from 5 patients with
liver cirrhosis and 3 healthy volunteers. Written consent was
obtained from all of the study participants, and the project was
approved by the committee of Ditan Hospital (Beijing, China) for
the screening, inspection and data collection from the
patients.
RNA preparation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using the RNAiso Plus
reagent (Takara Bio, Inc., Otsu, Japan) according to the
manufacturer's protocol. Total RNA (1 µg) was reverse-transcribed
using a cDNA Synthesis kit (Beijing Transgen Biotech Co., Ltd.,
Beijing, China), and the resulting cDNA was used as the template
for qPCR. qPCR was performed in a BIO-RAD IQ5 Real-Time PCR system
with a PCR SYBR Green Master mix reagent kit (Beijing Transgen
Biotech Co., Ltd.). Following an initial denaturing for 2 min at
94°C, qPCR was performed with 45 cycles of 94°C for 20 sec and 60°C
for 34 sec. All reactions were performed in triplicate. The results
were analyzed using the Cq method, using Data Assist Software
version 3.0 (Applied Biosystems; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). The human β-actin gene was used as an endogenous
control. The sequences of the PCR primers used are provided in
Table I. The universal primer used
for miR-21, miR-199a-3p and U6 was obtained from the miRcute Plus
miRNA qPCR Detection kit (SYBR-Green) (cat. no. FP411; Tiangen
Biotech Co., Ltd.). Data were analyzed with a one-way analysis of
variance and Tukey's test using Microsoft Excel 2010 (Microsoft
Corporation, Redmond, WA, USA). P<0.05 was considered to
indicate a statistically significant difference.
 | Table I.Primers used in the present
study. |
Table I.
Primers used in the present
study.
| Primers | Sequences
(5′-3′) |
|---|
| KIAA1524 |
|
|
Forward |
GCCACTCTGGGAAGCCATACTAAA |
|
Reverse |
GCAGCAGAAGGGTCACAAAACG |
| β-actin |
|
|
Forward |
ACTTAGTTGCGTTACACCCTT |
|
Reverse |
GTCACCTTCACCGTTCCA |
| hsa-miR-21
forward |
TAGCTTATCAGACTGATGTTG |
| hsa-miR-199a-3p
forward |
CCCAGTGTTCAGACTACCTGTTC |
| U6 forward |
CTGCGCAAGGATGACACGCAAATT |
Results
Differentially expressed miRNAs and
mRNAs in liver cirrhosis tissues
In total, 4 mRNA expression profiling studies and 2
miRNA expression profiling studies that met the inclusion criteria
were included. The characteristics of each study are summarized in
Table II. Of these studies, 166
cases of liver cirrhosis and 64 controls were analyzed.
 | Table II.Characteristics of mRNA and miRNA
expression profiles of liver cirrhosis. |
Table II.
Characteristics of mRNA and miRNA
expression profiles of liver cirrhosis.
| Author | Year | Country | GEO ID | Platform | Samples (N:C) | PMID | (Refs.) |
|---|
| mRNA expression
profile |
|
|
|
|
|
|
|
| Yildiz
et al | 2013 | Turkey | GSE17548 | GPL570
[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | 0:20 | 23691139 | (50) |
| Archer
et al | 2009 | USA | GSE17967 | GPL571 [HG-U133A_2]
Affymetrix Human Genome U133A 2.0 Array | 0:47 | 19861515 | (51) |
| Mas
et al | 2009 | USA | GSE14323 | GPL96 [HG-U133A]
Affymetrix Human Genome U133A Array | 19:41 | 19098997 | (52) |
| Caillot
et al | 2008 | France | GSE10356 | GPL5215 INSERM
Homo sapiens 14K array Liverpool3 | 24:21 | 19140229 | (53) |
| miRNA expression
profile |
|
|
|
|
|
|
|
|
Wojcicka et al | 2014 | Poland | GSE63046 | GPL11154 Illumina
HiSeq 2000 | 9:15 | 24875649 | (54) |
|
Vuppalanchi et al | 2013 | USA | GSE49012 | GPL17470 Thermo
Scientific Dharmacon microRNA human array | 12:22 | 24058572 | (55) |
Following normalization of the original miRNA and
mRNA expression datasets, a total of 48 differentially expressed
miRNAs were identified, including 5 that were upregulated and 43
that were downregulated (Table
III). Using integrated analysis, a set of 1,773 differentially
expressed genes (DEGs) were identified in liver cirrhosis tissues
compared with normal tissues, including 1,037 upregulated and 736
downregulated genes.
 | Table III.List of differentially expressed
miRNAs in liver cirrhosis. |
Table III.
List of differentially expressed
miRNAs in liver cirrhosis.
| miRNAs | Log
(fold-change) | P-value |
|---|
| Upregulated
miRNAs |
|
|
|
hsa-miR-212 | 4.45E-01 |
6.29×10−4 |
|
hsa-miR-142-5p | 9.23E-01 |
1.88×10−3 |
|
hsa-miR-142-3p | 9.39E-01 |
2.38×10−3 |
|
hsa-miR-199a-3p | 1.07E+00 |
8.38×10−4 |
|
hsa-miR-21 | 1.09E+00 |
4.15×10−4 |
| Downregulated
miRNAs |
|
|
|
hsa-miR-132 | −9.37E-01 |
7.20×10−4 |
|
hsa-miR-139-5p | −9.18E-01 |
4.80×10−4 |
|
hsa-miR-181b | −8.57E-01 |
2.22×10−4 |
|
hsa-miR-21* | −8.36E-01 |
3.55×10−4 |
|
hsa-miR-18a | −8.31E-01 |
4.50×10−4 |
|
hsa-let-7d | −8.30E-01 |
2.10×10−4 |
|
hsa-miR-20b | −8.28E-01 |
4.43×10−4 |
|
hsa-miR-660 | −8.26E-01 |
4.15×10−4 |
|
hsa-miR-376c | −8.16E-01 |
1.65×10−3 |
|
hsa-miR-192; | −7.74E-01 |
1.99×10−4 |
|
hsa-miR-377 | −7.67E-01 |
3.60×10−3 |
|
hsa-miR-18b | −7.67E-01 |
6.73×10−4 |
|
hsa-miR-29c* | −7.66E-01 |
1.21×10−3 |
|
hsa-miR-375 | −7.66E-01 |
6.19×10−4 |
|
hsa-miR-148b | −7.63E-01 |
5.17×10−4 |
|
hsa-miR-127-3p | −7.62E-01 |
1.64×10−3 |
|
hsa-miR-136* | −7.59E-01 |
5.34×10−4 |
|
hsa-miR-200c | −7.59E-01 |
1.93×10−3 |
|
hsa-miR-342-3p | −7.45E-01 |
5.11×10−3 |
|
hsa-miR-923 | −7.31E-01 |
8.61×10−4 |
|
hsa-miR-151a-5p | −7.30E-01 |
7.01×10−4 |
|
hsa-miR-191 | −7.27E-01 |
1.06×10−3 |
|
hsa-miR-450a | −7.14E-01 |
1.15×10−3 |
|
hsa-miR-151a-3p | −7.13E-01 |
8.95×10−4 |
|
hsa-miR-454 | −6.99E-01 |
3.24×10−3 |
|
hsa-miR-141 | −6.97E-01 |
4.59×10−3 |
|
hsa-let-7f-1* | −6.89E-01 |
2.58×10−3 |
|
hsa-miR-204 | −6.87E-01 |
2.84×10−4 |
|
hsa-miR-128 | −6.71E-01 |
3.74×10−3 |
|
hsa-miR-30a* | −6.59E-01 |
4.98×10−3 |
|
hsa-miR-92b | −6.46E-01 |
8.45×10−3 |
|
hsa-miR-103 | −6.30E-01 |
6.62×10−4 |
|
hsa-miR-582-5p | −6.26E-01 |
2.76×10−3 |
|
hsa-miR-455-5p | −6.23E-01 |
3.55×10−3 |
|
hsa-miR-519e* | −6.13E-01 |
3.10×10−4 |
|
hsa-miR-99a | −6.12E-01 |
7.53×10−3 |
|
hsa-miR-22 | −6.05E-01 |
2.12×10−3 |
|
hsa-miR-122* | −5.79E-01 |
5.61×10−3 |
|
hsa-miR-99b | −5.73E-01 |
1.43×10−3 |
|
hsa-miR-552 | −5.53E-01 |
5.33×10−3 |
|
hsa-miR-219-1-3p | −4.92E-01 |
3.57×10−3 |
|
hsa-miR-548a-3p | −3.93E-01 |
7.99×10−3 |
|
hsa-miR-491-5p | −3.52E-01 |
4.61×10−3 |
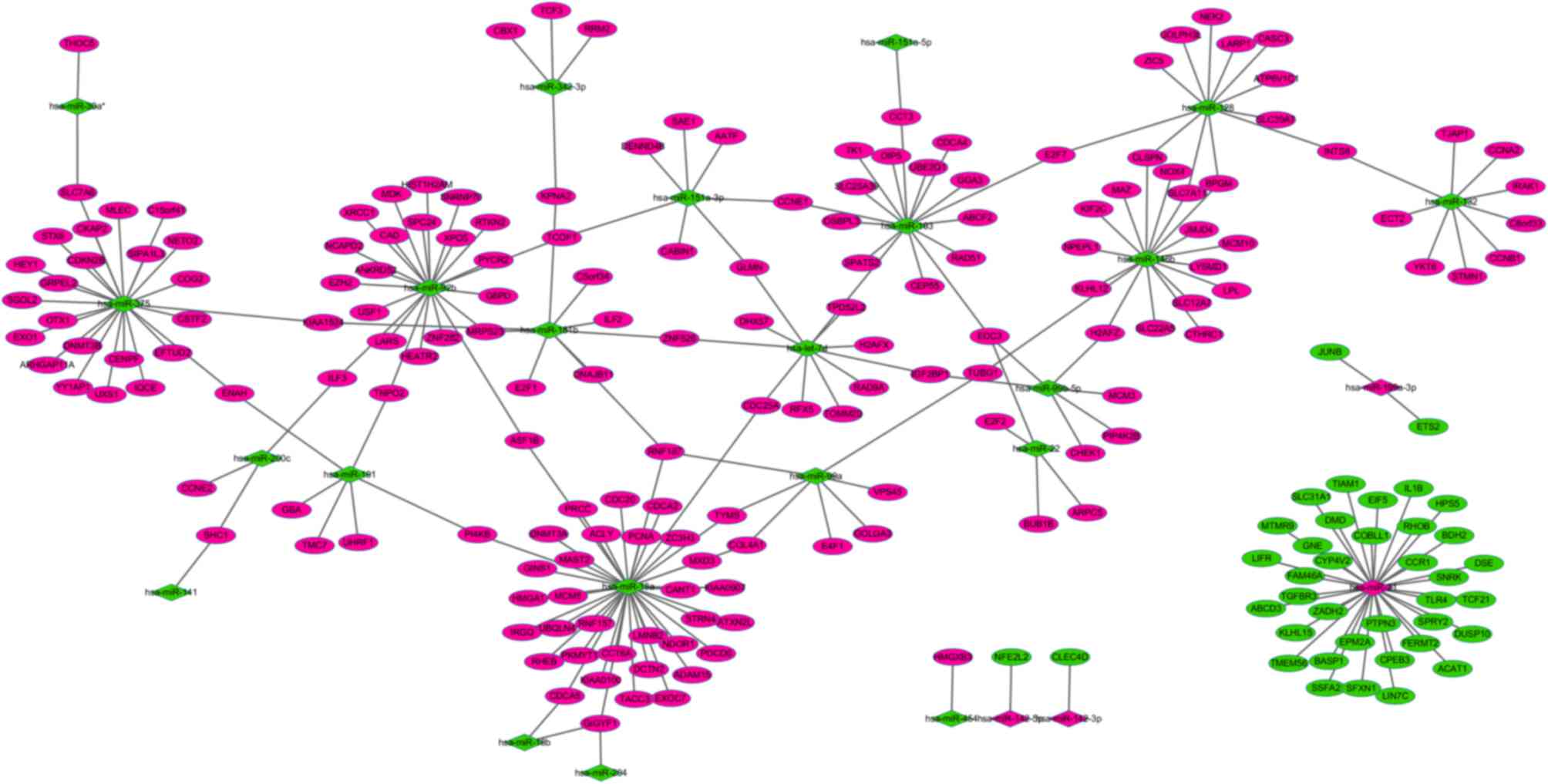
Regulatory network of miRNAs and
targets in liver cirrhosis
The miRTarBase database was used to predict the
putative transcriptional targets of upregulated or downregulated
miRNAs in liver cirrhosis tissues. Subsequent to comparing the
putative targets with DEGs in liver cirrhosis tissues, a total of
240 miRNA-target gene pairs, whose expression was inversely
correlated, were obtained. According to the miRNA-target gene
pairs, a miRNA-target gene regulatory network was constructed
(Fig. 1). In the network, the top
three miRNAs (miR-21, miR-18a and miR-375) regulated the majority
of the target genes. Leukemia inhibitory factor (LIFR) was
identified as a target of miR-21, whereas cancerous inhibitor of
protein phosphatase 2A (KIAA1524) and E2F transcription factor 1
(E2F1) were identified as targets of miR-181b.
Functional enrichment analysis
GO enrichment analysis of all target genes was
performed to understand their biological functions. It was
identified that the significantly enriched GO terms for molecular
functions were protein binding, nucleotide binding and DNA binding.
The significantly enriched GO terms for cellular components were
nucleus, cytoplasm and cytosol, while those for biological
processes were mitotic cell cycle, cell division and DNA
replication (Table IV).
 | Table IV.Top 15 GO terms of differentially
expressed microRNA target genes. |
Table IV.
Top 15 GO terms of differentially
expressed microRNA target genes.
| GO ID | GO term | Count | P-value | False discovery
rate |
|---|
| Biological
process |
|
|
|
|
|
GO:0000278 | Mitotic cell
cycle | 25 |
5.14×10−21 |
4.38×10−18 |
|
GO:0051301 | Cell division | 20 |
1.17×10−15 |
4.99×10−13 |
|
GO:0006260 | DNA
replication | 14 |
5.92×10−13 |
1.68×10−10 |
|
GO:0000082 | G1/S transition of
mitotic cell cycle | 13 |
5.73×10−12 |
1.22×10−9 |
|
GO:0007067 | Mitosis | 13 |
1.42×10−10 |
2.42×10−8 |
|
GO:0000075 | Cell cycle
checkpoint | 11 |
5.14×10−10 |
7.29×10−8 |
|
GO:0007049 | Cell cycle | 16 |
1.16×10−8 |
1.42×10−6 |
|
GO:0000086 | G2/M transition of
mitotic cell cycle protein kinase activity | 9 |
3.69×10−8 |
3.93×10−6 |
|
GO:0000079 | Regulation of
cyclin-dependent | 7 |
4.26×10−8 |
4.03×10−6 |
|
GO:0000077 | DNA damage
checkpoint | 5 |
4.59×10−7 |
3.91×10−5 |
|
GO:0000236 | Mitotic
prometaphase | 7 |
8.88×10−7 |
6.31×10−5 |
|
GO:0031100 | Organ
regeneration | 6 |
8.27×10−7 |
6.41×10−5 |
|
GO:0000087 | M phase of mitotic
cell cycle | 7 |
1.89×10−6 |
1.24×10−4 |
|
GO:0051726 | Regulation of cell
cycle | 6 |
3.35×10−6 |
2.04×10−4 |
|
GO:0000083 | Regulation of
transcription involved in G1/S phase of mitotic cell cycle | 4 |
5.85×10−6 |
3.32×10−4 |
| Cellular
component |
|
|
|
|
|
GO:0005634 | Nucleus | 99 |
4.92×10−27 |
1.02×10−24 |
|
GO:0005737 | Cytoplasm | 96 |
7.53×10−26 |
7.79×10−24 |
|
GO:0005829 | Cytosol | 51 |
1.68×10−17 |
1.16×10−15 |
|
GO:0005654 | Nucleoplasm | 32 |
6.06×10−16 |
3.14×10−14 |
|
GO:0005730 | Nucleolus | 30 |
5.18×10−9 |
2.14×10−7 |
|
GO:0005813 | Centrosome | 14 |
1.28×10−8 |
4.41×10−7 |
|
GO:0000785 | Chromatin | 8 |
1.87×10−8 |
5.53×10−7 |
|
GO:0005694 | Chromosome | 11 |
7.36×10−7 |
1.90×10−5 |
|
GO:0005856 | Cytoskeleton | 19 |
1.10×10−6 |
2.28×10−5 |
|
GO:0000776 | Kinetochore | 6 |
1.03×10−6 |
2.38×10−5 |
|
GO:0000775 | Chromosome,
centromeric region | 6 |
1.57×10−6 |
2.96×10−5 |
|
GO:0000794 | Condensed nuclear
chromosome | 4 |
2.38×10−5 |
4.10×10−4 |
|
GO:0000777 | Condensed
chromosome kinetochore | 5 |
3.49×10−5 |
5.56×10−4 |
|
GO:0048471 | Perinuclear region
of cytoplasm | 11 |
5.25×10−5 |
7.76×10−4 |
|
GO:0000922 | Spindle pole | 5 |
9.46×10−5 |
1.31×10−3 |
| Molecular
function |
|
|
|
|
|
GO:0005515 | Protein
binding | 100 |
6.85×10−35 |
2.18×10−32 |
|
GO:0000166 | Nucleotide
binding | 41 |
3.15×10−11 |
5.00×10−9 |
|
GO:0003677 | DNA binding | 31 |
1.26×10−7 |
1.34×10−5 |
|
GO:0005524 | Adenosine
triphosphate binding | 26 |
1.32×10−6 |
1.05×10−4 |
|
GO:0019899 | Enzyme binding | 9 |
2.50×10−6 |
1.59×10−4 |
|
GO:0019904 | Protein domain
specific binding | 8 |
6.99×10−6 |
3.70×10−4 |
|
GO:0016301 | Kinase
activity | 9 |
1.10×10−5 |
5.01×10−4 |
|
GO:0019901 | Protein kinase
binding | 8 |
1.05×10−4 |
4.19×10−3 |
|
GO:0043425 | Basic
helix-loop-helix transcription factor binding | 3 |
1.39×10−4 |
4.93×10−3 |
|
GO:0042393 | Histone
binding | 4 |
3.25×10−4 |
1.03×10−2 |
|
GO:0003700 | Sequence-specific
DNA binding transcription factor activity | 15 |
4.07×10−4 |
1.08×10−2 |
|
GO:0042826 | Histone deacetylase
binding | 4 |
4.00×10−4 |
1.16×10−2 |
|
GO:0051082 | Unfolded protein
binding | 5 |
8.39×10−4 |
2.05×10−2 |
|
GO:0008565 | Protein transporter
activity | 4 |
9.22×10−4 |
2.09×10−2 |
|
GO:0008139 | Nuclear
localization sequence binding | 2 |
1.28×10−3 |
2.54×10−2 |
The results of KEGG pathway enrichment analysis
indicated that the most significantly enriched pathway was the cell
cycle, which may have a significant effect on liver cirrhosis
(Table V).
 | Table V.KEGG pathways of differentially
expressed microRNA target genes. |
Table V.
KEGG pathways of differentially
expressed microRNA target genes.
| KEGG ID | KEGG term | Count | False discovery
rate |
|---|
| hsa04110 | Cell cycle | 15 |
1.47×10−13 |
| hsa05222 | Small cell lung
cancer | 6 |
7.27×10−4 |
| hsa04115 | Tumor protein 53
signaling pathway | 5 |
2.13×10−3 |
| hsa00240 | Pyrimidine
metabolism | 5 |
7.56×10−3 |
| hsa04114 | Oocyte meiosis | 5 |
1.31×10−2 |
| hsa00072 | Synthesis and
degradation of ketone bodies | 2 |
1.85×10−2 |
| hsa03030 | DNA
replication | 3 |
2.05×10−2 |
| hsa05215 | Prostate
cancer | 4 |
2.15×10−2 |
| hsa03013 | RNA transport | 5 |
2.25×10−2 |
| hsa05162 | Measles | 5 |
2.32×10−2 |
| hsa04914 |
Progesterone-mediated oocyte
maturation | 4 |
2.42×10−2 |
| hsa05200 | Pathways in
cancer | 7 |
3.61×10−2 |
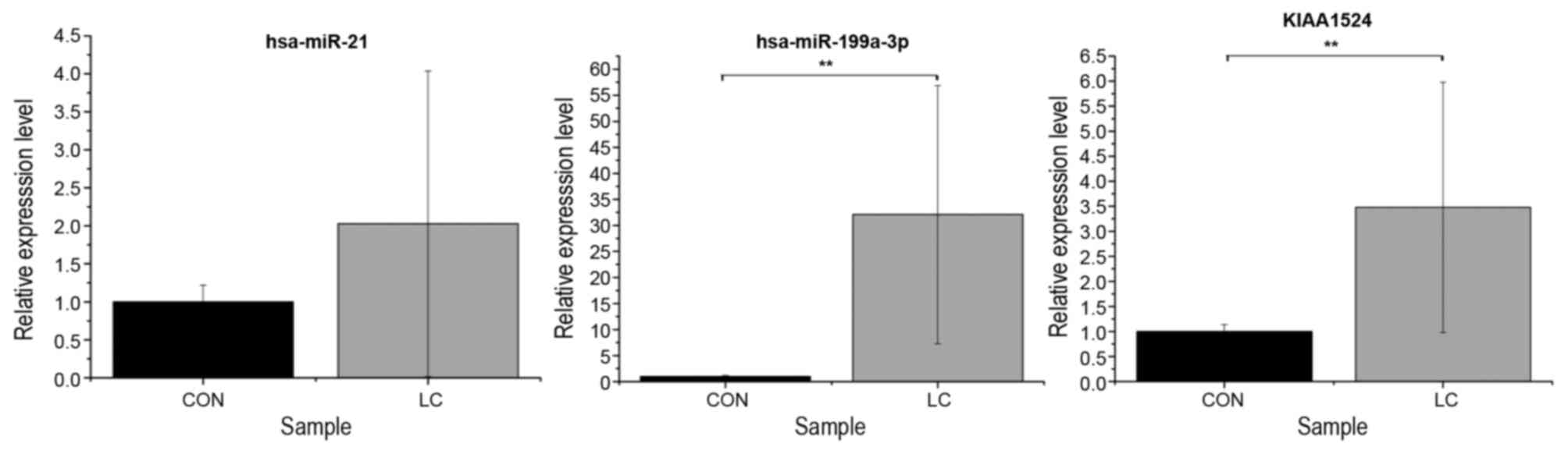
RT-qPCR validation
To validate the associated between miRNAs and their
target genes in the regulatory network, the blood samples from 5
pairs of patients with liver cirrhosis and normal controls were
used. hsa-miR-21 (P=0.1678), hsa-miR-199a-3p (P=0.0001) and
KIAA1524 (P=0.0028) were selected for RT-qPCR. The RT-qPCR results
indicated that the expression was consistent with the integrated
analysis data (Fig. 2).
Discussion
An increasing number of studies have focused on the
role of miRNAs in regulating cancer progression and metastasis,
including proliferation, invasion, migration, angiogenesis and
apoptosis (28,29). Emerging evidence suggests that
circulating miRNAs may function as stable, reliable and
non-invasive diagnostic biomarkers for cancer (30). In the present study, 2 miRNA
expression profiling studies and 4 mRNA expression profiling
studies of liver cirrhosis tissues were integrated, and a set of 48
differentially expressed miRNA and 1,773 DEGs were identified. By
bioinformatics prediction, a total of 240 miRNA-target gene pairs
were obtained, whose expression was inversely correlated. The
miRNA-targets regulatory network was constructed for liver
cirrhosis tissues. In the miRNA-targets regulatory network, it was
identified that the top ten upregulated and downregulated miRNAs
were hsa-miR-212, hsa-miR-21, hsa-miR-199a-3p, hsa-miR-142-5p,
hsa-miR-142-3p, hsa-miR-132, hsa-miR-139-5p, hsa-miR-181b,
hsa-miR-21* and hsa-miR-18a, and the majority of them had
previously been identified to be closely associated with alcoholic
liver disease, hepatic fibrogenesis, liver cirrhosis or
hepatocellular carcinoma (31–34).
Several miRNAs are involved in hepatic fibrogenesis
(20,35,36). It
was previously suggested that miR-21 was upregulated at the onset
of fibrosis in the human liver, and that it may promote fibrogenic
activation of fibroblasts (37,38) and be
involved in the amplification of certain important cellular
signaling pathways (39,40). A recent study also indicated that a
significant elevation of hepatic miR-21 expression is associated
with mitogen-activated protein kinase 3 signaling and
epithelial-mesenchymal transition in liver fibrosis (34). The miR-199 family is associated with
liver fibrosis. By comprehensive analysis, Murakami et al
(41) identified that the high
expression of 4 miRNAs (miR-199a, miR-199a*, miR-200a and miR-200b)
were closely associated with the progression of liver fibrosis in
humans and mice. Consistent with this, the present study revealed
that miR-21 and miR-199a-3p were also significantly upregulated in
liver cirrhosis tissues, implying that they may serve an important
role in the progression of liver cirrhosis and that they may be
promising biomarkers for the early diagnosis of liver
cirrhosis.
A previous study demonstrated that the other two
miRNAs, miR-142-3p and miR-142-5p, were significantly downregulated
in HCC, and that they may cooperatively regulate cell movement
(42). Conversely, the results of the
present study indicated that miR-142-5p and miR-142-3p were
significantly upregulated in liver cirrhosis tissues compared with
normal tissues, implying that the expression pattern of these 2
miRNAs may be particular to liver cirrhosis, and that they may
serve different roles in liver cirrhosis and HCC by a currently
unknown mechanism.
In addition, Wang et al (43) identified that the expression of
miR-181b may be induced by transforming growth factor-β1, which
serves an important role in liver cirrhosis. miR-132 expression may
be induced following alcohol intake, and it was previously shown to
be increased in alcoholic liver disease (31). It was indicated that serum miR-18a was
significantly higher in patients infected with hepatitis B virus
(HBV) with HCC compared with healthy controls, and it may be used
in discriminating HBV-associated HCC from chronic hepatitis or
cirrhosis (32). Notably, in the
present study, the aforementioned 3 miRNAs (miR-181b, miR-132 and
miR-18a) were significantly downregulated in liver cirrhosis
tissues compared with normal tissues, indicating that they may
exhibit potential for diagnostic and therapeutic applications in
liver cirrhosis.
In order to additionally investigate the role of
miRNAs in liver cirrhosis, the transcriptional targets of the
identified miRNAs in liver cirrhosis tissues were predicted, and
several targets were noted, including LIFR, KIAA1524 and E2F1. It
was observed that the LIFRβ subunit is intensely expressed in
reactive bile ductductuules in the cirrhotic liver, in which bile
duct proliferation occurs frequently (44). Previous studies have demonstrated that
KIAA1524 is a cellular inhibitor of protein phosphatase 2A, which
is an important tumor-suppressor protein, and that the inhibition
of KIAA1524 may determine the apoptotic effects of erlotinib on HCC
cells (45–47). An additional important target was
E2F1, which is considered a novel fibrogenic gene, and was
identified to be highly upregulated in human liver cirrhotic
specimens (48). Additionally,
experiments in mice demonstrated that E2F1-deficiency may largely
inhibit the development of biliary liver fibrosis (48). In the present study, the dysregulation
of LIFR, KIAA1524 and E2F1 in the cirrhotic liver clearly suggested
their importance in the development of liver cirrhosis. The results
of the function enrichment analysis of the target genes indicated
that the cell cycle was the most significantly enriched pathway.
This result is consistent with the importance of the cell cycle in
various types of cancer (49).
In conclusion, a set of 48 differentially expressed
miRNAs and 1,773 DEGs were identified in liver cirrhosis tissues
compared with normal tissues. A total of 240 miRNA-target gene
pairs whose expression was inversely correlated were identified.
Additionally, a global miRNAs-target regulatory network was
constructed, which was expected to improve the understanding of the
regulatory mechanisms of liver cirrhosis pathology, and to provide
a clearer biomarker for diagnosis and safer therapeutic strategies
of liver cirrhosis. Additional functional studies are required to
confirm the regulative associations between the miRNAs and their
predictive targets.
References
|
1
|
Schuppan D and Afdhal NH: Liver cirrhosis.
Lancet. 371:838–851. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siddique I, El-Naga HA, Madda JP, Memon A
and Hasan F: Sampling variability on percutaneous liver biopsy in
patients with chronic hepatitis C virus infection. Scand J
Gastroenterol. 38:427–432. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bedossa P, Dargere D and Paradis V:
Sampling variability of liver fibrosis in chronic hepatitis C.
Hepatology. 38:1449–1457. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Regev A, Berho M, Jeffers LJ, Milikowski
C, Molina EG, Pyrsopoulos NT, Feng ZZ, Reddy KR and Schiff ER:
Sampling error and intraobserver variation in liver biopsy in
patients with chronic HCV infection. Am J Gastroenterol.
97:2614–2618. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ratziu V, Charlotte F, Heurtier A, Gombert
S, Giral P, Bruckert E, Grimaldi A, Capron F and Poynard T; LIDO
Study Group, : Sampling variability of liver biopsy in nonalcoholic
fatty liver disease. Gastroenterology. 128:1898–1906. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Povero D, Busletta C, Novo E, di Bonzo LV,
Cannito S, Paternostro C and Parola M: Liver fibrosis: A dynamic
and potentially reversible process. Histol Histopathol.
25:1075–1091. 2010.PubMed/NCBI
|
|
7
|
Pinzani M and Vizzutti F: Fibrosis and
cirrhosis reversibility: Clinical features and implications. Clin
Liver Dis. 12:901–913, x. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gieling RG, Burt AD and Mann DA: Fibrosis
and cirrhosis reversibility-molecular mechanisms. Clin Liver Dis.
12:915–937, xi. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA. 105:pp.
10513–10518. 2008; View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Elfimova N, Schlattjan M, Sowa JP, Dienes
HP, Canbay A and Odenthal M: Circulating microRNAs: Promising
candidates serving as novel biomarkers of acute hepatitis. Front
Physiol. 3:4762012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang XW, Heegaard NH and Orum H: MicroRNAs
in liver disease. Gastroenterology. 142:1431–1443. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Borel F, Konstantinova P and Jansen PL:
Diagnostic and therapeutic potential of miRNA signatures in
patients with hepatocellular carcinoma. J Hepatol. 56:1371–1383.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Giordano S and Columbano A: MicroRNAs: New
tools for diagnosis, prognosis, and therapy in hepatocellular
carcinoma? Hepatology. 57:840–847. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Varnholt H, Drebber U, Schulze F,
Wedemeyer I, Schirmacher P, Dienes HP and Odenthal M: MicroRNA gene
expression profile of hepatitis C virus-associated hepatocellular
carcinoma. Hepatology. 47:1223–1232. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gupta A, Swaminathan G, Martin-Garcia J
and Navas-Martin S: MicroRNAs, hepatitis C virus and HCV/HIV-1
co-infection: New insights in pathogenesis and therapy. Viruses.
4:2485–2513. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lanford RE, Hildebrandt-Eriksen ES, Petri
A, Persson R, Lindow M, Munk ME, Kauppinen S and Ørum H:
Therapeutic silencing of microRNA-122 in primates with chronic
hepatitis C virus infection. Science. 327:198–201. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Janssen HL, Reesink HW, Lawitz EJ, Zeuzem
S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A,
Zhou Y, et al: Treatment of HCV infection by targeting microRNA. N
Engl J Med. 368:1685–1694. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Noetel A, Kwiecinski M, Elfimova N, Huang
J and Odenthal M: microRNA are central players in anti- and
profibrotic gene regulation during liver fibrosis. Front Physiol.
3:492012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wong CM, Wong CC, Lee JM, Fan DN, Au SL
and Ng IO: Sequential alterations of microRNA expression in
hepatocellular carcinoma development and venous metastasis.
Hepatology. 55:1453–1461. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41(Database Issue):
D991–D995. 2013.PubMed/NCBI
|
|
23
|
Law CW, Chen Y, Shi W and Smyth GK: Voom:
Precision weights unlock linear model analysis tools for RNA-seq
read counts. Genome Biol. 15:R292014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Young MD, Wakefield MJ, Smyth GK and
Oshlack A: Gene ontology analysis for RNA-seq: Accounting for
selection bias. Genome Biol. 11:R142010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Visone R and Croce CM: MiRNAs and cancer.
Am J Pathol. 174:1131–1138. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mirnezami AH, Pickard K, Zhang L, Primrose
JN and Packham G: MicroRNAs: Key players in carcinogenesis and
novel therapeutic targets. Eur J Surg Oncol. 35:339–347. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ajit SK: Circulating microRNAs as
biomarkers, therapeutic targets, and signaling molecules. Sensors
(Basel). 12:3359–3369. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bala S and Szabo G: MicroRNA signature in
alcoholic liver disease. Int J Hepatol. 2012:4982322012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li L, Guo Z, Wang J, Mao Y and Gao Q:
Serum miR-18a: A potential marker for hepatitis B virus-related
hepatocellular carcinoma screening. Dig Dis Sci. 57:2910–2916.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Brockhausen J, Tay SS, Grzelak CA,
Bertolino P, Bowen DG, D'Avigdor WM, Teoh N, Pok S, Shackel N,
Gamble JR, et al: miR-181a mediates TGF-β-induced hepatocyte EMT
and is dysregulated in cirrhosis and hepatocellular cancer. Liver
Int. 35:240–253. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao J, Tang N, Wu K, Dai W, Ye C, Shi J,
Zhang J, Ning B, Zeng X and Lin Y: miR-21 simultaneously regulates
ERK1 signaling in HSC activation and hepatocyte EMT in hepatic
fibrosis. PLoS One. 9:e1080052014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang T, Zhang L, Shi C, Sun H, Wang J, Li
R, Zou Z, Ran X and Su Y: TGF-β-induced miR-21 negatively regulates
the antiproliferative activity but has no effect on EMT of TGF-β in
HaCaT cells. Int J Biochem Cell Biol. 44:366–376. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lakner AM, Steuerwald NM, Walling TL,
Ghosh S, Li T, McKillop IH, Russo MW, Bonkovsky HL and Schrum LW:
Inhibitory effects of microRNA 19b in hepatic stellate
cell-mediated fibrogenesis. Hepatology. 56:300–310. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Thum T, Gross C, Fiedler J, Fischer T,
Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, et
al: MicroRNA-21 contributes to myocardial disease by stimulating
MAP kinase signalling in fibroblasts. Nature. 456:980–984. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pandit KV, Milosevic J and Kaminski N:
MicroRNAs in idiopathic pulmonary fibrosis. Transl Res.
157:191–199. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Niu J, Shi Y, Tan G, Yang CH, Fan M,
Pfeffer LM and Wu ZH: DNA damage induces NF-κB-dependent
microRNA-21 up-regulation and promotes breast cancer cell invasion.
J Biol Chem. 287:21783–21795. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bakirtzi K, Hatziapostolou M,
Karagiannides I, Polytarchou C, Jaeger S, Iliopoulos D and
Pothoulakis C: Neurotensin signaling activates microRNAs-21 and
−155 and Akt, promotes tumor growth in mice, and is increased in
human colon tumors. Gastroenterology. 141:1749–1761 e1. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Murakami Y, Toyoda H, Tanaka M, Kuroda M,
Harada Y, Matsuda F, Tajima A, Kosaka N, Ochiya T and Shimotohno K:
The progression of liver fibrosis is related with overexpression of
the miR-199 and 200 families. PLoS One. 6:e160812011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tsang FH, Au SL, Wei L, Fan DN, Lee JM,
Wong CC, Ng IO and Wong CM: MicroRNA-142-3p and microRNA-142-5p are
downregulated in hepatocellular carcinoma and exhibit synergistic
effects on cell motility. Front Med. 9:331–343. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang B, Li W, Guo K, Xiao Y, Wang Y and
Fan J: miR-181b promotes hepatic stellate cells proliferation by
targeting p27 and is elevated in the serum of cirrhosis patients.
Biochem Biophys Res Commun. 421:4–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Znoyko I, Sohara N, Spicer SS, Trojanowska
M and Reuben A: Expression of oncostatin M and its receptors in
normal and cirrhotic human liver. J Hepatol. 43:893–900. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yu HC, Chen HJ, Chang YL, Liu CY, Shiau
CW, Cheng AL and Chen KF: Inhibition of CIP2A determines
erlotinib-induced apoptosis in hepatocellular carcinoma. Biochem
Pharmacol. 85:356–366. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yu HC, Hung MH, Chen YL, Chu PY, Wang CY,
Chao TT, Liu CY, Shiau CW and Chen KF: Erlotinib derivative
inhibits hepatocellular carcinoma by targeting CIP2A to reactivate
protein phosphatase 2A. Cell Death Dis. 5:e13592014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Junttila MR, Puustinen P, Niemelä M, Ahola
R, Arnold H, Böttzauw T, Ala-aho R, Nielsen C, Ivaska J, Taya Y, et
al: CIP2A inhibits PP2A in human malignancies. Cell. 130:51–62.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang Y, Xu N, Xu J, Kong B, Copple B, Guo
GL and Wang L: E2F1 is a novel fibrogenic gene that regulates
cholestatic liver fibrosis through the Egr-1/SHP/EID1 network.
Hepatology. 60:919–930. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Swiatkowska A, Zydowicz P, Sroka J and
Ciesiolka J: The role of the 5′ terminal region of p53 mRNA in the
p53 gene expression. Acta Biochim Pol. 63:645–651. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yildiz G, Arslan-Ergul A, Bagislar S, Konu
O, Yuzugullu H, Gursoy-Yuzugullu O, Ozturk N, Ozen C, Ozdag H,
Erdal E, et al: Genome-wide transcriptional reorganization
associated with senescence-to-immortality switch during human
hepatocellular carcinogenesis. PLoS One. 8:e640162013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Archer KJ, Mas VR, David K, Maluf DG,
Bornstein K and Fisher RA: Identifying genes for establishing a
multigenic test for hepatocellular carcinoma surveillance in
hepatitis C virus-positive cirrhotic patients. Cancer Epidemiol
Biomarkers Prev. 18:2929–2932. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mas VR, Maluf DG, Archer KJ, Yanek K, Kong
X, Kulik L, Freise CE, Olthoff KM, Ghobrial RM, McIver P and Fisher
R: Genes involved in viral carcinogenesis and tumor initiation in
hepatitis C virus-induced hepatocellular carcinoma. Mol Med.
15:85–94. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Caillot F, Derambure C, Bioulac-Sage P,
Francois A, Scotte M, Goria O, Hiron M, Daveau M and Salier JP:
Transient and etiology-related transcription regulation in
cirrhosis prior to hepatocellular carcinoma occurrence. World J
Gastroenterol. 15:300–309. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wojcicka A, Swierniak M, Kornasiewicz O,
Gierlikowski W, Maciag M, Kolanowska M, Kotlarek M, Gornicka B,
Koperski L, Niewinski G, et al: Next generation sequencing reveals
microRNA isoforms in liver cirrhosis and hepatocellular carcinoma.
Int J Biochem Cell Biol. 53:208–217. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Vuppalanchi R, Liang T, Goswami CP,
Nalamasu R, Li L, Jones D, Wei R, Liu W, Sarasani V, Janga SC and
Chalasani N: Relationship between differential hepatic microRNA
expression and decreased hepatic cytochrome P450 3A activity in
cirrhosis. PLoS One. 8:e744712013. View Article : Google Scholar : PubMed/NCBI
|