Introduction
Breast cancer (BC) is the most common malignancy and
the first leading cause of tumor-associated mortality in females
worldwide (1), and the frequency of
BC is increasing in developed and developing countries (2). Tumor metastasis is the primary reason
for the poor prognosis or high mortality of BC (3), therefore the identification of
biomarkers for tumor metastasis may aid in developing novel
treatments for, and in the diagnosis of, BC.
MicroRNAs (miRNAs/miRs) are a class of small,
non-coding, single-stranded RNAs that regulate multiple target
mRNAs, generally through perfect or imperfect binding to the
3′-untranslated region (UTR) of the mRNA. Previous studies have
indicated that miRNAs serve critical roles in various
tumor-associated biological processes, including proliferation,
metastasis, apoptosis and differentiation (4–6).
Dysregulation of miRNAs leads to a tumor suppressive or oncogenic
effect depending on the target mRNAs, and ultimately contributes to
the initiation and progression of human malignancies (7–9).
Therefore, miRNAs may have potential as diagnostic or prognostic
biomarkers for cancer.
In the present study, miR-124-3p was investigated as
a metastasis inhibitor and its expression in BC tissues was
determined. PDCD6 was also investigated as target of miR-124-3p.
This data expands our knowledge of the role of miR-124-3p in BC
development and considers the miR-124-3p/PDCD6 signaling axis as an
important underlying molecular mechanism and effective therapeutic
strategy for advanced BC.
Materials and methods
Tissue specimens and cell lines
Primary BC tissues and corresponding non-tumor
tissues were obtained during surgery from 40 female patients (mean
age, 54.52 years; range, 42–68 years) who were diagnosed and
treated at Shandong Provincial Hospital Affiliated to Shandong
University (Jinan, China) between February 2010 and August 2011.
Tumor stage was recorded according to the classification criteria
of the Union for International Cancer Control (10). All tumors and normal tissues were
confirmed by two independent pathologists. Matched non-cancerous
tissues from the macroscopic tumor margin were obtained and used as
normal controls. The clinicopathological characteristics were
obtained from medical records. The present study was approved by
the Ethics Committee of Shandong Provincial Hospital and written
informed consent was obtained from patients prior to surgery.
Human BC MCF-7 and MDA-MB-231 cells were purchased
from the American Type Culture Collection (Manassas, VA, USA), and
cultured at 37°C with 5% CO2 in a humidified incubator.
The MCF-7 cells were cultured in RPMI-1640 medium, and the
MDA-MB-231 cells were cultured in Dulbecco's modified Eagle's
medium (both Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis of miRNA and mRNA
levels
RT-qPCR assays were performed to determine the
expression level of miR-124-3p and PDCD6 mRNA in cells or tissues.
Total RNA was extracted using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol,
and the RNA concentration was measured using a NanoDrop™ 2000
spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE,
USA). For miRNA detection, total RNA was used to generate
complementary (c)DNA using the PrimeScript RT reagent kit (Clontech
Laboratories, Inc., Mountainview, CA, USA), and the
SYBR® Premix ExTaq (Clontech Laboratories Inc.) was used
to perform RT-qPCR. For mRNA detection, 500 ng total RNA was
subjected to first-strand cDNA synthesis using M-MLV Reverse
Transcriptase (Promega Corporation), and RT-qPCR assay was
performed using the SYBR-Green PCR master mix (Roche Diagnostics,
Basel, Switzerland) on the Fast Real-Time ABI 7500 PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The following
PCR conditions were as follows: 95°C for 30 sec, followed by 42
cycles at 95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec. The
sequences of the primers were as follows: PDCD6 (sense),
5′-CGGCAAGTTGTCGGAGACG-3′; PDCD6 (antisense),
5′-CCTGGAGGTTGGGATGCTCT-'; β-actin (sense),
5′-AGGGAAATCGTGCGTGAC-3′; β-actin (antisense),
5′-CGCTCATTGCCGATAGTG-3′; miR-124-3p (sense),
5′-TAAGGCACGCGGTGAATGCC-3′; miR-124-3p (antisense),
5′-GATTGAATCGAGCACCAGTTAC-3′; U6 (sense),
5′-CGCTTCGGCAGCACATATACTA-3′; U6 (antisense)
5′-GATTGAATCGAGCACCAGTTAC-3′. The relative expression of miR-124-3p
and PDCD6 mRNA was normalized to U6 spliceosomal RNA and β-actin,
respectively, using the 2−ΔΔCq method (11). RT-PCR was performed in triplicate.
Transfection and plasmid
construction
miR-124-3p mimics (5′-UAAGGCACGCGGUGAAUGCC-3′),
inhibitor (5′-UAAGGCACGCGGUGAAUGCC-3′), mimic negative control (con
M; 5′-UUCUCCATCGUGCCUCUAT-3′), and inhibitor negative control (con
I; 5′-CCGUACUUCGCUAGAUCA-3′) were synthesized by Guangzhou RiboBio
Co., Ltd. (Guangzhou, China) and transfected into cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. For the
luciferase reporter assay, the 3′-UTR of PDCD6 containing the
putative binding sites for miR-124-3p was amplified from genomic
DNA by PCR and cloned into the pGL3-luciferase reporter plasmid
(Promega Corporation, Madison, WI, USA). The Platinum™ Taq DNA
Polymerase (Invitrogen; Thermo Fisher Scientific, Inc.) was used in
PCR reaction and conditions were as followed: 95°C for 1 min,
followed by 45 cycles of 95°C for 30 sec, 56°C for 20 sec and 72°C
for 20 sec. The primers used to amplify the 3′-UTR of PDCD6 were as
follows: Forward, 5′-GCTACCTGACTAAGCAT-3′ and reverse,
5′-CCTAGATCGGATAGCTAGC-3′. Mutations in the binding site of the
PDCD6 3′-UTR were generated using the QuikChange™ Site-Directed
Mutagenesis kit (Agilent Technologies, Inc., Santa Clara, CA, USA)
according to the manufacturer's protocol. The sequence lacking the
PDCD6 3′-UTR was introduced into cells to rescue PDCD6 expression
using pcDNA3.1(+) (Invitrogen; Thermo Fisher Scientific, Inc.)
plasmid. The primers used to amplify PDCD6 without the 3′-UTR were
as follows: Forward, 5′-TGCGTAGCCTAGTACAG-3′ and reverse,
5′-GTCGACTGACAGGCTA-3′.
Wound-healing and Transwell
assays
To determine the effect of upregulated or
downregulated miR-124-3p on cell migratory behavior, a
wound-healing assay was performed and the closure of the wounds in
each well was evaluated. The two cell lines were seeded onto 35-mm
dishes coated with fibronectin, and the cells were allowed to reach
100% confluency. An artificial wound was created on a confluent
cell monolayer without FBS using a 200-ml pipette tip 24 h after
transfection. The culture medium was refreshed with serum-free
medium, and the cells were incubated for additional 24 h at 37°C.
Serial images were obtained at 0 and 24 h using a light microscope
(Olympus, Tokyo, Japan).
For the invasion assay, 2×104 transfected
cells were seeded into the upper chamber of the
Matrigel® Transwell chamber (BD Biosciences, Franklin
Lakes, NJ, USA). Medium containing 10% FBS in the lower chamber
acted as the chemoattractant. Following incubation for a further 48
h at 37°C, non-invading cells were removed from the upper surface
with a cotton swab, and the invaded cells attached to the lower
surface of the membrane were fixed with methanol, stained with 0.1%
crystal violet and then counted using an Olympus light microscope
(Olympus Corporation, Tokyo, Japan).
Western blotting analysis
Western blotting analysis was performed according to
a standard protocol. Briefly, the cells were washed twice with ice
cold PBS and lysed on ice in RIPA buffer (Beyotime Institute of
Biotechnology, Haimen, China) and the protein concentration was
determined using the Pierce BCA protein assay kit (Thermo Fisher
Scientific, Inc.). Equal amounts of proteins (20 µg) were separated
on a 10% SDS-PAGE gel and then transferred to polyvinylidene
fluoride membranes (EMD Millipore, Billerica, MA, USA). The
membranes were blocked with 5% skimmed milk and incubated at room
temperature for 2 h with an anti-PDCD6 antibody (catalog no.
ab109181; dilution, 1:2,000; Abcam, Cambridge, MA, USA),
anti-E-cadherin antibody (catalog no. ab15148; dilution, 1:500;
Abcam), anti-N-cadherin antibody (catalog no. ab15148; dilution,
1:500; Abcam), anti-Vimentin antibody (catalog no. ab137321;
dilution, 1:1,000; Abcam) and β-actin (catalog no. A5316; dilution,
1:5,000; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Following
washing with Tris-buffered saline with Tween-20 [10 mM Tris (pH
8.0), 150 mM NaCl and 0.1% Tween-20], the membranes were then
incubated with a secondary goat anti-rabbit horseradish
peroxidase-conjugated antibody (catalog no. sc-2004; dilution,
1:5,000; Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h at
37°C. The protein bands were detected using an enhanced ECL
chemiluminescent Substrate kit (Pierce; Thermo Fisher Scientific,
Inc.).
Dual luciferase reporter assay
Recombinant plasmids of pGL3-PDCD6-3′-UTR-wild-type
(WT) or pGL3-PDCD6-3′-UTR-Mutation (Mut) were constructed as
aforementioned. Cells (1×105 cells/well) were seeded in
a 24-well plate and co-transfected with 40 nM of either miR-124-3p
mimic or mimic negative control, 20 ng of either
pGL3-PDCD6-3′-UTR-WT or pGL3-PDCD6-3′-UTR-Mut, and 2 ng of pRL-TK
(Promega Corporation) using Lipofectamine 2000, according to the
manufacturer's protocol. Following incubation for 48 h, the cells
were lysed and the luciferase reporter assay was performed using
the Dual-Luciferase® Reporter assay system (Promega
Corporation), according to the manufacturer's protocol.
Target prediction
To identify the potential targets of miR-124-3p, a
bioinformatics analysis was performed using three publically
available databases: TargetScan (http://www.targetscan.org/vert_71/), miRDB (http://www.mirdb.org/miRDB/) and microRNA (http://www.microrna.org/microrna/home.do). miRDB is an
online database for miRNA target prediction and functional
annotations.
Statistical analysis
All data were analyzed using SPSS software (version
13.0; SPSS, Inc., Chicago, IL, USA) or GraphPad Prism (version 5;
GraphPad Software, Inc., La Jolla, CA, USA). The results obtained
from in vitro assays are presented as the mean ± standard
error of the mean and the data were analyzed by Student's t-test. A
paired samples t-test was performed to investigate the differences
in miR-124-3p and PDCD6 mRNA expression between normal and
cancerous tissues. A Mann-Whitney test was used to compare
differences in miR-124-3p levels between two groups of patients
with certain clinicopathological characteristics, while a
Kruskal-Wallis test was performed to compare more than two groups,
followed by Dunn's multiple comparison post-hoc test between
groups. The correlation between miR-124-3p and PDCD6 was analyzed
using a Spearman's rank correlation test. A Kaplan-Meier estimator
analysis and a log-rank test were performed for the overall
survival analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
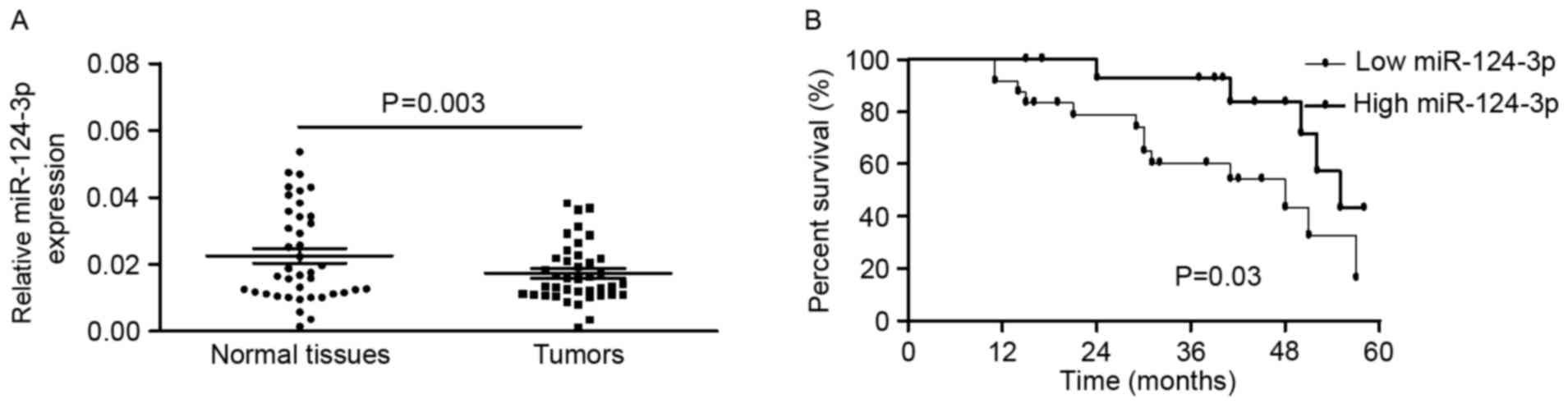
miR-124-3p is downregulated in BC
tissue compared with non-tumor tissue in patients with BC
To further study the association between miR-124 and
BC pathogenesis, the expression of miR-124-3p in 40 patients with
BC was detected using RT-qPCR. The expression of miR-124-3p was
significantly decreased in BC tissues compared with the
corresponding non-tumor tissues (0.0174±0.0088 vs. 0.0226±0.0139;
P=0.003, paired t-test; Fig. 1A). In
addition, the associations between the expression level of
miR-124-3p and different clinicopathological characteristics of
patients with BC were investigated. As illustrated in Table I, the expression of miR-124-3p was
markedly lower in patients with lymph node metastasis compared with
that in patients without lymph node metastasis (P=0.006,
Mann-Whitney test). When comparing other subgroups, including age
(<55 and ≥55 years old), differentiation (well/moderate/poor),
tumor stage (I/II/III/IV), and estrogen receptor status
(negative/positive), the differences in miR-124-3p levels were not
statistically significant. The mean level of miR-124-3p expression
in the BC tissues was used as a threshold to divide patients into
two groups (low or high expression groups). Kaplan-Meier estimator
analyses indicated that patients with BC with high miR-124-3p
expression were likely to survive longer than those with low
miR-124-3p expression (Fig. 1B).
 | Table I.Clinicopathological characteristics of
patients with breast cancer and correlation with miR-124-3p
expression. |
Table I.
Clinicopathological characteristics of
patients with breast cancer and correlation with miR-124-3p
expression.
| Clinicopathological
characteristic | No. of patients
(n=40) | miR-124-3p level
(mean ± standard deviation) | P-value |
|---|
| Age, years |
|
| 0.652a |
|
<55 | 23 |
0.0161±0.0074 |
|
| ≥55 | 17 |
0.0190±0.0103 |
|
| Differentiation |
|
| 0.911b |
| Well | 6 |
0.0124±0.0066 |
|
|
Moderate | 20 |
0.0179±0.0100 |
|
| Poor | 14 |
0.0170±0.0081 |
|
| Tumor stage |
|
| 0.192b |
| I | 8 |
0.0189±0.0089 |
|
| II | 8 |
0.0200±0.0053 |
|
| III | 22 |
0.0163±0.0099 |
|
| IV | 2 |
0.0118±0.0052 |
|
| Lymph node |
|
| 0.006a |
| (−) | 17 |
0.0216±0.0098 |
|
| (+) | 23 |
0.0142±0.0064 |
|
| Estrogen receptor
status |
|
| 0.419a |
|
Negative | 26 |
0.0183±0.0093 |
|
|
Positive | 14 |
0.0155±0.0076 |
|
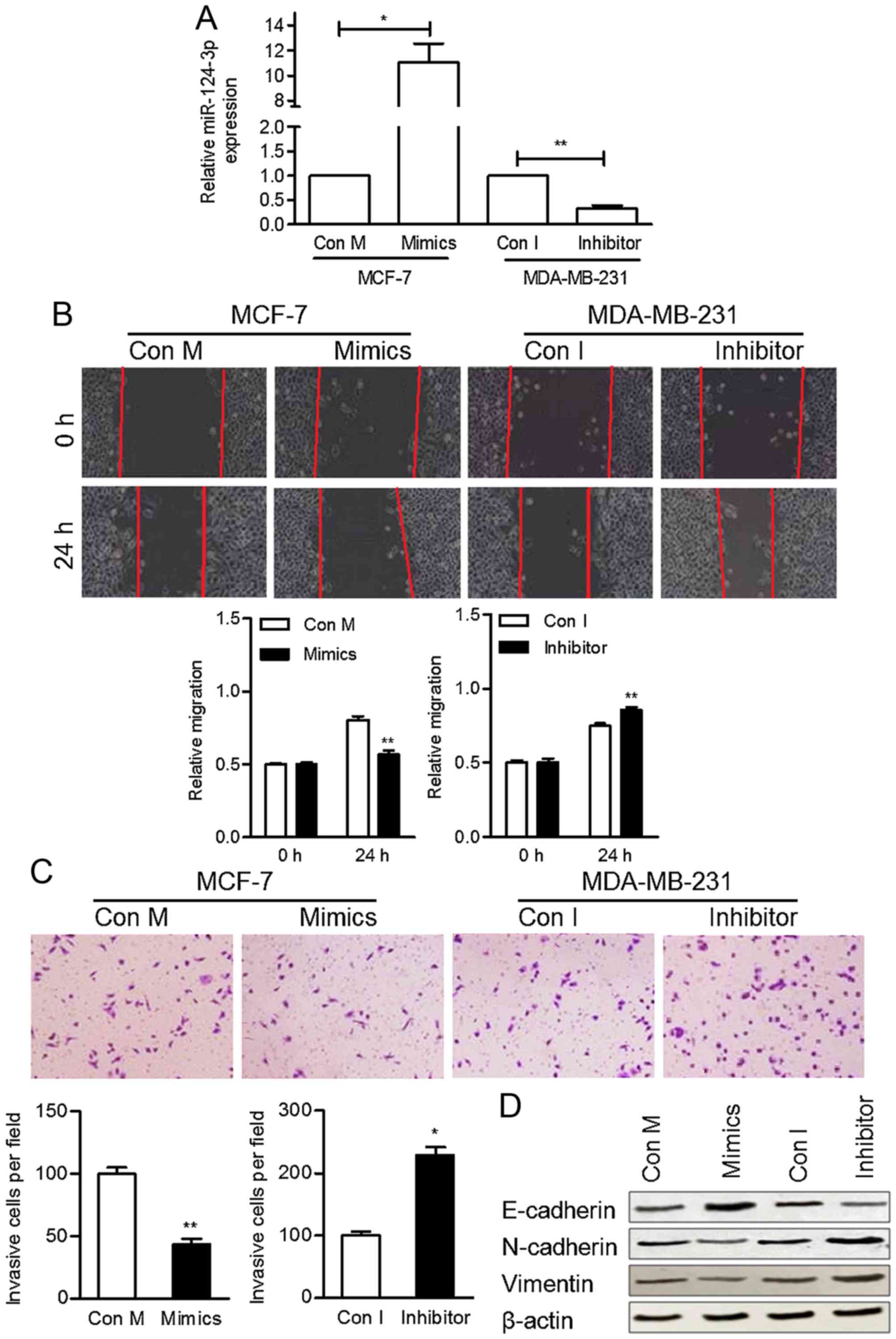
miR-124-3p inhibits cell migration and
invasion in vitro
As downregulated miR-124-3p expression was
associated with lymph node metastasis, the effect of miR-124-3p on
cell motility was investigated. miR-124-3p was overexpressed or
inhibited by transfecting MCF-7 and MDA-MB-231 cells with
miR-124-3p mimics and a miR-124-3p inhibitor, respectively. The
expression of miR-124-3p in transfected cells was confirmed using
RT-qPCR (Fig. 2A).
The migratory ability of these cells was
subsequently determined using a wound-healing assay. MCF-7 cells
treated with miR-124-3p mimics were significantly less migratory
compared with the control cells at 24 h (P<0.05), whereas
MDA-MB-231 cells transfected with the miR-124-3p inhibitor were
significantly more migratory compared with the control cells
(P<0.05) (Fig. 2B). In addition, a
Transwell invasion assay was performed to determine the invasive
ability of BC cells transfected with miR-124-3p mimics or an
inhibitor. Transfection of cells with miR-124-3p mimics
significantly decreased the invasion ability of MCF-7 cells
compared with the control cells (P<0.01), whereas transfection
with the miR-124-3p inhibitor significantly increased the invasion
ability of MDA-MB-231 cells (P<0.05) (Fig. 2C). These results suggest that
miR-124-3p regulates cell motility and that the inhibition of
miR-124-3p may promote breast tumor metastasis.
Western blotting analyses were performed to
investigate whether the observed changes in cell motility were
accompanied by the epithelial-mesenchymal transition (EMT) of BC
cells. The protein level of E-cadherin was increased by miR-124-3p
mimics and suppressed by the inhibitor. The protein levels of
Vimentin and N-cadherin were promoted by inhibitor and decreased in
cells transfected with miR-124-3p mimics (Fig. 2D). These results suggest that
miR-124-3p inhibits BC cell EMT and tumor metastasis.
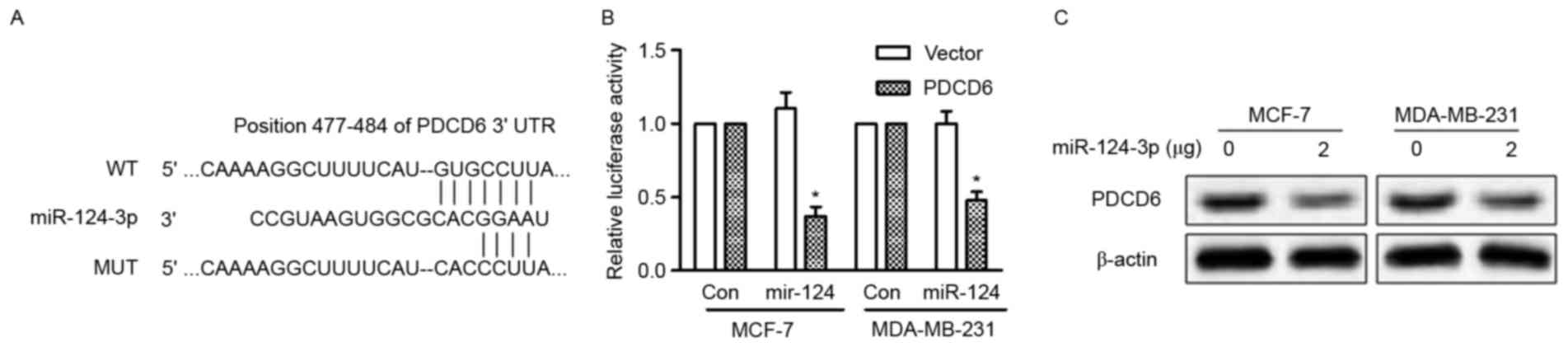
PDCD6 is a direct target of
miR-124-3p
All three databases (TargetScan, miRDB and microRNA)
predicted PDCD6 as a target of miR-124-3p. PDCD6 has been
demonstrated to be overexpressed in metastatic ovarian cancer
(12) and the 3′-UTR of PDCD6 mRNA
possesses a highly conserved sequence (position 477–484) that is
complementary to the seed sequence of miR-124-3p (Fig. 3A). To validate that PDCD6 is a direct
target of miR-124-3p, luciferase reporter vectors containing the
binding site of WT PDCD6 3′-UTR or the mutant type 3′-UTR were
constructed and transfected into MCF-7 and MDA-MB-231 cells along
with miR-124-3p mimic or mimic control. The overexpression of
miR-124-3p significantly reduced the activity of the WT PDCD6
3′-UTR (P<0.05), but the activity of the mutant 3′-UTR was
unchanged (Fig. 3B). In addition,
overexpression of miR-124-3p in MCF-7 and MDA-MB-231 cells reduced
the protein expression of PDCD6 (Fig.
3C). These results indicate that miR-124-3p directly targets
PDCD6 and inhibits its expression.
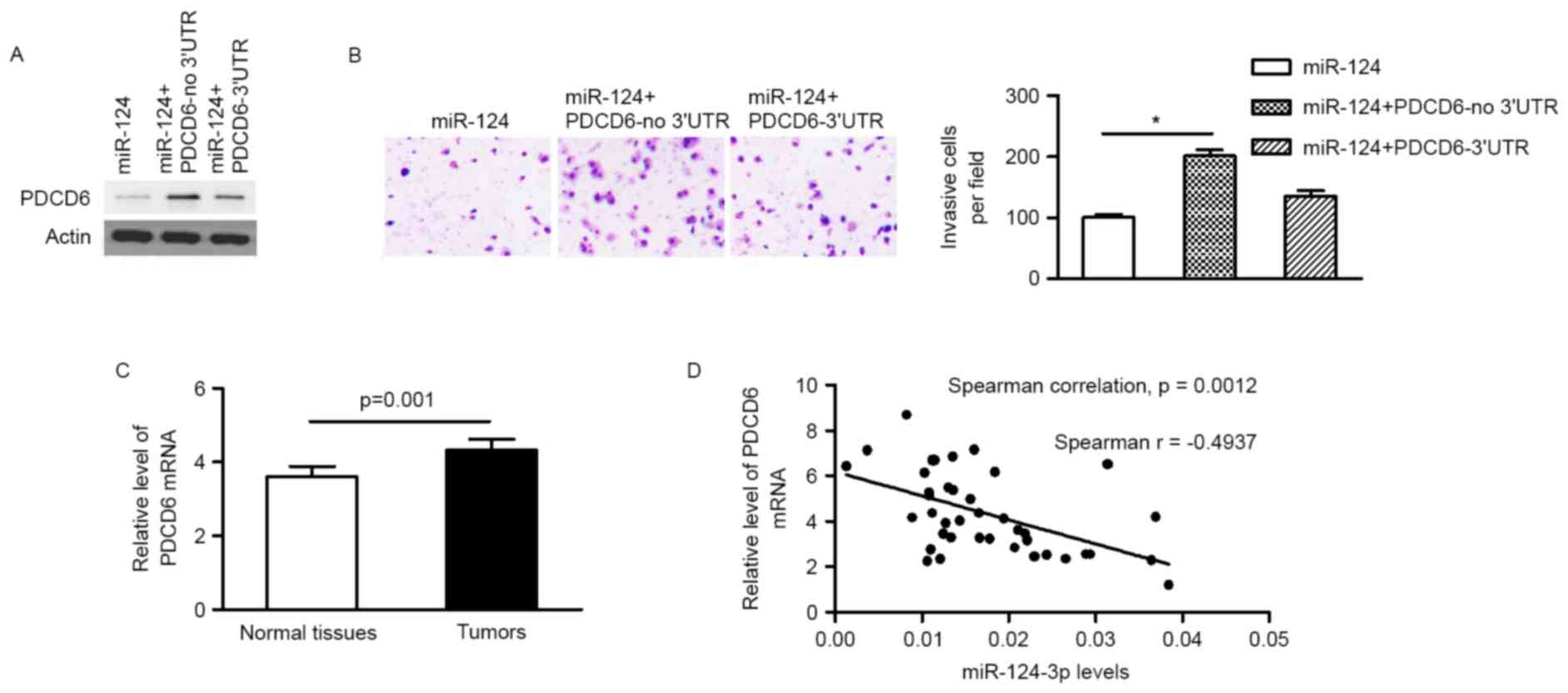
Restoration of PDCD6 expression
rescues the effect of miR-124-3p on BC cells
In order to demonstrate the functional connection
between miR-124-3p and PDCD6, a PDCD6 vector lacking the PCDC6
3′-UTR (PCDC6-no 3′UTR) was transfected into MCF-7 cells stably
expressing miR-124-3p. Protein expression levels of PDCD6, and the
invasion ability of MCF-7 cells, were restored 48 h after
transfection (Fig. 4A and B). The
subsequent reintroduction of PDCD6 with an intact 3′UTR partly
restored the inhibitory effect of miR-124-3p mimics on the invasion
ability of MCF-7 cells (Fig. 4B),
indicating that PDCD6 acts as a functional target of miR-124-3p in
BC cells.
miR-124-3p expression negatively
correlates with PDCD6 expression in BC tumors
The expression of PDCD6 in primary tumor tissue and
adjacent non-tumor tissue from patients with BC was quantified
using RT-qPCR. The mRNA expression level of PDCD6 was significantly
increased in primary tumors in comparison with adjacent non-tumor
tissues (4.334±1.779 vs. 3.600±1.750; P=0.001; Fig. 4C). A Spearman's rank correlation test
revealed that the expression of miR-124-3p was inversely correlated
with PDCD6 expression level (Fig.
4D), indicating that the miR-124-3p/PDCD6 signaling axis serves
a role in BC progression and may be a potential target of novel
treatments for BC.
Discussion
Previous studies have demonstrated that the
dysregulation of miRNA contributes to carcinogenesis by suppressing
target gene expression; therefore, miRNAs could serve as effective
biomarkers for the prognosis and treatment of patients with BC
(13,14). Data from the present study
demonstrated that miR-124-3p inhibits cell motility and EMT, and
that decreased miR-124-3p expression correlated with lymph node
metastasis in patients with BC. In addition, PDCD6 was revealed to
be a direct target of miR-124-3p. miR-124-3p inhibited the
expression of PDCD6 in the BC cells in vitro and PDCD6
expression was negatively correlated with miR-124-3p expression in
BC tissues.
In the present study, the expression of miR-124-3p
in BC tumor tissue was compared with that in matched normal tissue.
The levels of miR-124-3p were significantly reduced in tumor tissue
compared with matched normal tissue, which was consistent with
previous studies (15–17). These results suggest that the
dysregulation of miR-124-3p is associated with the pathogenesis of
BC. In addition, downregulation of miR-124-3p was correlated with
lymph node metastasis and decreased overall survival probability,
which is consistent with previous BC studies (16–18) and
studies on other types of tumor (19,20).
Therefore miR-124-3p is a promising prognostic marker for various
tumors.
In vitro analyses in the present study
demonstrated that miR-124-3p inhibits EMT and cell motility. In
addition, PDCD6 was identified as a direct target of miR-124-3p.
Reintroduction of PDCD6 impaired the tumor suppressor role of
miR-124-3p by promoting cell invasion. This suggests that the
miR-124-3p/PDCD6 signaling axis serves a role in the increased
invasiveness of BC cells. Previous studies have demonstrated that
that miR-124-3p directly targets ρ-associated protein kinase 1
(21), sphingosine kinase 1 (22), forkhead box protein Q1 (23) and talin 1 (24) to inhibit migration and invasion in
numerous types of tumor. Therefore, restoring the expression of
miR-124-3p may be an effective therapeutic strategy for advanced
cancer.
Overall, the mRNA levels of PDCD6, which is highly
expressed in mesenchymal tumors (25)
and metastatic ovarian cancer (12),
were significantly increased in the BC tissues in the present
study. To the best of our knowledge, this is the first study to
demonstrate that miR-124-3p mediates its effects on BC metastasis
through the inhibition of PDCD6. The results from the present study
revealed a negative correlation between PDCD6 and miR-124-3p
expression in BC tissues, demonstrating a functional connection
between miR-124 and PDCD6 in primary tumors. The findings from the
present study further our understanding of the role of miR-124-3p
in breast tumorigenesis and demonstrate that the miR-124-3p/PDCD6
signaling axis could be a target of novel treatments for patients
with BC.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ha R, Chow D and Wynn R: Global trend in
breast cancer imaging research 1992–2012: Bibliometric study. AJR
Am J Roentgenol. 202:696–697. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Allemani C, Weir HK, Carreira H, Harewood
R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A,
et al: Global surveillance of cancer survival 1995–2009: Analysis
of individual data for 25,676,887 patients from 279
population-based registries in 67 countries (CONCORD-2). Lancet.
385:977–1010. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ge L, Zheng B, Li M, Niu L and Li Z:
MicroRNA-497 suppresses osteosarcoma tumor growth in vitro and in
vivo. Oncol Lett. 11:2207–2212. 2016.PubMed/NCBI
|
|
5
|
Liu Z, Gersbach E, Zhang X, Xu X, Dong R,
Lee P, Liu J, Kong B, Shao C and Wei JJ: miR-106a represses the Rb
tumor suppressor p130 to regulate cellular proliferation and
differentiation in high-grade serous ovarian carcinoma. Mol Cancer
Res. 11:1314–1325. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
He J, Zhang W, Zhou Q, Zhao T, Song Y,
Chai L and Li Y: Low-expression of microRNA-107 inhibits cell
apoptosis in glioma by upregulation of SALL4. Int J Biochem Cell
Biol. 45:1962–1973. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shen B, Zhang Y, Yu S, Yuan Y, Zhong Y, Lu
J and Feng J: MicroRNA-339, an epigenetic modulating target is
involved in human gastric carcinogenesis through targeting NOVA1.
FEBS Lett. 589:3205–3211. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Y, Xu D, Bao C, Zhang Y, Chen D, Zhao
F, Ding J, Liang L, Wang Q, Liu L, et al: MicroRNA-135b, a HSF1
target, promotes tumor invasion and metastasis by regulating RECK
and EVI5 in hepatocellular carcinoma. Oncotarget. 6:2421–2433.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kan H, Guo W, Huang Y and Liu D:
MicroRNA-520g induces epithelial-mesenchymal transition and
promotes metastasis of hepatocellular carcinoma by targeting SMAD7.
FEBS Lett. 589:102–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Simbrich A, Wellmann I, Heidrich J,
Heidinger O and Hense HW: Trends in advanced breast cancer
incidence rates after implementation of a mammography screening
program in a German population. Cancer Epidemiol. 44:44–51. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Su D, Xu H, Feng J, Gao Y, Gu L, Ying L,
Katsaros D, Yu H, Xu S and Qi M: PDCD6 is an independent predictor
of progression free survival in epithelial ovarian cancer. J Transl
Med. 10:312012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang Q, Wang Y, Lu X, Zhao Z, Zhu L, Chen
S, Wu Q, Chen C and Wang Z: MiR-125b regulates
epithelial-mesenchymal transition via targeting Sema4C in
paclitaxel-resistant breast cancer cells. Oncotarget. 6:3268–3279.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sahlberg K Kleivi, Bottai G, Naume B,
Burwinkel B, Calin GA, Børresen-Dale AL and Santarpia L: A serum
microRNA signature predicts tumor relapse and survival in
triple-negative breast cancer patients. Clin Cancer Res.
21:1207–1214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liang YJ, Wang QY, Zhou CX, Yin QQ, He M,
Yu XT, Cao DX, Chen GQ, He JR and Zhao Q: MiR-124 targets Slug to
regulate epithelial-mesenchymal transition and metastasis of breast
cancer. Carcinogenesis. 34:713–722. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dong LL, Chen LM, Wang WM and Zhang LM:
Decreased expression of microRNA-124 is an independent unfavorable
prognostic factor for patients with breast cancer. Diagn Pathol.
10:452015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li L, Luo J, Wang B, Wang D, Xie X, Yuan
L, Guo J, Xi S, Gao J, Lin X, et al: Microrna-124 targets
flotillin-1 to regulate proliferation and migration in breast
cancer. Mol Cancer. 12:1632013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Arabkheradmand A, Safari A, Seifoleslami
M, Yahaghi E and Gity M: Down-regulated microRNA-124 expression as
predictive biomarker and its prognostic significance with
clinicopathological features in breast cancer patients. Diagn
Pathol. 10:1782015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li H, Xie S, Liu M, Chen Z, Liu X, Wang L,
Li D and Zhou Y: The clinical significance of downregulation of
mir-124-3p, mir-146a-5p, mir-155-5p and mir-335-5p in gastric
cancer tumorigenesis. Int J Oncol. 45:197–208. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li X, Yu Z, Li Y, Liu S, Gao C, Hou X, Yao
R and Cui L: The tumor suppressor miR-124 inhibits cell
proliferation by targeting STAT3 and functions as a prognostic
marker for postoperative NSCLC patients. Int J Oncol. 46:798–808.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
An L, Liu Y, Wu A and Guan Y: microRNA-124
inhibits migration and invasion by down-regulating ROCK1 in glioma.
PLoS One. 8:e694782013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang H, Wang Q, Zhao Q and Di W: MiR-124
inhibits the migration and invasion of ovarian cancer cells by
targeting SphK1. J Ovarian Res. 6:842013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Peng XH, Huang HR, Lu J, Liu X, Zhao FP,
Zhang B, Lin SX, Wang L, Chen HH, Xu X, et al: MiR-124 suppresses
tumor growth and metastasis by targeting Foxq1 in nasopharyngeal
carcinoma. Mol Cancer. 13:1862014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang W, Mao YQ, Wang H, Yin WJ, Zhu SX
and Wang WC: MiR-124 suppresses cell motility and adhesion by
targeting talin 1 in prostate cancer cells. Cancer Cell Int.
15:492015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
la Cour JM, Høj BR, Mollerup J, Simon R,
Sauter G and Berchtold MW: The apoptosis linked gene ALG-2 is
dysregulated in tumors of various origin and contributes to cancer
cell viability. Mol Oncol. 1:431–439. 2008. View Article : Google Scholar : PubMed/NCBI
|