Introduction
Osteosarcoma is the most common type of malignant
tumor arising from bone in children and adolescents, with high
biologic aggressiveness and comprising 2.4% of all malignancies in
pediatric patients (1,2). Although modern treatment methods combine
surgery, multiagent chemotherapy, and sometimes radiotherapy, the
5-year survival rate for patients diagnosed with osteosarcoma is
60–70% (3). The past decade has
revealed the molecular pathogenesis of osteosarcoma expands, and
that potential therapeutic targets are being identified (4,5). It is
essential to comprehensively understand molecular mechanisms in
order to improve the prognosis of patients with osteosarcoma
through tumour-targeted therapies.
MicroRNAs (miRNAs) are a class of non-coding, small
regulatory RNAs, 21–23 nucleotides in length (6), transcribed from introns or
non-protein-coding genes, which cleavage of their target mRNAs by
binding to complementary sites in their 3′-untranslated regions
(UTR) or mediate translational suppression (7). miRNAs play crucial roles in cell cycle
regulation, differentiation, apoptosis, tumorigenesis, migration
and invasion (8–10). Accumulating evidences have shown the
aberrant expression of numerous miRNAs is associated with the
development and metastasis of cancer (11,12),
including human osteosarcoma (13).
Previous studies have researched the role of miRNAs in osteosarcoma
using miRNA profile (14,15). Pan et al and Salah et al
have reported that miR-27a promoted the proliferation, migration
and invasion of human osteosarcoma cells (16,17).
However, the miR-27a expression pattern and clinical value in human
osteosarcoma remain to be determined.
Cyclin G1 (CCNG1) was primarily identified as a
novel member of the cyclin family with homology to c-src (18), and importantly, it was first
identified as a p53-regulated transcript induced by DNA damage
(19). CCNG1 is transcriptionally
activated by p53, and negatively regulates p53 family proteins.
CCNG1, a master tumor suppressor, is directly regulated by
miR-27b/miR-508-5p (20), miR122
(21), and miR1271 (22).
In the present study, we investigated whether
miR-27a expression in osteosarcoma tissues and cells was
upregulated, as compared to normal non-cancer tissue and a normal
human osteoblastic cell line. miR-27a significantly promoted
osteosarcoma cell migration and invasion in vitro.
Additionally, CCNG1 was identified as a direct target of miR-27a.
miR-27a/CCNG1 axis is a potential therapeutic target for human
osteosarcoma.
Materials and methods
Tissue samples
A total of 48 paired primary osteosarcoma and their
matched adjacent non-cancerous bone tissue samples were collected
from patients who underwent surgery at the Department of
Orthopedics, Jinan Central Hospital Affiliated to Shandong
University (Jinan, China), between 2013 and 2016. The patients had
not received chemotherapy prior to surgery. All the tissue samples
were instantly snap-frozen in liquid nitrogen at the time of
surgery, and stored at −80°C for using in the subsequent tests. The
diagnosis of osteosarcoma was confirmed pathologically.
Ethics approval
The study was approved by the Research Ethics
Committee of Jinan Central Hospital Affiliated to Shandong
University, and informed consent was obtained from all the
patients. Procedures performed in the studies involving human
participants were in accordance with the ethical standards of the
Institutional and/or National Research Committee and with the 1964
Helsinki Declaration and its later amendments or comparable ethical
standards.
Cell culture and transfection
Four osteosarcoma cell lines (HOS, SaOS2, 143B and
MG63), and the osteoblastic cell line (hFOB1.19) were obtained from
the Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). Cells were cultured in RPMI-1640 (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
fetal bovine serum (FBS), 100 mg/ml streptomycin and 100 U/ml
penicillin (Invitrogen; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) at 37°C in a humidified incubator containing 5%
CO2. The cells were transfected with the indicated
nucleotides or plasmid using Lipofectamine 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNAs were extracted from the cultured tissue
samples and cells using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. miRNA
cDNA was synthesized using the One-Step PrimeScript miRNA cDNA
synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China).
RT-qPCR was performed with SYBR-Green Premix Ex Taq II (Takara
Biotechnology Co., Ltd.) with a StepOne-Plus real-time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). The level of mature miR-27a was normalized relative to U6
endogenous control and CCNG1 expression was normalized relative to
β-actin (endogenous control) using the 2−∆∆Cq method and
at least 3 independent experiments were performed to generate each
data set. Primers used were purchased from Guangzhou RiboBio Co.,
Ltd. (Guangzhou, China).
Cell transwell assay
Cell transwell assay was performed in a 24-well
plate with 8 mm pore size chamber inserts (Corning Incorporated,
Corning, NY, USA). For the migration and invasion assays, after
transfection with either the mimic/inhibitor or control miR,
1×105 cells/well were placed into the upper chamber with
or without membrane Matrigel (BD Biosciences, Franklin Lakes, San
Jose, CA, USA), respectively. In transwell assay, the upper chamber
contained 200 µl serum-free medium, and the lower chambers
contained 800 µl of medium with 15% FBS. After 36 h of incubation
at 37°C in 5% CO2, the cells on the upper surface of the
membrane were removed, and the cells that had moved to the bottom
of the chamber were fixed with 100% methanol for 30 min and stained
with 0.1% crystal violet for 30 min. The stained cells were imaged
and counted using an inverted microscope (Olympus Corporation,
Tokyo, Japan).
Western blot analysis
Cells were lysed in RIPA buffer. The proteins were
extracted and separated by 10% sodium dodecyl
sulphate-polyacrylamide gel electrophoresis. The separated proteins
were electro-transferred to polyvinylidene difluoride membrane
(Millipore, Boston, MA, USA). The membrane was blocked with 5%
non-fat milk and incubated overnight with primary anti-AEG-1
(Invitrogen; Thermo Fisher Scientific, Inc.) at 4°C. The membranes
were incubated for 1 h at 25°C with horseradish
peroxidase-conjugated secondary antibodies (1:2,000; Bosis,
Beijing, China). Relative protein expression was obtained by
normalization against glyceraldehyde 3-phosphate dehydrogenase. The
proteins of interest were revealed by the ECL western blotting kit
(PSC Biotech Pte., Ltd., Shanghai, China). Densitometry analysis of
the protein blots was performed by Lab Works™ software (UVP, Inc.,
Upland, CA, USA).
Dual-luciferase reporter assay
For dual-luciferase reporter assays, the 3′-UTR of
CCNG1 containing miR-27a binding sites were cloned into a pmirGLO
dual-luciferase vector (Promega Corporation, Madison, WI, USA) to
generate wild-type (WT) pmirGLO-CCNG1-1 3′-UTR. The mutant (MUT)
3′-UTR of CCNG1 gene with miR-27a target sites were
generated using a Site-Directed Mutagenesis kit (Agilent
Technologies, Inc., Santa Clara, CA, USA), and cloned into a
pmirGLO dual-luciferase vector (Promega Corporation) to generate
MUT pmirGLO-CCNG1-1 3′-UTR. The WT pmirGLO-CCNG1-1 3′-UTR and MUT
pmirGLO-CCNG1-1 3′-UTR were co-transfected with miR-27a mimics,
inhibitor or negative control (NC) by using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). After incubation for
two days, luciferase activity was detected by using a dual-Glo
luciferase assay system (Promega Corporation) according to the
manufacturer's protocol.
Statistical analysis
The results are reported as mean ± SD and data
analysis was performed using SPSS 17.0 software (SPSS, Inc.,
Chicago, IL, USA). The relationship between the expression of
miR-27a and CCNG1 were confirmed by Spearman's rho correlation.
Differences between the two groups were calculated using the
Student's t-test or Chi-square test. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-27a expression is upregulated and
inversely correlates with CCNG1 in human osteosarcoma tissues and
cells
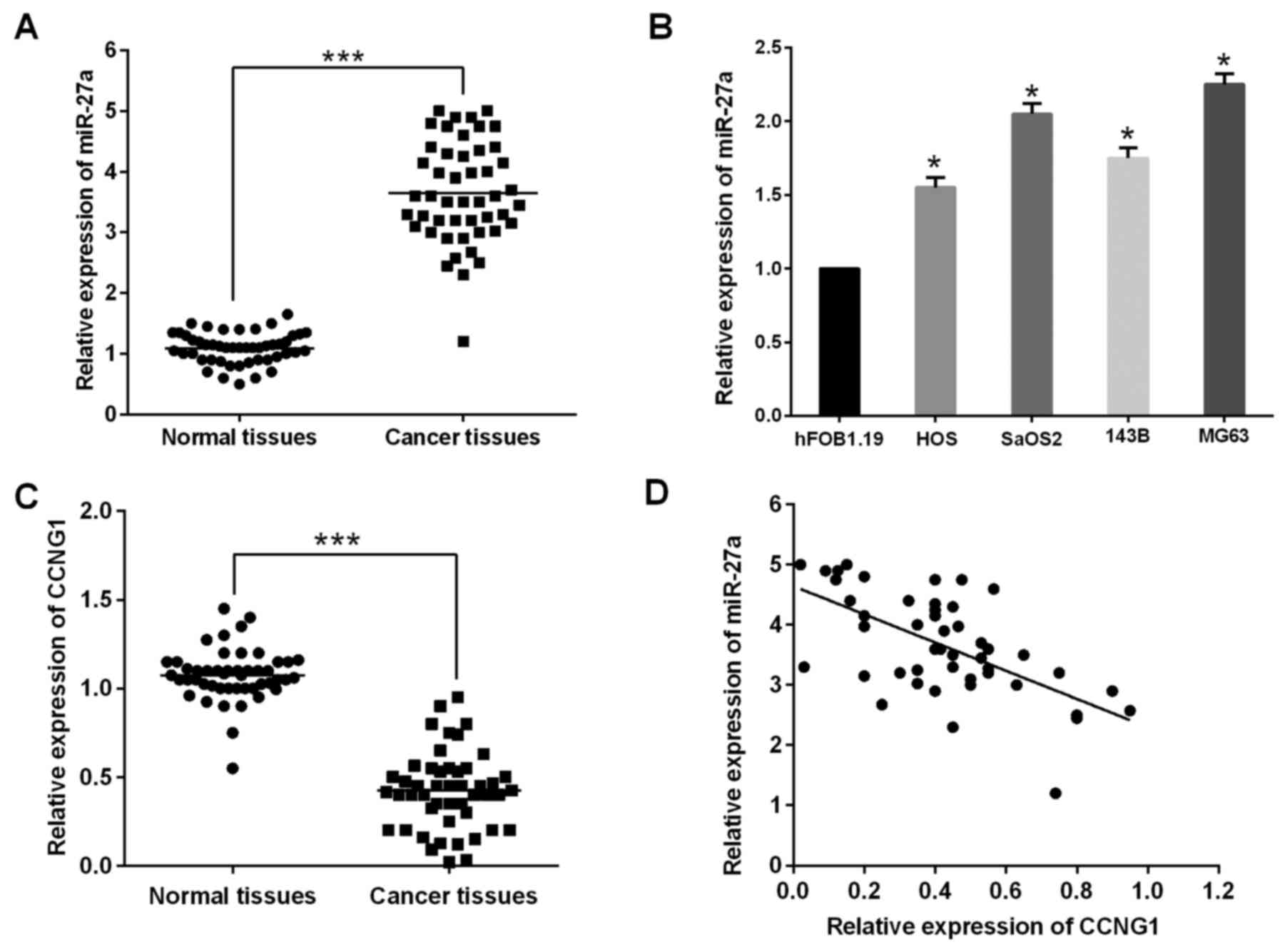
To investigate the expression and significance of
miR-27a in human osteosarcoma, the study first evaluated miR-27a
expression in 48 paired primary osteosarcoma and their matched
adjacent non-cancerous bone tissue samples using RT-qPCR. miR-27a
expression was clearly upregulated in osteosarcoma tissues compared
with non-cancerous bone tissue samples (Fig. 1A). Furthermore, miR-27a was also
significantly increased in the four osteosarcoma cell lines (HOS,
SaOS2, 143B and MG63) compared with that of the osteoblastic cell
line hFOB1.19 (Fig. 1B).
The study further investigated the expression of
CCNG1 in 48 pairs of osteosarcoma tissue and adjacent normal
tissues. The results showed that CCNG1 was significantly
downregulated in osteosarcoma tissue compared with the
paired-adjacent normal tissues and inversely correlated with the
miR-27a level in cervical cancer tissues (Fig. 1C and D), suggesting that miR-27a may
have a critical role as an oncogene in human osteosarcoma. However,
the specific function of miR-27a in human osteosarcoma remained to
be determined.
Effect of miR-27a on human
osteosarcoma cell migration and invasion
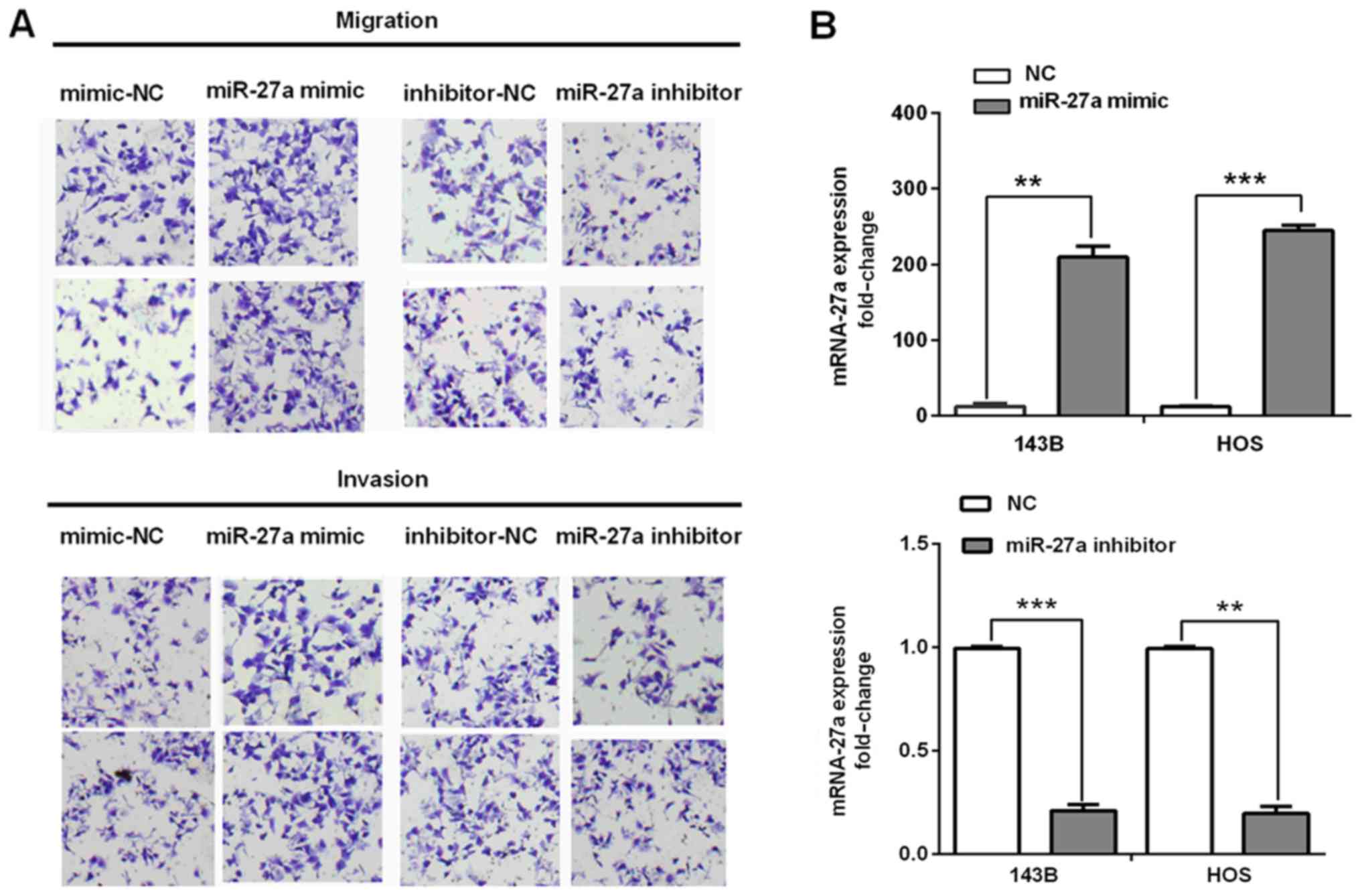
To reveal the biological effects of miR-27a on the
migration and invasion of human osteosarcoma cells, cell migration
and invasion abilities were determined in the 143B and HOS cells
transfected with the miR-27a mimic, miR-27a inhibitor, or NC by
transwell assays. The re-expression levels of miR-27a in
transfected and normal cells were detected by RT-qPCR. It was found
that overexpression of miR-27a significantly promoted the migration
and invasion capacities in the 143B cells and HOS cells (Fig. 2A). Similarly, Fig. 2B shows the re-expression levels of
miR-27a in 143B and HOS cells transfected with the miR-27a mimic or
inhibitor compared with corresponding NC. These results suggested
that miR-27a acts as an oncomiR and promotes cell migration and
invasion during osteosarcoma progression.
CCNG1 as a direct binding target of
miR-27a
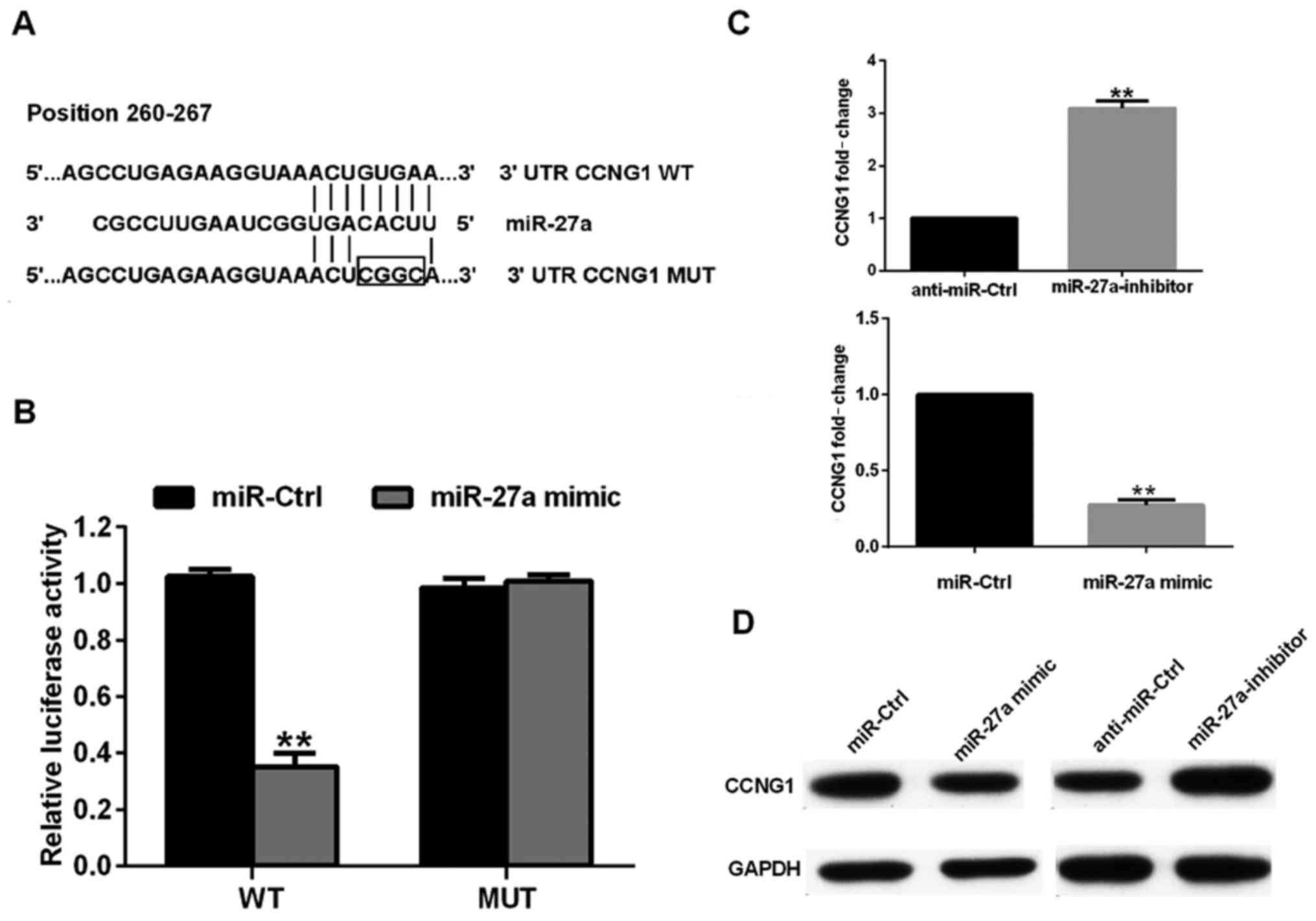
To determine the mechanisms underlying how miR-27a
functions as an oncogene of human osteosarcoma pathology, we used
the bioinformatics algorithms TargetScan to search for the
potential target genes of miR-27a. Our analysis revealed that CCNG1
was a potential target of miR-27a based on putative target
sequences at position 260–267 bp of the CCNG1 3′UTR (Fig. 3A). To confirm CCNG1 as a direct target
of miR-27a, we engineered luciferase reporter constructs,
containing the WT or MUT 3′UTR of the CCNG1 gene. Luciferase
reporter assay showed that miR-27a significantly decreased the
luciferase activity of the CCNG1 3′UTR but not that of the MUT in
143B cells (Fig. 3B). We further
examined whether CCNG1 levels are negatively regulated by miR-27a
in osteosarcoma cell lines. As shown in Fig. 3C and D, mRNA and protein levels of
CCNG1 were significantly enhanced or reduced in response to miR-27a
mimics/inhibitor in HOS cells, compared to the corresponding
controls. Taken together, these data strongly suggest that CCNG1 is
a direct target of miR-27a in human osteosarcoma.
Discussion
Osteosarcoma is the most common type of malignant
tumor arising from bone in children and adolescents that threatens
human life. The past decade has revealed the developement of
molecular pathogenesis of osteosarcoma; thus, there is a search for
potential therapeutic targets (4,5). miRNAs
are a type of small-molecule non-coding RNA, that play vital roles
in regulating expression levels of post-transcriptional gene. The
functions of miRNAs vary in different clinical diseases and may
regulate all aspects of bioactivities, including differentiation
and development, proliferation, metabolism, viral infection,
apoptotic cell death and tumorigenesis (23). Increasing evidence has shown the
abnormal expression of miRNAs is associated with the development
and metastasis of human osteosarcoma (13). Examination of the relationships
between miRNAs and human osteosarcoma possibly provide a new
orientation for the diagnosis and therapy of osteosarcoma. In
previous studies, partial miRNAs were reported to inhibit
osteosarcoma tumorigenicity and to suppress cell proliferation; and
partially were reported to promote osteosarcoma cell proliferation
and induce cell survival (24–26).
Previous studies reported miR-27a functions
differentially in various cancer types, including oral squamous
cell carcinoma (27), breast
(28), gastric (29), colorectal (30), and colonic cancer (31). Furthermore, Pan et al have
reported that miR-27a could promote proliferation, migration and
invasion by targeting MAP2K4 in human osteosarcoma cells (16). Salah et al reported miR-27a
contributes to the metastatic properties of osteosarcoma cells
(17). CCNG1 was primarily identified
as a novel member of cyclin family with homology to c-src (18). CCNG1, a master tumor suppressor, has
been shown to be directly regulated by various miRNAs in different
tumors (20–22). In a previous study it was reported
that CCNG1 inhibited osteosarcoma tumor growth in nude mice
(32).
Our results have found that miR-27a acts as an
oncogene in human osteosarcoma. The results showed miR-27a
expression was upregulated in human osteosarcoma tissues and cells,
which is similar to those obtained in previous studies (14,16). The
overexpression of miR-27a significantly promoted the migration and
invasion capacities in the human osteosarcoma cells. Moreover, the
study demonstrated that CCNG1 was a directed target, and that
miR-27a expression inversely correlated with CCNG1 in human
osteosarcoma tissues and cells. The western blot analysis and
Luciferase reporter assay also showed that CCNG1 was regulated by
miR-27a.
In conclusion, results of the present study have
shown that miR-27a is significantly upregulated and inversely
correlates with CCNG1 in osteosarcoma. These results demonstrate
that miR-27a has powerful oncogenic, metastatic and invasive
regulatory effects that are mediated by CCNG1. The study indicates
that miR-27a acts as an oncogene and is a promising therapeutic
target in human osteosarcoma.
References
|
1
|
Geller DS and Gorlick R: Osteosarcoma: A
review of diagnosis, management, and treatment strategies. Clin Adv
Hematol Oncol. 8:705–718. 2010.PubMed/NCBI
|
|
2
|
Raymond AK and Jaffe N: Osteosarcoma
multidisciplinary approach to the management from the pathologist's
perspective. Cancer Treat Res. 152:63–84. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferrari S, Mercuri M, Bacci G, Bielack SS
and Jürgens H: Comment on ‘Prognostic factors in high-grade
osteosarcoma of the extremities or trunk: An analysis of 1,702
patients treated on neoadjuvant Cooperative Osteosarcoma Study
Group protocols’. J Clin Oncol. 20:2910–2911. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Broadhead ML, Clark JC, Myers DE, Dass CR
and Choong PF: The molecular pathogenesis of osteosarcoma: A
review. Sarcoma. 2011:9592482011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He JP, Hao Y, Wang XL, Yang XJ, Shao JF,
Guo FJ and Feng JX: Review of the molecular pathogenesis of
osteosarcoma. Asian Pac J Cancer Prev. 15:5967–5976. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fang Z, Du R, Edwards A, Flemington EK and
Zhang K: The sequence structures of human microRNA molecules and
their implications. PLoS One. 8:e542152013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nicoloso MS, Spizzo R, Shimizu M, Rossi S
and Calin GA: MicroRNAs - the micro steering wheel of tumour
metastases. Nat Rev Cancer. 9:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bueno MJ, Pérez de Castro I and Malumbres
M: Control of cell proliferation pathways by microRNAs. Cell Cycle.
7:3143–3148. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Saumet A and Lecellier CH: microRNAs and
personalized medicine: Evaluating their potential as cancer
biomarkers. Adv Exp Med Biol. 888:5–15. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nugent M: microRNA and bone cancer. Adv
Exp Med Biol. 889:201–230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jones KB, Salah Z, Del Mare S, Galasso M,
Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL, et
al: miRNA signatures associate with pathogenesis and progression of
osteosarcoma. Cancer Res. 72:1865–1877. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gougelet A, Pissaloux D, Besse A, Perez J,
Duc A, Dutour A, Blay JY and Alberti L: Micro-RNA profiles in
osteosarcoma as a predictive tool for ifosfamide response. Int J
Cancer. 129:680–690. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pan W, Wang H, Jianwei R and Ye Z:
MicroRNA-27a promotes proliferation, migration and invasion by
targeting MAP2K4 in human osteosarcoma cells. Cell Physiol Biochem.
33:402–412. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salah Z, Arafeh R, Maximov V, Galasso M,
Khawaled S, Abou-Sharieha S, Volinia S, Jones KB, Croce CM and
Aqeilan RI: miR-27a and miR-27a* contribute to metastatic
properties of osteosarcoma cells. Oncotarget. 6:4920–4935. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tamura K, Kanaoka Y, Jinno S, Nagata A,
Ogiso Y, Shimizu K, Hayakawa T, Nojima H and Okayama H: Cyclin G: A
new mammalian cyclin with homology to fission yeast Cig1. Oncogene.
8:2113–2118. 1993.PubMed/NCBI
|
|
19
|
Okamoto K and Beach D: Cyclin G is a
transcriptional target of the p53 tumor suppressor protein. EMBO J.
13:4816–4822. 1994.PubMed/NCBI
|
|
20
|
Shang Y, Feng B, Zhou L, Ren G, Zhang Z,
Fan X, Sun Y, Luo G, Liang J, Wu K, et al: The
miR27b-CCNG1-P53-miR-508-5p axis regulates multidrug resistance of
gastric cancer. Oncotarget. 7:538–549. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fornari F, Gramantieri L, Giovannini C,
Veronese A, Ferracin M, Sabbioni S, Calin GA, Grazi GL, Croce CM,
Tavolari S, et al: MiR-122/cyclin G1 interaction modulates p53
activity and affects doxorubicin sensitivity of human
hepatocarcinoma cells. Cancer Res. 69:5761–5767. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu X, Ma L, Rao Q, Mao Y, Xin Y, Xu H, Li
C and Wang X: MiR-1271 inhibits ovarian cancer growth by targeting
cyclin G1. Med Sci Monit. 21:3152–3158. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM
and Zhang GZ: Biological functions of microRNAs: A review. J
Physiol Biochem. 67:129–139. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kushlinskii NE, Fridman MV and Braga EA:
Molecular mechanisms and microRNAs in osteosarcoma pathogenesis.
Biochemistry (Mosc). 81:315–328. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ram Kumar RM, Boro A and Fuchs B:
Involvement and clinical aspects of microRNA in osteosarcoma. Int J
Mol Sci. 17:E8772016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma C, Zhan C, Yuan H, Cui Y and Zhang Z:
MicroRNA-603 functions as an oncogene by suppressing BRCC2 protein
translation in osteosarcoma. Oncol Rep. 35:3257–3264. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zeng G, Xun W, Wei K, Yang Y and Shen H:
MicroRNA-27a-3p regulates epithelial to mesenchymal transition via
targeting YAP1 in oral squamous cell carcinoma cells. Oncol Rep.
36:1475–1482. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Morales S, Gulppi F, Gonzalez-Hormazabal
P, Fernandez-Ramires R, Bravo T, Reyes JM, Gomez F, Waugh E and
Jara L: Association of single nucleotide polymorphisms in
Pre-miR-27a, Pre-miR-196a2, Pre-miR-423, miR-608 and Pre-miR-618
with breast cancer susceptibility in a South American population.
BMC Genet. 17:1092016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Danza K, Silvestris N, Simone G, Signorile
M, Saragoni L, Brunetti O, Monti M, Mazzotta A, De Summa S, Mangia
A, et al: Role of miR-27a, miR-181a and miR-20b in gastric cancer
hypoxia-induced chemoresistance. Cancer Biol Ther. 17:400–406.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu F, Dear K, Huang L, Liu L, Shi Y, Nie
S, Liu Y, Lu Y and Xiang H: Association between microRNA-27a
rs895819 polymorphism and risk of colorectal cancer: A
meta-analysis. Cancer Genet. 209:388–394. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gao Y, Li BD and Liu YG: Effect of miR27a
on proliferation and invasion in colonic cancer cells. Asian Pac J
Cancer Prev. 14:4675–4678. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen DS, Zhu NL, Hung G, Skotzko MJ,
Hinton DR, Tolo V, Hall FL, Anderson WF and Gordon EM: Retroviral
vector-mediated transfer of an antisense cyclin G1 construct
inhibits osteosarcoma tumor growth in nude mice. Hum Gene Ther.
8:1667–1674. 1997. View Article : Google Scholar : PubMed/NCBI
|