Introduction
Esophageal cancer is a notable global health
problem, although its incidence varies throughout the world
(1–6).
According to histological classification, esophageal cancer is
categorized into esophageal squamous cell carcinoma (ESCC) and
esophageal adenocarcinoma (EAC), each of which possesses different
risk factors and pathogenesis (6).
ESCC frequently occurs in China and other Asian countries, as well
as in certain parts of Iran and South Africa, whereas the incidence
of EAC is markedly increased in Western countries, including the
USA and a number of European countries (6). EAC accounts for ~66% of esophageal
cancer cases in the USA, whereas ESCC currently accounts for 90% of
worldwide esophageal cancer cases (6). Thus far, the majority of esophageal
cancer cases are diagnosed at advanced stages of the disease, which
makes curative surgery unfeasible, leading to a poor overall
prognosis in patients with esophageal cancer. Thus, novel
strategies and biomarkers are necessary in order to provide early
detection, prevention or prediction of treatment responses and the
prognosis of esophageal cancer.
In all nucleated cells mitochondria are powerhouses,
converting oxygen and glucose into adenosine triphosphate (ATP)
through oxidative phosphorylation (7). The mitochondrion possesses its own
mitochondrial DNA (mtDNA), which is ring-like and double-stranded.
Human mtDNA contains non-coding and coding regions, and comprises
~16,569 base pairs (7). The 13
polypeptides encoded by the coding region constitute respiratory
enzyme complexes, two ribosomal RNAs and a set of 22 transfer RNAs,
all of which are necessary for the synthesis of certain
mitochondrial peptides (8). The
displacement loop (D-loop) is a non-coding region serving a
function in mtDNA replication and transcription. Thus, D-loop
mutations may lead to alterations in mtDNA content or gene
expression (9,10). Cells typically contain high levels of
mtDNA, and the majority of the copies are determinated at birth
(11).
Compared with nuclear DNA, the mitochondrial genome
is more susceptible to mutation and is sensitive to oxidative
damage (8,12). Thus, mitochondrial dysfunction is
hypothesized to have a marked influence on tumorigenesis and
progression. The copy number of mtDNA is increased during
progression of ESCC, glioma, lung and laryngeal cancer, whereas the
copy number of mtDNA is decreased in bladder, breast and gastric
cancer, and hepatocellular carcinomas, compared with non-cancerous
tissues (8,12–16).
Furthermore, accumulated evidence has demonstrated that in
peripheral blood lymphocytes, altered mtDNA content is closely
associated with the risk of certain types of cancer (17–24).
However, the association between various copy numbers of mtDNA and
prognosis in ESCC remains unresolved. In the present study, altered
mtDNA content in normal esophageal tissues and ESCC tissues was
investigated using reverse transcription-quantitative polymerase
chain reaction (RT-qPCR), and the associations between mtDNA
content and clinicopathological features, and the prognosis of
patients with ESCC were also explored.
Materials and methods
Patients and tissue samples
Approval for the present study was obtained from
Human Ethics Committee and Institutional Review Board, Xi'an
Jiaotong University (Xi'an, China). A total of 141
paraffin-embedded ESCC tissue samples were collected from the First
Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China)
between January 2002 and September 2010. Additionally, 45 samples
were obtained from esophagitis patients that underwent endoscopy
for biopsy, and were used as control subjects. Patients with
chemoradiotherapy were excluded. Written informed consent was
obtained from each patient prior to surgery. The diagnosis of
esophageal diseases was based on histological examination according
to the World Health Organization diagnosis standard of esophageal
diseases (25). The
clinicopathological data of the patients was collected from their
medical records and the results are presented in Table I.
 | Table I.Clinicopathological characteristics of
patients with ESCC. |
Table I.
Clinicopathological characteristics of
patients with ESCC.
| Characteristics | Value |
|---|
| Gender, n (%) |
|
| Male | 111 (78.7) |
|
Female | 30 (21.3) |
| Age, years |
|
| Mean | 58.3 |
| SD | 8.5 |
| Tumor localization,
n (%) |
|
| Upper
esophagus | 32 (22.7) |
| Middle
esophagus | 75 (53.2) |
| Lower
esophagus | 34 (24.1) |
| Tumor size, n
(%) |
|
| <3
cm3 | 36 (25.5) |
| 3-5
cm3 | 72 (51.1) |
| >5
cm3 | 33 (23.4) |
| Differentiation, n
(%) |
|
|
Well/moderate | 112 (79.4) |
|
Poor/undifferentiated | 29 (20.6) |
| Tumor invasion, n
(%) |
|
| T1 | 15 (10.6) |
| T2 | 26 (18.4) |
| T3 | 72 (51.1) |
| T4 | 28 (19.9) |
| TNM stage, n
(%) |
|
| I | 15 (10.6) |
| II | 73 (51.8) |
|
III | 50 (35.5) |
| IV | 3 (2.1) |
| Lymph node
metastasis, n (%) |
|
|
Yes | 58 (41.1) |
| No | 83 (58.9) |
| No. of LNM, n
(%) |
|
| N0 | 83 (58.9) |
| N1
(1–6) | 51 (36.2) |
| N2
(7–15) | 7 (4.9) |
| N3
(≥16) | 0 (0.0) |
| Survival status, n
(%) |
|
|
Deceased | 61 (43.3) |
|
Alive | 80 (56.7) |
DNA preparation
Serial sections were cut at 5 µm from paraffin
embedded tumor and control tissues. One section from each sample
was stained by hematoxylin and eosin (H&E) staining as
described below. Briefly after deparaffinizing and hydrating the
tissue sections, slides were incubated with hematoxylin solution in
a staining jar for 10 min to stain the nuclei. Then the slides were
transferred to a staining jar with running water until the water
ran clear. Slides were then transferred to a staining jar with
Eosin solution for 3 min. Successively transfer the slides into
staining jars with 70% ethanol for 20 sec, 90% ethanol for 20 sec,
100% ethanol for 1 min and xylene for 3 min. Slides were mounted
with xylene-based mounting media and covered with cover slides. The
representative tumor tissue was examined by an expert surgical
pathologist at The Department of Pathology of the First Affiliated
Hospital of Xi'an Jiaotong University (Xi'an, China) for esophageal
cancer. The tumor tissues were isolated through manual
microdissection using an inverted microscope according to the
H&E marker. DNA was then isolated from the tumor tissues as
described previously (26). Briefly,
after a treatment for 12 h at room temperature with xylene to
remove paraffin, the tissues were then subjected to digestion with
1% sodium dodecyl sulfate (SDS) and proteinase K at 48°C for 48 h,
with addition of several spiking aliquots of concentrated
proteinase K to facilitate digestion. DNA was subsequently isolated
using a standard phenol-chloroform extraction and ethanol
precipitation protocol, and kept at −80°C until use.
mtDNA content analysis
The relative mtDNA content of the patients with ESCC
and the control subjects was measured using RT-qPCR as described
previously (15). The specific
primers and TaqMan probes for MT-ND1 (mitochondrially
encoded NADH dehydrogenase 1) and the β-actin gene were designed
using Primer Express software (version 3.0; Applied Biosystems;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) (Table II). Each sample was prepared in
triplicate. To standardize the input DNA, β-actin was measured in
parallel as a reference gene. Normal leukocyte DNA was used in
serial dilutions to establish standard curves. The relative mtDNA
content of each sample was measured as described previously
(18).
 | Table II.Primer and TaqMan probe sequences
used in this study. |
Table II.
Primer and TaqMan probe sequences
used in this study.
| Genes | Forward primer
sequence (5′-3′) | Probe sequence
(5′-3′) | Reverse primer
sequence (5′-3′) |
|---|
| MT-ND1 |
CCCCTAAAACCCGCCACATCCGCCCCGACC-TAMRA |
6FAM-ACCCTCTACATCA |
GTAGAAGAGCGATGGTGAGAGC |
| β-actin |
TCACCCACACTGTGCCCATCTACGA |
6FAM-ATGCCCTCCC |
TCGGTGAGGATCTTCATGAGGTACCATGCCATCC-TAMRA |
Statistical analysis
The copy number of mtDNA between patients with ESCC
and control subjects was compared using a Mann-Whitney U
test. The association of mtDNA content with clinicopathological
features was assessed using univariate analysis. Multivariate
models were developed and adjusted according to age, tumor size,
differentiation and lymph node metastasis. Survival time was
determined from the day of primary tumor resection to the day of
cancer-associated mortality or last clinical follow-up. The
Kaplan-Meier estimator method was used to analyze patient survival
stratified by mtDNA content variations and the difference between
the Kaplan-Meier curves was assessed using a log-rank test. A
multivariate Cox's proportional hazards regression model was
applied to calculate the hazard ratio (HR) with 95% confidence
intervals (CIs) for prognosis evaluation. A multivariate Cox's
regression analysis was performed to assess the influence of mtDNA
content on patient prognosis, which was independent of the number
of lymph node metastases, tumor invasion and differentiation.
Statistical analyses were performed using SPSS software (version
11.5; SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Relative mtDNA content in ESCC
RT-qPCR was performed to analyze the mtDNA copy
number in 141 patients with ESCC and 45 control subjects. As
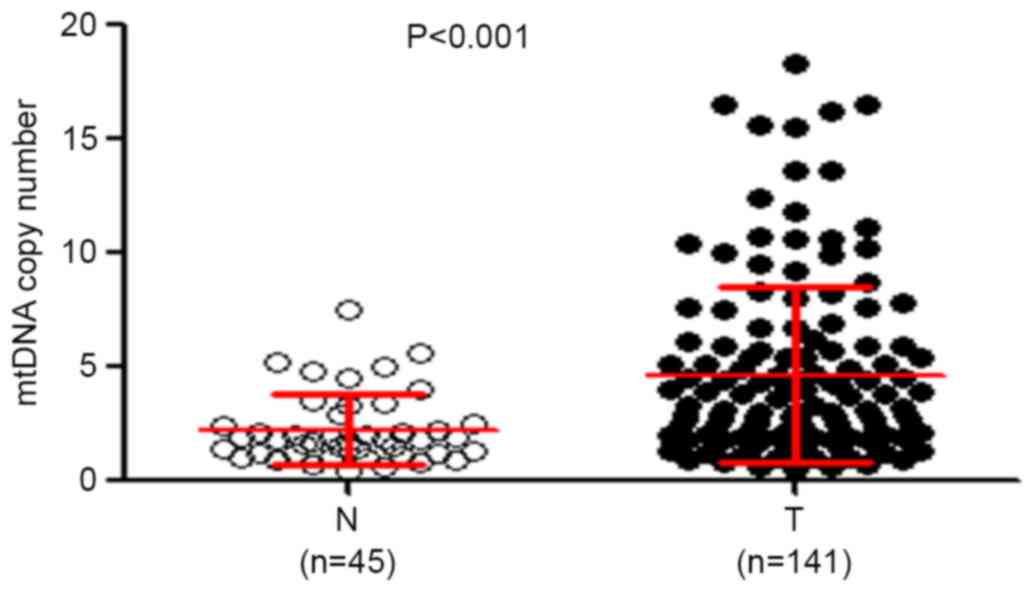
presented in Fig. 1, the relative
mean mtDNA content was significantly increased in patients with
ESCC (4.67±3.88 copies) compared with that in control subjects
(2.24±1.80 copies) (P<0.001). The median values among patients
with ESCC and control subjects were 3.15 copies (interquartile
range between 0.53 and 18.28 copies) and 1.80 copies (interquartile
range between 0.47 and 7.50 copies), respectively, suggesting that
the majority of patients with ESCC had increased mtDNA content
compared with control subjects, which was consistent with a
previous study (5). The possible
difference in mtDNA content in tumor tissues with regard to
selected clinicopathological features was also evlaluated. As
presented in Fig. 2, no significant
differences were identified in mtDNA copy number with reagrd to
sex, age, tumor localization, tumor size, differentiation, tumor
invasion, Tumor-Node-Metastasis stage (27) and lymph node metastasis. The group of
patients who sucumbed to the disease exhibited an increased mtDNA
copy number compared with the group of patients who did not
succumb. Although no statistically significant difference was
observed, there was a notable trend toward a positive association
between mtDNA copy number and survival status of patients with ESCC
(P=0.054).
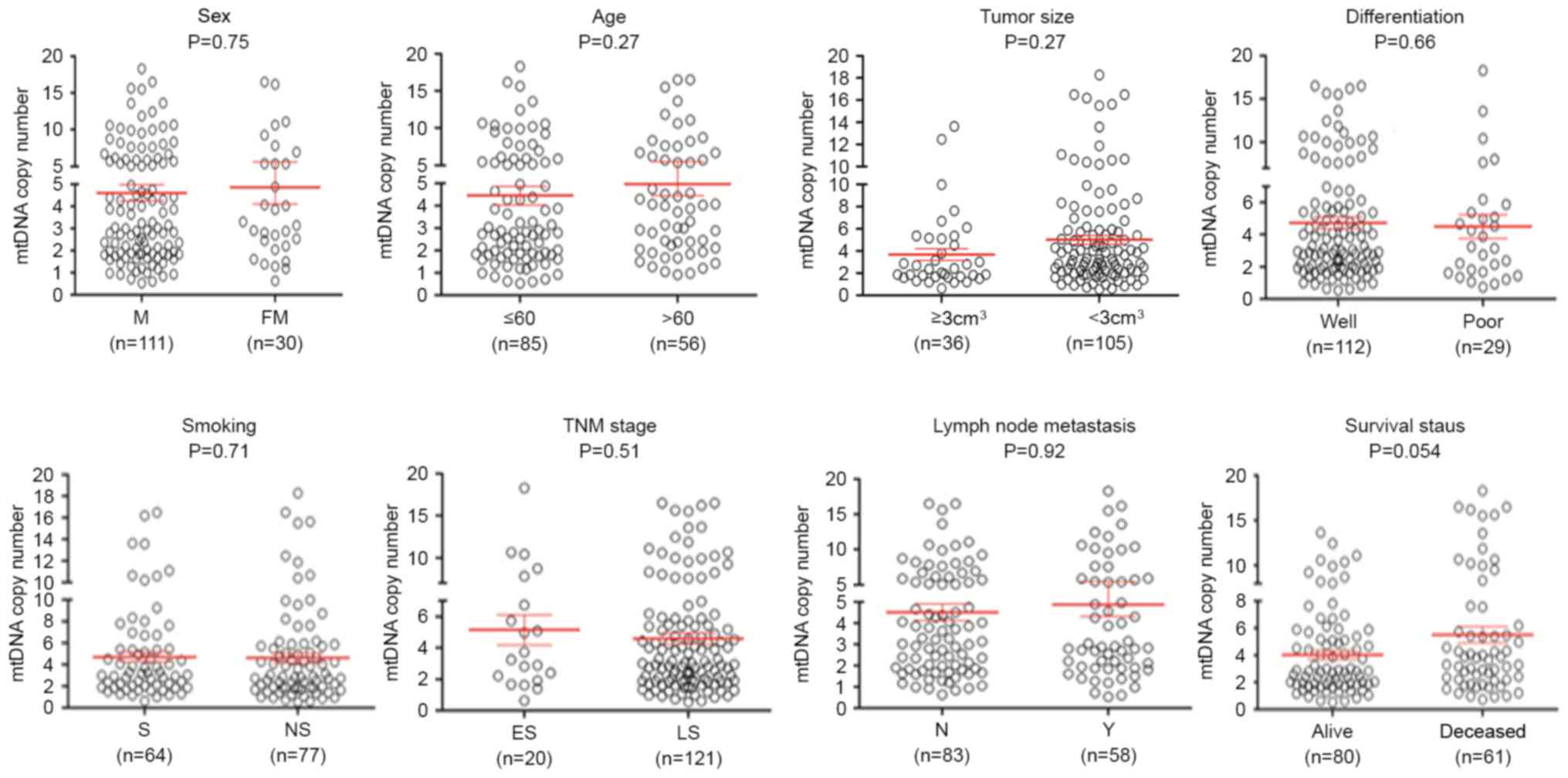 | Figure 2.Association of mtDNA copy number with
clinicopathological characteristics in ESCC. The copy number of
mtDNA was analyzed using reverse transcription-quantitative
polymerase chain reaction. Circles represent the mtDNA copy number
of each case. Horizontal lines represent mean ± standard error of
the mean. Sample means were compared using the Mann-Whitney U test.
mtDNA, mitochondrial DNA; ESCC, esophageal squamous cell carcinoma;
FM, female; M, male; well, well/moderate differentiation; poor,
poor/undifferentiated; ES, early-stage; LS, late-stage; S, smoking;
NS, non-smoking; N, no metastasis present; Y, yes metastasis
present; TNM, Tumor-Node-Metastasis. |
Association between altered mtDNA
content and clinicopathological features of ESCC
To investigate the association between mtDNA copy
number and the clinicopathological features of ESCC, the median
mtDNA copy number (3.14 copies) in the ESCC samples was selected as
a threshold value. Using this threshold value, the ESCC samples
were classified into two groups, namely increased mtDNA content
(≥3.14 copies) and decreased mtDNA content (<3.14 copies). As
presented in Table III, mtDNA copy
number variations was significantly associated with survival status
in patients with ESCC. Compared with the control, the patients with
ESCC exhibiting increased mtDNA content had an increased mortality
rate [odds ratio (OR), 1.95; 95% confidence interval (CI),
1.04–3.64; P=0.04]. Multivariable logistic regression was performed
to evaluate the independent association of variable mtDNA copy
number with age, tumor size, differentiation and lymph node
metastasis. As presented in Table IV
the patients with increased mtDNA copy number had a 2.41 times
higher risk of mortality than those with decreased mtDNA copy
number. The data identified that increased mtDNA copy number
remained positively associated with survival status following
adjustment (OR, 2.41; 95% CI, 1.13–5.13; P=0.02).
 | Table III.Copy number variations of
mitochondrial DNA in esophageal squamous cell carcinoma: Univariate
associations with clinicopathological characteristics. |
Table III.
Copy number variations of
mitochondrial DNA in esophageal squamous cell carcinoma: Univariate
associations with clinicopathological characteristics.
|
Characteristics | Odds ratio (95%
CI) | P-value |
|---|
| Sex | 0.98
(0.44–2.20) | 0.97 |
| Age,
yearsa | 1.37
(0.71–2.66) | 0.35 |
| Tumor
localizationb | 0.90
(0.57–1.41) | 0.64 |
| Tumor size,
cm3c | 1.87
(0.83–4.20) | 0.13 |
|
Differentiationd | 1.33
(0.76–2.33) | 0.33 |
| Tumor
invasione | 1.31
(0.48–3.55) | 0.59 |
| TNM
stagef | 0.78
(0.30–2.01) | 0.61 |
| Lymph node
metastasis | 0.74
(0.38–1.46) | 0.39 |
| Survival
statusg | 1.95
(1.04–3.64) | 0.04 |
 | Table IV.Copy number variations of
mitochondrial DNA in esophageal squamous cell carcinoma:
Multivariable associations with clinicopathological
characteristics. |
Table IV.
Copy number variations of
mitochondrial DNA in esophageal squamous cell carcinoma:
Multivariable associations with clinicopathological
characteristics.
|
Characteristics | Odds Ratio (95%
CI) | P-value |
|---|
| Sex | 1.07
(0.43–2.64) | 0.89 |
| Agea | 1.01
(0.97–1.06) | 0.55 |
| Tumor
sizeb | 1.58
(0.90–2.77) | 0.11 |
|
Differentiationc | 1.72
(0.90–2.77) | 0.99 |
| Tumor
invasiond | 0.67
(0.16–2.81) | 0.58 |
| TNM
stagee | 0.69
(0.23–2.02) | 0.50 |
| Lymph node
metastasis | 0.68
(0.16–2.86) | 0.59 |
| Survival
statusf | 2.41
(1.13–5.13) | 0.02 |
|
Smokingg | 1.15
(0.47–2.79) | 0.75 |
Effect of increased mtDNA content on
prognosis of ESCC
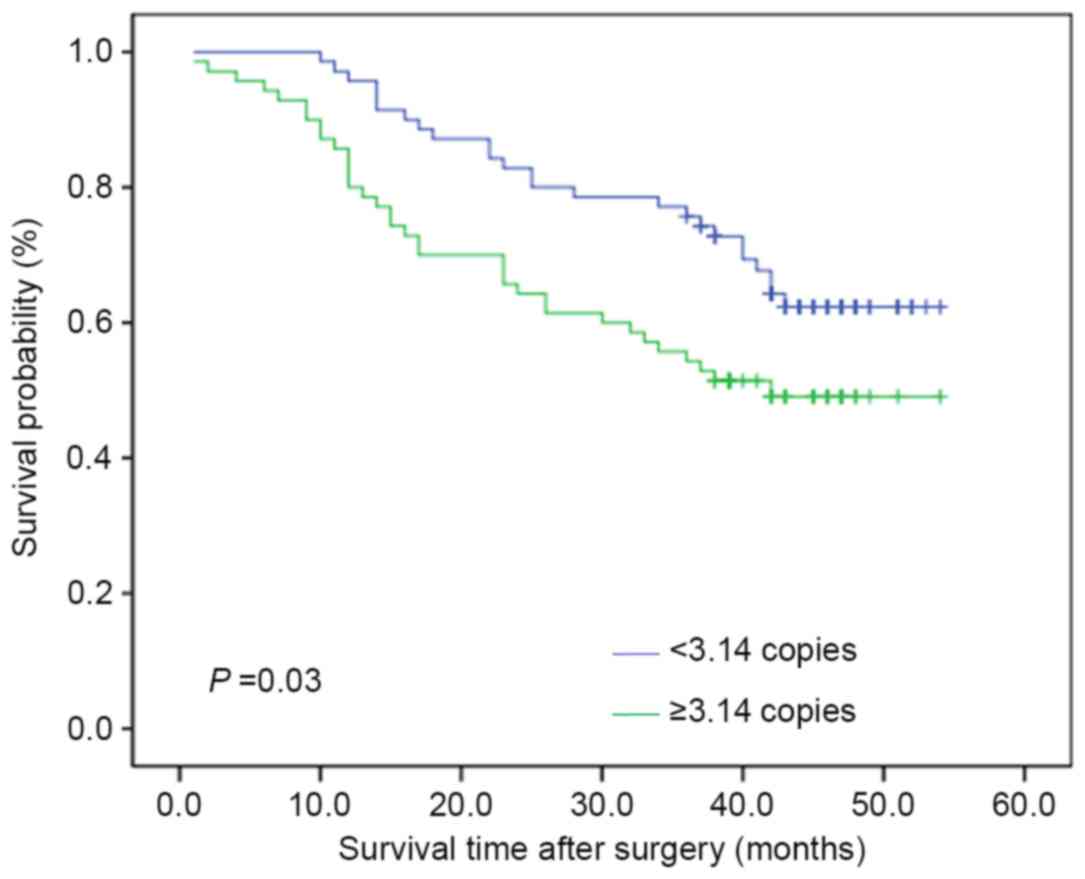
The Kaplan-Meier estimator survival curves and
log-rank test were used to assess the effect of altered mtDNA
content on the survival time of patients with ESCC. As presented in
Fig. 3, the patients with increased
mtDNA copy number experienced shorter survival times than those
with decreased mtDNA copy number (median survival time: 42.00 vs.
38.00 months; P=0.03). Similarly, Cox's unvariate regression
analysis demonstrated a marked association of increased mtDNA copy
number with poor patient survival [hazard ratio (HR), 1.76; 95% CI,
1.06–2.97, P=0.03; Table V] as well
as age [HR, 1.75; 95% CI, 1.05–2.91, P=0.03] and TNM stage [HR,
2.40; 95% CI, 1.44–3.99, P<0.01; Table
V]. Additionally, Cox's multivariate regression analysis
indicated that increased mtDNA content is a predictor of increased
mortality for patients with ESCC (HR, 1.91; 95% CI, 1.13–3.22;
P=0.02), and is an independent variable with regard to the age and
tumor stage (Table V).
 | Table V.Prognostic value of
clinicopathological factors and copy number variation of
mitochondrial DNA in univariate and multivariate Cox's regression
analysis (n =141). |
Table V.
Prognostic value of
clinicopathological factors and copy number variation of
mitochondrial DNA in univariate and multivariate Cox's regression
analysis (n =141).
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | Hazard ratio (95%
CI) | P-value | Hazard ratio (95%
CI) | P-value |
|---|
| Copy number |
|
0.03 |
|
0.02 |
|
<3.14 | 1.00 |
| 1.00 |
|
|
≥3.14 | 1.76
(1.06–2.97) |
| 1.91
(1.13–3.22) |
|
| Age,
yearsa |
|
0.03 |
|
0.02 |
|
<60 | 1.00 |
| 1.00 |
|
|
≥60 | 1.75
(1.05–2.91) |
| 1.89
(1.13–3.17) |
|
| Tumor
invasionb |
|
0.06 |
|
0.18 |
|
T1/T2 | 1.00 |
| 1.00 |
|
|
T3/T4 | 2.65
(0.96–7.30) |
| 2.03
(0.72–5.68) |
|
| TNM
stagec |
| <0.01 |
| <0.01 |
|
I/II | 1.00 |
| 1.00 |
|
|
III/IV | 2.40
(1.44–3.99) |
| 2.54
(1.50–4.30) |
|
Discussion
The primary function of the mitochondria is to
produce the energy source ATP. Additionally, the mitochondria also
serve functions in numerous cell processes, including cell
differentiation, growth, death, cell cycle regulation and signal
transduction (28). A previous study
identified that extranuclear DNA, including mtDNA, and intranuclear
genetic materials may have an effect on ESCC and the development of
other tumors (8). It was identified
that mtDNA content was significantly increased in patients with
ESCC compared with control subjects, as supported by the results of
a previous study (5). These data
suggest that increased mtDNA content may be associated with
esophageal tumorigenesis and the high mortality rate of patients
with ESCC.
Changes in the mtDNA or mitochondrial dysfunction
are frequently observed in tumor cells (8,12–16). Several prior studies indicated that
mtDNA content is increased in different types of cancer, including
ESCC (5,8,14–16). This proliferation of mtDNA is possibly
a compensation mechanism induced by an energy shortage in tumor
cells (29,30). This is supported by a previous study,
which demonstrated that relative mtDNA copy number increased
progressively in normal esophageal mucosa, ESCC and metastatic
lymph nodes, and that increased mtDNA content contributed to the
increased bioenergetic capbility of the mitochondria, further
promoting tumor invasion of ESCC (5).
In addition to its function supplying energy,
increased mtDNA content may be associated with mtDNA escape in
cancer. It has been identified that mtDNA escape is an essential
exchange between the nuclear and mitochondrial genomes in yeast
cells (31,32). Similar to yeast, mammalian cells may
require increased mtDNA content for the communication of genetic
material between mitochondrial and nuclear genomes (33,34). In
line with this, a previous study utilizing fluorescence in
situ hybridization demonstrated that mitochondrial sequence
hybridization is observed in the nuclei more frequently in glioma
tissues compared with normal brain tissues (33). Furthermore, it has been demonstrated
that mtDNA sequences are able to interrupt intronic regions of
certain cancer-associated genes, including the tumor suppressor
gene mothers against decapentaplegic homolog 2 (35). mtDNA is able to cross with the nuclear
genome in intronic and exonic regions (36,37). Thus,
increased mtDNA content may be symptomatic of an early genetic
event contributing to tumorigenesis by supplying the energy for
tumor cell growth and interrupting the function of specific tumor
suppressor genes.
Since increased mtDNA content may contribute to
tumorigenesis, a number of studies have been performed in order to
explore the association between altered mtDNA content and clinical
outcomes of patients with cancer (38,39). In
the present study, it was revealed that increased mtDNA content was
associated with the survival status of patients with ESCC, and that
the patients with high mtDNA content experienced significantly
shorter survival times than those with low mtDNA content. When
multivariate survival analysis was performed using Cox's regression
model, it was once again identified that increased mtDNA content
was associated with poorer patient survival rates. Taken together,
these findings suggest that an altered mtDNA copy number may be a
prognostic signature in ESCC.
A number of novel systems for copy number variation
analysis have been developed, including magnetic
nanoparticles-chemiluminescence detection (a combination of
magnetic separation and chemiluminescence) (40), which has been used to analyze the copy
numbers of porcine endogenous retroviruses and proviruses from
Chinese Bama minipigs. This system possessess the advantages of
being low-cost, simple, and providing high specificity and
sensitivity compared with RT-qPCR, and as a result it, may be used
in future studies (41).
In summary, the mtDNA content in patients with ESCC
was investigated and it was demonstrated that increased mtDNA
content may have an important impact on the tumorigenic phenotype
of ESCC. The results of the present study also revealed that
increased mtDNA copy number is an independent prognostic factor in
ESCC, which may serve as a biomarker for predicting poor prognosis
in patients with ESCC.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant nos. 81402340 and 81472622), and
the Fundamental Research Funds for the Central Universities (grant
no. xjj2014147) and the Natural Science Basic Research Plan in
Shaanxi Province of China (Program no. 2015JM8390).
References
|
1
|
Li JY, Liu BQ, Li GY, Chen ZL, Sun XI and
Rong SD: Atlas of cancer mortality in the People's Republic of
China. An aid for cancer control and research. Int J Epidemiol.
10:127–133. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li JY: Cancer mapping as an epidemiologic
research resource in China. Recent Results Cancer Res. 114:115–136.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wu IC, Lu CY, Kuo FC, Tsai SM, Lee KW, Kuo
WR, Cheng YJ, Kao EL, Yang MS and Ko YC: Interaction between
cigarette, alcohol and betel nut use on esophageal cancer risk in
Taiwan. Eur J Clin Invest. 36:236–241. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lin CS, Chang SC, Wei YH, Chou TY, Wu YC,
Lin HC, Wang LS and Hsu WH: Prognostic variables in thoracic
esophageal squamous cell carcinoma. Ann Thorac Surg. 87:1056–1065.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin CS, Chang SC, Wang LS, Chou TY, Hsu
WH, Wu YC and Wei YH: The role of mitochondrial DNA alterations in
esophageal squamous cell carcinomas. J Thorac Cardiovascular Surg.
139:189–197.e4. 2010. View Article : Google Scholar
|
|
6
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global Cancer Statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Iborra FJ, Kimura H and Cook PR: The
functional organization of mitochondrial genomes in human cells.
BMC Biol. 2:92004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wallace DC: Mitochondria and cancer. Nat
Rev Cancer. 12:685–698. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shadel GS: Expression and maintenance of
mitochondrial DNA: New insights into human disease pathology. Am J
Pathol. 172:1445–1456. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chinnery PF and Hudson G: Mitochondrial
genetics. Br Med Bull. 106:135–159. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lightowlers RN, Chinnery PF, Turnbull DM
and Howell N: Mammalian mitochondrial genetics: Heredity,
heteroplasty and disease. Trends Genet. 13:450–455. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Copeland WC, Wachsman JT, Johnson FM and
Penta JS: Mitochondrial DNA alterations in cancer. Cancer Invest.
20:557–569. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang G, Qu Y, Dang S, Yang Q, Shi B and
Hou P: Variable copy number of mitochondrial DNA (mtDNA) predicts
worse prognosis in advanced gastric cancer patients. Diagn Pathol.
8:1732013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dang S, Qu Y, Wei J, Shao Y, Yang Q, Ji M,
Shi B and Hou P: Low copy number of mitochondrial DNA (mtDNA)
predicts worse prognosis in early-stage laryngeal cancer patients.
Diagn Pathol. 9:282014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Qu Y, Gao K, Yang Q, Shi B, Hou P
and Ji M: High copy number of mitochondrial DNA (mtDNA) predicts
good prognosis in glioma patients. Am J Cancer Res. 5:1207–1216.
2015.PubMed/NCBI
|
|
16
|
Reznik E, Miller ML, Şenbabaoğlu Y, Riaz
N, Sarungbam J, Tickoo SK, Al-Ahmadie HA, Lee W, Seshan VE, Hakimi
AA and Sander C: Mitochondrial DNA copy number variation across
human cancers. Elife. 5:pii:e107692016. View Article : Google Scholar
|
|
17
|
Lan Q, Lim U, Liu CS, Weinstein SJ,
Chanock S, Bonner MR, Virtamo J, Albanes D and Rothman N: A
prospective study of mitochondrial DNA copy number and risk of
non-Hodgkin lymphoma. Blood. 112:4247–4249. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xing J, Chen M, Wood CG, Lin J, Spitz MR,
Ma J, Amos CI, Shields PG, Benowitz NL, Gu J, et al: Mitochondrial
DNA content: Its genetic heritability and association with renal
cell carcinoma. J Natl Cancer Inst. 100:1104–1112. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bonner MR, Shen M, Liu CS, Divita M, He X
and Lan Q: Mitochondrial DNA content and lung cancer risk in Xuan
Wei, China. Lung Cancer. 63:331–334. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shen J, Platek M, Mahasneh A, Ambrosone CB
and Zhao H: Mitochondrial copy number and risk of breast cancer: A
pilot study. Mitochondrion. 10:62–68. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liao LM, Baccarelli A, Shu XO, Gao YT, Ji
BT, Yang G, Li HL, Hoxha M, Dioni L, Rothman N, et al:
Mitochondrial DNA copy number and risk of gastric cancer: A report
from the Shanghai Women's Health Study. Cancer Epidemiol Biomarkers
Prev. 20:1944–1949. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lynch SM, Weinstein SJ, Virtamo J, Lan Q,
Liu CS, Cheng WL, Rothman N, Albanes D and Stolzenberg-Solomon RZ:
Mitochondrial DNA copy number and pancreatic cancer in the
alpha-tocopherol beta-carotene cancer prevention study. Cancer Prev
Res (Phila). 4:1912–1919. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shen L, Wei J, Chen T, He J, Qu J, He X,
Jiang L, Qu Y, Fang H, Chen G, et al: Evaluating mitochondrial DNA
in patients with breast cancer and benign breast disease. J Cancer
Res Clin Oncol. 4:669–675. 2011. View Article : Google Scholar
|
|
24
|
Hosgood HD III, Liu CS, Rothman N,
Weinstein SJ, Bonner MR, Shen M, Lim U, Virtamo J, Cheng WL,
Albanes D and Lan Q: Mitochondrial DNA copy number and lung cancer
risk in a prospective cohort study. Carcinogenesis. 31:847–849.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hamiltion SR and Aaltonen LA: Pathology
and genetics of tumours of the digestive system. Lyon: IARCPress;
pp. 11–36. 2000
|
|
26
|
Shi J, Yao D, Liu W, Wang N, Lv H, Zhang
G, Ji M, Xu L, He N, Shi B and Hou P: Highly frequent PIK3CA
amplification is associated with poor prognosis in gastric cancer.
BMC Cancer. 12:502012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Stahl M, Mariette C, Haustermans K,
Cervantes A and Arnold D; ESMO Guidelines Working Group, :
Oesophageal cancer: ESMO clinical practice guidelines for
diagnosis, treatment and follow-up. Ann Oncol. 24 Suppl
6:vi51–vi56. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
McBride HM, Neuspiel M and Wasiak S:
Mitochondria: More than just a powerhouse. Curr Biol. 16:R551–R560.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gasparre G, Romeo G, Rugolo M and Porcelli
AM: Learning from oncocytic tumors: Why choose inefficient
mitochondria? Biochim Biophys Acta. 1807:633–642. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Iommarini L, Calvaruso MA, Kurelac I,
Gasparre G and Porcelli AM: Complex impairment in mitochondrial
diseases and cancer: Parallel roads leading to different outcomes.
Int J Biochem Cell Biol. 45:47–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Campbell CL and Thorsness PE: Escape of
mitochondrial DNA to the nucleus in yme1 yeast is mediated by
vacuolar-dependent turnover of abnormal mitochondrial compartments.
J Cell Sci. 111:2455–2564. 1998.PubMed/NCBI
|
|
32
|
Thorsness PE and Weber ER: Escape and
migration of nucleic acids between chloroplasts, mitochondria, and
the nucleus. Int Rev Cytol. 165:207–234. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liang BC and Hays L: Mitochondrial DNA
copy number changes in human gliomas. Cancer Lett. 2:167–173. 1996.
View Article : Google Scholar
|
|
34
|
Netter P and Robineau S: The differential
over amplification of short sequences in the mitochondrial DNA of
rho-petites in Saccharomyces cerevisiae stimulates recombination.
Gene. 83:25–38. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Eppert K, Scherer SW, Ozcelik H, Pirone R,
Hoodless P, Kim H, Tsui LC, Bapat B, Gallinger S, Andrulis IL, et
al: MADR2 maps to 18q21 and encodes a TGFbeta-regulated MAD-related
protein that is functionally mutated in colorectal carcinoma. Cell.
86:543–552. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Borensztajn K, Chafa O, Alhenc-Gelas M,
Salha S, Reghis A, Fischer AM and Tapon-Bretaudière J:
Characterization of two novel splice site mutations in human factor
VII gene causing severe plasma factor VII deficiency and bleeding
diathesis. Br J Haematol. 117:168–171. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Turner C, Killoran C, Thomas NS, Rosenberg
M, Chuzhanova NA, Johnston J, Kemel Y, Cooper DN and Biesecker LG:
Human genetic disease caused by de novo mitochondrial-nuclear DNA
transfer. Hum Genet. 112:303–309. 2003.PubMed/NCBI
|
|
38
|
Zhang G, Qu Y, Dang S, Yang Q, Shi B and
Hou P: Variable copy number of mitochondrial DNA (mtDNA) predicts
worse prognosis in advanced gastric cancer patients. Diagn Pathol.
8:1732013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dang S, Qu Y, Wei J, Shao Y, Yang Q, Ji M,
Shi B and Hou P: Low copy number of mitochondrial DNA (mtDNA)
predicts worse prognosis in early-stage laryngeal cancer patients.
Diagn Pathol. 9:282014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu M, Hu P, Zhang G, Zeng Y, Yang H, Fan
J, Jin L, Liu H, Deng Y, Li S, et al: Copy number variation
analysis by ligation-dependent PCR based on magnetic nanoparticles
and chemiluminescence. Theranostics. 4:71–85. 2015. View Article : Google Scholar
|
|
41
|
Yang H, Liu M, Zhou B, Deng Y, He N, Jiang
H, Guo Y, Lan G, Jiang Q, Yang X and Li Z: Chemiluminescent
detection for estimating relative copy numbers of porcine
endogenous retrovirus proviruses from Chinese Minipigs based on
magnetic nanoparticles. J Nanosci Nanotechnol. 16:6505–6510. 2016.
View Article : Google Scholar : PubMed/NCBI
|