Introduction
Oral squamous cell carcinoma (OSCC) is an aggressive
type of cancer that can exhibit a variable degree of malignant
behavior. Although advances have been made in conventional
treatment, the mortality rate caused by OSCC has not markedly
improved for the past several decades (1), and the biological characteristics of
OSCCs are not yet well understood. The poor prognosis of patients
with OSCC has now been attributed to recurrence, cervical lymph
node metastasis, and resistance to radiotherapy and chemotherapy
(2). An adapter-based differential
display method was previously employed to elucidate the wide range
of genetic events occurring during OSCC development from
pre-cancerous leukoplakia (LP) (3). A
comprehensive gene expression profile was also generated to
discriminate between LPs and OSCCs (4). Following therapy, metastasis has proven
to be a main cause of local relapse in patients with OSCC. Of the
conventional staging and grading systems that are used for the
assessment of OSCC tissues, the Yamamoto-Kohama (YK) mode of
invasion (5) system can be largely
associated with prognosis, particularly with regard to lymph node
metastases (6,7). The YK mode of invasion was devised from
the grading of the mode of invasion originally described by
Jacobson et al (8); in this
histological grading, grade 4 is sub-classified into grades 4C and
D, whereas evaluation of other grades is the same as defined by
Jacobson's classifier (8). Using
primary OSCCs, molecular events associated with the YK mode of
invasion were evaluated previously and prediction models for the
invasion status were constructed (9).
Clinical specimens generally originate from
different genetic backgrounds, which may have considerable
influence on the variation of gene effects. Consequently, there is
little opportunity to directly compare primary and metastatic OSCC
specimens from the same patient. As an alternative approach to
elucidate genetic events intimately associated with the metastatic
potentials of OSCC cells, the establishment of metastatic
sub-clones (L cells) was attempted from the primary mouse OSCC
Sq-1979 cell line in the present study. Next, comprehensive gene
expression was compared between Sq-1979 and L cells to identify
differentially expressed mRNAs. Our previous studies demonstrated
that the expression of certain mRNAs, such as keratin1 and
transglutaminase 3, exhibit continual changes from pre-cancerous to
cancerous tissues and to further malignant OSCCs (4,9). In the
present study, it was revealed that the expression of certain
marker mRNAs could be an index for evaluating the histological
grading of precancerous and OSCC tissues obtained from
patients.
Materials and methods
Experimental animals
Male, 5-week-old, 160 C3H/HeN mice were purchased
from Chubu Kagaku Shizai Co., Ltd. (Nagoya, Japan) and mice were
housed one per cage in a room at 22–23°C under standard atmospheric
pressure with a 12 h light/dark cycle with ad libitum access
to Oriental MF solid chow (Oriental Yeast Co., Tokyo, Japan) and
water; the domestication was continued for 2 weeks before the start
of each experiment. The present study was approved by the Animal
Ethics Committee of Asahi University (Mizuho, Gifu, Japan).
Cells and establishment of
sub-clones
The C3H mouse OSCC Sq1979 cell line was obtained
from the Riken BioResource Center (Ibaraki, Japan). Cells were
grown at 37°C in 5% CO2 in Eagle's minimum essential
medium (E-MEM; Wako Pure Chemical Industries, Ltd., Osaka, Japan)
supplemented with 10% fetal bovine serum (FBS; Nichirei
Biosciences, Inc., Tokyo, Japan) and 1% penicillin/streptomycin
(10,000 U/ml penicillin, 10,000 µg/ml streptomycin; Gibco; Thermo
Fisher Scientific Inc., Waltham, MA, USA). A total of
1×107 Sq-1979 cells were suspended in 0.1 ml saline,
then subcutaneously injected into the posterior neck area of five
7-week-old male C3H/HeN mice. After 3 months, metastasized regional
lymph nodes were dissected into E-MEM supplemented with 10% FBS and
minced to isolate attached cells. Next, metastasized sub-clones,
termed L2-3, L3-5, L5-11, L6-8 and L6-9 cells, were isolated by a
limiting serial-dilution method, as described previously (10). Using the same procedure, for later
experiments, 233-1 and 233-11 independent cell clones were isolated
from primary OSCC tissues of Sq-1979-implanted mice.
Proliferation properties
To examine the cell proliferation rate in
vitro, 2×104 cells were seeded into each well of a
6-well plate. Doubling times (DTs) were calculated by counting cell
numbers using a Burker-Turk hemocytometer after 24, 48, 72 and 96
h. To evaluate the in vivo proliferation of OSCC cells,
1×106-1×107 cells suspended in 0.1 ml saline
were injected subcutaneously into the lateroabdominal area of 5
male, 6-week-old, C3H/HeN mice. The tumor volume was measured using
a digital caliper, and calculated as follows: Tumor volume=(major
axis) × (minor axis)2 × 0.52, as described previously
(11). Tumor DT estimates used the
first and last available tumor volumes (Vo and
Vi), the time interval Ti (in days) between
the two exams, and the following formula: DT=log2
Ti/(logVi - logVo), as described
previously (12).
Transplantability
To examine the transplantability, 1×104,
1×105, 1×106 or 1×107 Sq-1979,
Sq-1979-1, 23-1, 233-11, L2-3, L3-5, L5-11, L6-8 and L6-9 cells
were injected subcutaneously into the lateroabdominal area of male,
6-week-old, C3H/HeN mice (n=5 per experiment). After 1 month, mice
bearing tumors (>4 mm) were counted as positive animals.
Transplantability was defined as: Transplantability=positive
animals/total animals ×100.
Survival rates
To examine survival rates, 1×106
Sq-1979-1, 23-1, 233-11, L2-3, L3-5, L5-11, L6-8 and L6-9 cells
were injected subcutaneously into the lateroabdominal area of male,
6-week-old, C3H/HeN mice (n=5 per experiment). 5 PBS-injected mice
were observed as controls. The survival time (days) was determined.
Mice were considered to have survived until they failed to eat or
drink for 24 h, or until tumors reached a maximum volume of 3510
mm3 at which they interfered with locomotion to eat or
drink, at which point they were euthanized by cervical dislocation,
in accordance with guidelines set out by Workman et al
(13).
RNA extraction and microarray
analysis
RNA extraction was performed using ISOGEN (Nippon
Gene Co., Ltd., Tokyo, Japan), according to the manufacturer's
protocols. Total RNA was extracted from Sq-1979 and L5-11 cells.
cDNA microarray analysis was performed (Oncomics, Nagoya, Japan)
using the Superscript G3 mouse GE Microarray 60K kit (Agilent
Technologies, Inc., Santa Clara, CA, USA). To ensure the
reliability of the data, genes were considered to be differentially
expressed using the following threshold criteria: P<0.001 (using
Student's t-test) and fold-change >2.0.
CDNA samples of precancerous and OSCC
tissues
cDNA samples from 18 patients with OSCC (median age,
65 years; 7 male, 11 female; age range, 38–91 years) and 18
patients with LP (median age, 72 years; age range, 57–98 years)
were incorporated into the present study. Tissue samples that had
been surgically resected in the Dental Hospital of Hokkaido
University (Sapporo, Hokkaido, Japan) between February 1998 and
April 2004, and cDNA synthesis had already been performed, as
described previously (9). All
procedures were undertaken after written informed consent had been
obtained from each patient, and the study adhered to the ethical
guidelines of the dental hospital of the Hokkaido University School
of Dentistry (Sapporo, Japan). The present study was also approved
by the Ethics Committee of Asahi University (no. 27007).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Whole-cell RNA extraction and quantitative PCR was
performed as previously described (4). Primer sequences were designed by Primer
Express software (version 2; Applied Biosystems; Thermo Fisher
Scientific, Inc.). The primers used are summarized in Table I. Each expression level of mRNA was
normalized to ribosomal protein S5 (RPS5).
 | Table I.Polymerase chain reaction primers
used in this experiment. |
Table I.
Polymerase chain reaction primers
used in this experiment.
| Symbol | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| Mouse |
|
Krt7 |
AACAGCCGCTCCCTGGACTTG |
GGTCATCCCCGTGCTTCCC |
|
Slc16a13 |
GGCTTCCTCAACCCTGGTAGTCC |
GCCGATACTCTCGATCATCTGCAC |
|
Slc44a3 |
ATGGATCGTCGGAGAAACCGTAC |
CCCTATCTCACAATGGGCTGGAG |
|
Fyb |
AAGTTGCAGGACAAAGCTCGCCT |
TCCTCGTAGGTAGGTTTCGCTGCC |
|
Tmem173 |
CCTCCGTACTGTCCCAAGAGCCA |
CCAACCATTGAAGGAAGGCTCAG |
|
Rps5 |
AGAAGACTCAACACGCATTGGGC |
GCACTCAGCGATGGTCTTGATGT |
| Human |
|
KRT7 |
GACATCTTTGAGGCCCAGATTGC |
CTGTGCGGCGGTTAATTTCATC |
|
SLC16A13 |
TCCTGGATCGCCTCCATAGGAATC |
AGGAAGTAGCAAAAGAGGCGAGCA |
|
SLC44A3 |
TTTGGCTATGACAGCTTTGGCAA |
TGCGGTTGAGCTGCGTACCTTT |
|
FYB |
CAGGAAGATCCACTAAAGGAGGCC |
CCCCGTGTTATATTTCGCCATGAG |
|
TMEM173 |
AAGGGAATTTCAACGTGGCCAT |
ATATACAGCCGCTGGCTCACTGC |
|
RPS5 |
GAGCGCCTCACTAACTCCATGATGA |
CACTGTTGATGATGGCGTTCACCA |
Statistical analysis
Data are expressed as the mean ± standard deviation.
A Mann-Whitney U test or Student's t-test (for microarray analysis)
was applied to determine the significance of differences between
two groups using Excel Statistics (2008; version 1) (SSRI Inc.,
Tokyo, Japan). P<0.05 was considered to indicate a statistically
significant difference.
Results
Proliferation properties of
Sq-1979-derived sub-clones
To evaluate the proliferation properties of OSCC
sub-clones, DTs in vitro and in vivo were compared.
As shown in Table II, the DTs of
tumor volume for Sq-1979 and 233 cells had a mean value of 14.7 and
14.8 days, respectively, whereas that of L cells was significantly
shorter, at 8.3 days. The in vitro population DT of Sq-1979
and 233 cells was a mean of 11.4 and 14.4 h, respectively, which
was shorter than in vivo and similar to that of L cells
(mean, 14.4 h). These results indicated that L cells have a
specific property that is advantageous for in vivo cell
proliferation.
 | Table II.Growth properties of Sq1979 cells and
the sub-clones. |
Table II.
Growth properties of Sq1979 cells and
the sub-clones.
|
| Doubling time |
|---|
|
|
|
|---|
| Cells | In vivo,
days | In vitro,
h |
|---|
| Original cells |
|
Sq-1979 | 20.1±9.8 | 10.1±0.2 |
| Sub-clones |
|
Sq-1979-1 |
11.6±7.3a | 11.0±3.3 |
|
Sq-1979-2 | 14.8 | 11.8±1.0 |
|
Sq-1979-3 | 12.3 | 11.6±0.8 |
| Primary tumors |
|
233-1 | 12.0±5.8 | 12.7±0.5 |
|
233-11 | 17.5±6.9 | 16.0 ±4.8 |
| Lymph node
metastases |
|
L2-3 |
8.3±5.6b |
12.1±0.3a |
|
L3-5 |
8.1±3.5b |
11.6±1.1a |
|
L5-11 |
7.3±5.7b |
15.4±1.2a |
|
L6-8 |
4.4±0.8b |
14.1±0.4a |
|
L6-9 |
5.6±1.7b |
18.6±2.5a |
Transplantability and survival
rates
As shown in Table
III, when 1×105 cells were inoculated into mice, the
transplantability of the majority of L cells, including L3-5, L5-11
and L6-9 cells, ranged between 20 and 100%; however, with the
exception of 233-11 cells, parental cells and non-metastasized
sub-clones, including Sq-1979, Sq-1979-1 and 233-1 cells, were not
transplantable, even though mice were inoculated with the same
number of cells. When mice were inoculated with 1×106
cells, all the cell types exhibited substantial transplantability
of >60%. These results demonstrated that the majority of L cells
exhibited markedly higher transplantability than parental Sq-1979
cells and the non-metastatic sub-clones, including Sq-1979-1 and
233-11 cells.
 | Table III.Transplantability of oral squamous
cell carcinoma cells. |
Table III.
Transplantability of oral squamous
cell carcinoma cells.
|
| Transplantability,
% |
|---|
|
|
|
|---|
| Cells/body | 104 | 105 | 106 | 107 |
|---|
| Sq-1979 | NP | 0 | 88 | 98 |
| Sq-1979-1 | NP | 0 | 100 | 92 |
| 233-1 | NP | 0 | 60 | 88 |
| 233-11 | NP | 20 | 100 | 100 |
| L2-3 | 0 | 0 | 83 | NP |
| L3-5 | 0 | 20 | 79 | NP |
| L5-11 | 40 | 100 | 100 | NP |
| L6-8 | NP | NP | 100 | NP |
| L6-9 | 100 | 80 | 100 | NP |
The present study also examined the mean survival
time of tumor-burdened mice. As shown in Table IV, mice transplanted with L5-11 and
L6-9 cells exhibited significantly shorter survival times than
those transplanted with Sq1979-1, 233-1, L2-3 and L3-5. The mice
transplanted with L6-8 cells also exhibited shorter survival times.
These results indicated that the majority of L cells possess highly
malignant and advanced phenotypes compared with parental Sq-1979
cells and their non-metastatic sub-clones.
 | Table IV.Survival rates of tumor-transplanted
mice. |
Table IV.
Survival rates of tumor-transplanted
mice.
| Cells | Mean survival time
± SD, days |
|---|
| Sq-1979-1 | 138±39 |
| 233-1 | 146±24 |
| L2-3 |
99±18 |
| L3-5 | 113±25 |
| L5-11 |
81±8a |
| L6-8 | 57 |
| L6-9 |
60±4b |
Isolation of mRNAs predominantly
expressed in L cells
To clarify the identity of expressed genes
associated with the malignant phenotypes of L cells, comprehensive
gene expression of L5-11 and Sq-1979-1 cells was compared using
microarray analysis. Of the 60 mRNAs more predominantly expressed
in L5-11 cells (≥3-fold) than in Sq-1979-1 cells (data not shown),
the expression among Sq-1979, 233 and L cells was further verified
using RT-PCR analysis. Consequently, 5 mRNAs were focused on,
including keratin 7 (Krt7), FYN binding protein
(Fyb), solute carrier family 16 member 13 (Slc16a13),
transmembrane protein 173 (Tmem173) and solute carrier
family 44 member 3 (Slc44a3) mRNA. As shown in Fig. 1, the expression of Krt7 mRNA
was markedly lower in Sq-1979-1, −2, −3, 233-1 and −11 cells
compared with in any other L cells, including L2-3, 3-5, 5–11, 6–8
and 6–9 cells. The expression of Fyb mRNA was also lower in
Sq-1979-1, −2, −3, 233-1 and −11 cells compared with L cells,
including L2-3, 5-11, 6–8 and 6–9 cells. The expression of
Slc16a13, Temem173 and Slc44a3 mRNA was almost
undetectable in the three Sq-1979 sub-clones and 233-1 cells,
whereas the expression was high overall in all 5 L cells. The
results of the present study demonstrated that the expression of
Krt7, Fyb, Slc16a13, Temem173 and
Slc44a3 mRNAs was significantly higher in metastatic
sub-clones, including 5 L cell types, than in original Sq-1979 and
primary 233 cells.
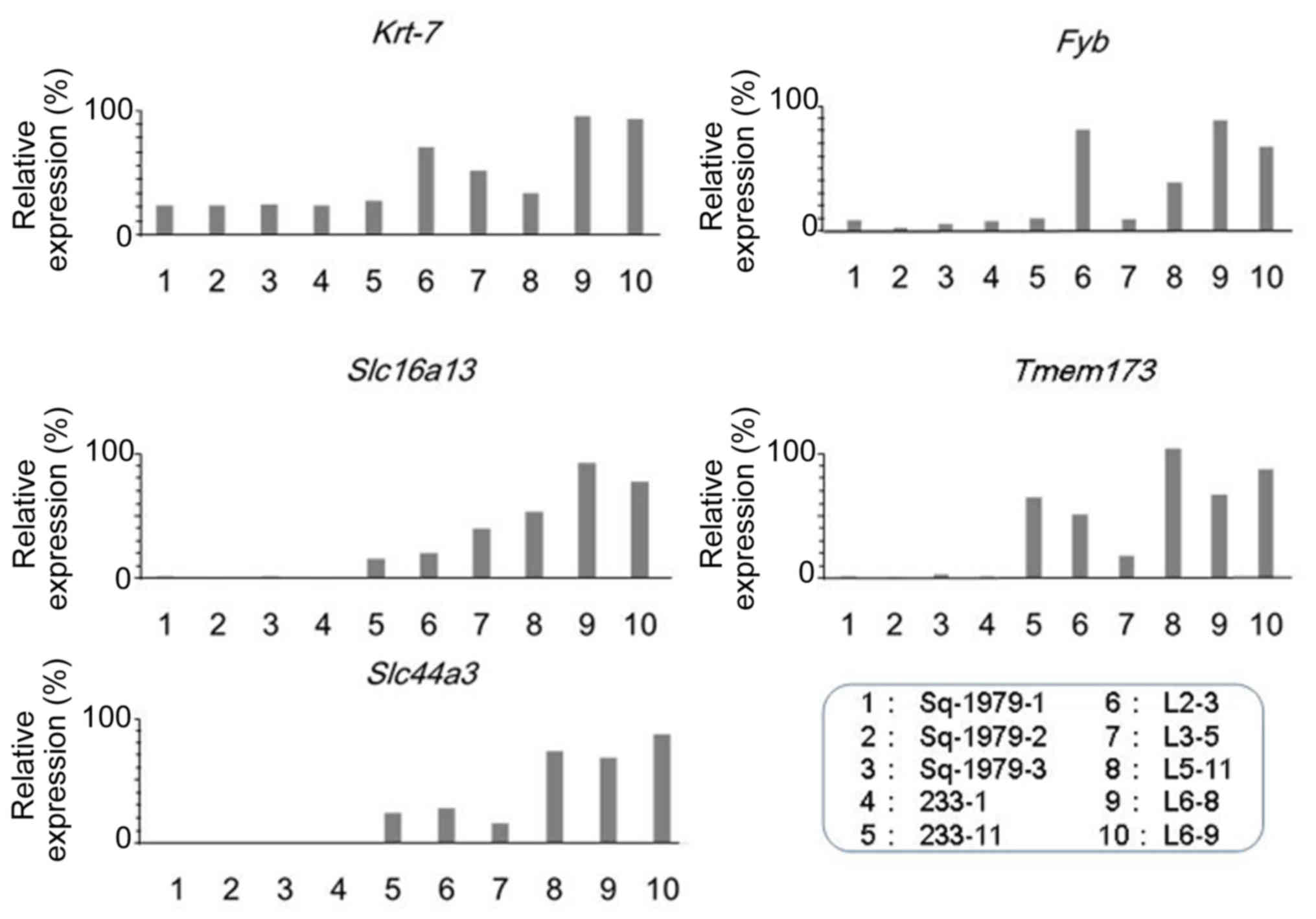 | Figure 1.Expression of Fyb,
Slc16a13, Krt7, Temem173 and Slc44a3
mRNAs in Sq1979 and the sub-clones. Relative expression levels,
expressed as percentages, are in ordinate, and cells are in
abscissa. Fyb, FYN-binding protein; Slc16a13, solute
carrier family 16 member 13; Krt7, keratin 7;
Temem173, transmembrane protein 173. |
Expression of mRNAs in human OSCC and
LP tissues
To further evaluate the expression of these 5 mRNAs
among oral malignancies, RT-PCR analyses using cDNAs derived from
18 LP (Table V) and 18 OSCC (Table VI) tissues were performed. The main
clinicopathological characteristics of each patient are summarized
in the Tables V and VI, respectively. The expression of
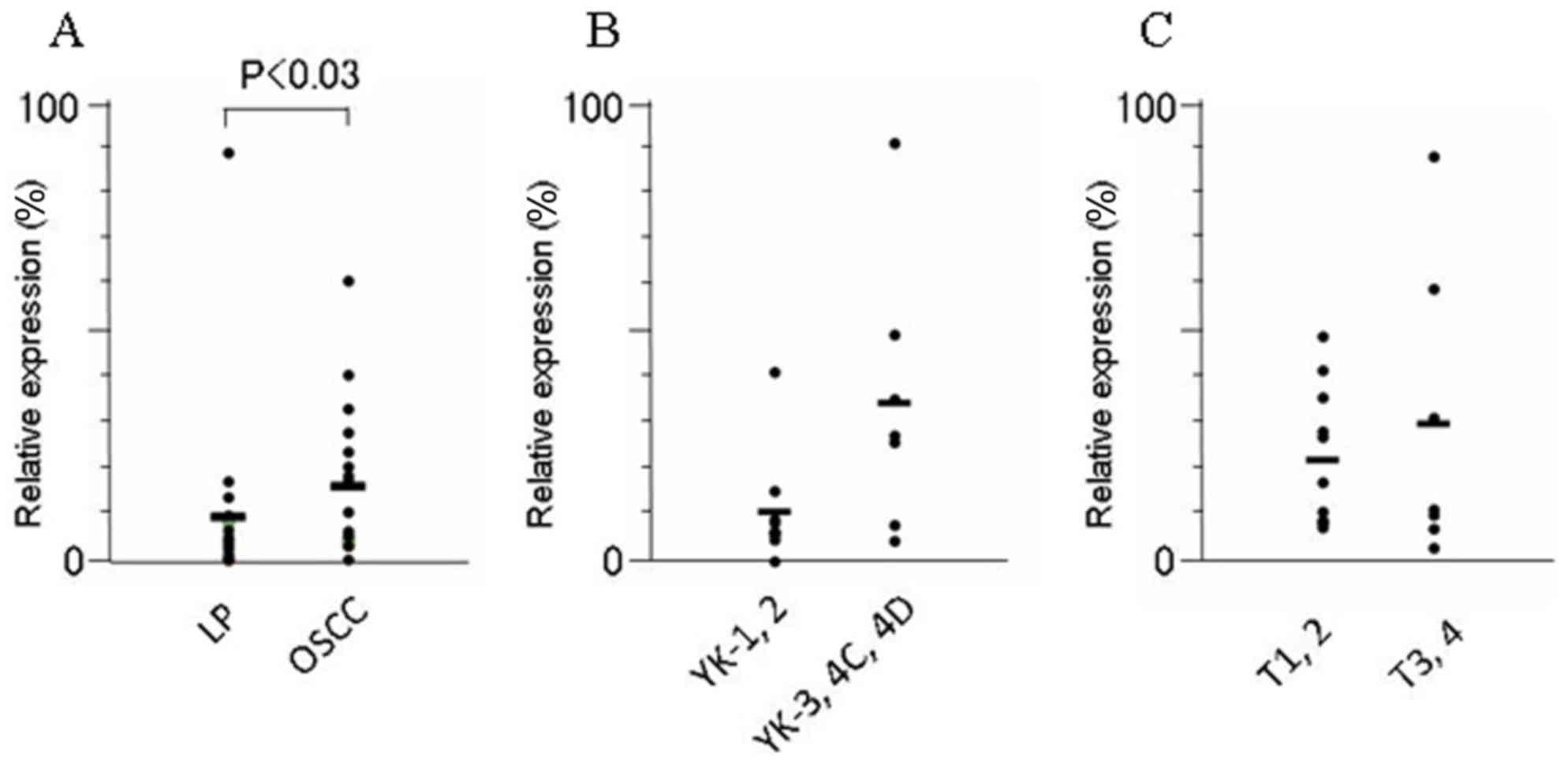
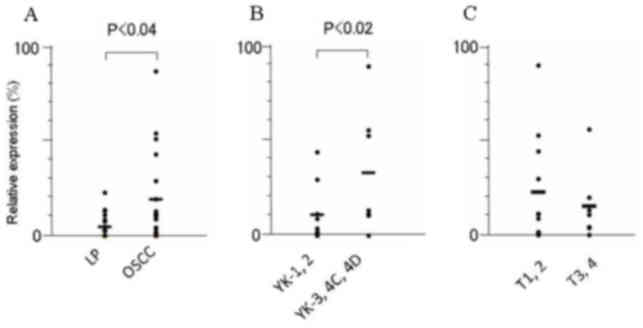
FYB and SLC16A13 mRNA was significantly higher in
OSCCs than in LPs tissues (P<0.03 and P<0.04, respectively)
(Figs. 2 and 3; Table
VII). Furthermore, among the OSCCs, the expression of
SLC16A13 mRNA was significantly elevated, in accordance with
the acquisition of invasion status; this expression was
significantly higher in OSCCs of higher YK grades (YK3-4D) than in
those of lower grades (YK1 and 2) (P<0.02). The level of
KRT7, SLC44A3 and TEMEM173 mRNA did not differ
significantly between human OSCC and LP tissues, nor between OSCCs
in different YK grades (Table VII).
The expression of none of these 5 mRNAs was regulated in
association with tumor size (T stage) according to the
tumor-node-metastasis classification of the Union for International
Cancer Control (14) among OSCC
tissues (Table VII).
 | Table V.Clinicopathological features of 18
leukoplakia patients. |
Table V.
Clinicopathological features of 18
leukoplakia patients.
| Characteristic | Patients, n |
|---|
| Total | 18 |
| Site |
|
|
Tongue | 8 |
|
Gingiva | 5 |
| Buccal
mucosa | 3 |
|
Other | 2 |
| Histology |
|
|
Hyperplasia | 3 |
| Mild
dysplasia | 3 |
|
Moderate dysplasia | 3 |
| Severe
dysplasia | 6 |
|
Unclassified | 3 |
 | Table VI.Clinicopathological features of 18
patients with oral squamous cell carcinoma. |
Table VI.
Clinicopathological features of 18
patients with oral squamous cell carcinoma.
| Characteristic | Patients, n |
|---|
| Total | 18 |
| Sex |
|
|
Male | 13 |
|
Female | 5 |
| Site |
|
|
Tongue | 9 |
| Gingiva
(upper) | 3 |
| Gingiva
(lower) | 3 |
| Buccal
mucosa | 2 |
| Floor
of mouth | 1 |
| T
classification |
|
| T1 | 6 |
| T2 | 5 |
| T3 | 3 |
| T4 | 4 |
| Metastasis |
|
|
Negative | 15 |
|
Positive | 3 |
| Mode of
invasion |
|
|
YK-1 | 2 |
|
YK-2 | 7 |
|
YK-3 | 4 |
|
YK-4C | 2 |
|
YK-4D | 2 |
|
Unidentified | 1 |
 | Table VII.Association between
clinicopathological factors and mRNA expression among oral squamous
cell carcinoma (n=18) and leukoplakia (n=18) tissues. |
Table VII.
Association between
clinicopathological factors and mRNA expression among oral squamous
cell carcinoma (n=18) and leukoplakia (n=18) tissues.
|
|
| P-value
(Mann-Whitney U test) |
|---|
|
|
|
|
|---|
| Factors | Patients, n | FYB |
SLC16A13 |
TEMEM173 | SLC44A3 | KRT-7 |
|---|
|
OSCC/leukoplakia |
|
<0.03a |
<0.04a | <1.00 | <0.12 | <0.28 |
|
OSCC | 18 |
|
|
|
|
|
|
Leukoplakia | 18 |
|
|
|
|
|
| T
classification |
| <1.00 | <0.74 | <0.23 | <0.23 | <0.14 |
|
T1-2 | 11 |
|
|
|
|
|
|
T3-4 | 7 |
|
|
|
|
|
| Mode of
invasion |
| <0.07 |
<0.02a | <0.25 | <1.00 | <0.17 |
|
YK1-2 | 9 |
|
|
|
|
|
|
YK3-4D | 8 |
|
|
|
|
|
Discussion
The present study established metastatic sub-clones
(L cells) from mouse oral squamous Sq-1979 cells. The aggressive
nature of the L cells was demonstrated by their higher in
vivo proliferation rates and transplantability, and the lower
survivability of tumor-burdened mice. Using these models,
preferentially expressed genes were screened for in L cells, and 5
mRNAs, Fyb, Slc16a13, Krt-7, Temem173
and Slc44a3, were isolated.
Of these mRNAs, FYB was significantly
elevated in aggressive human OSCC tissues compared with that in
LPs. FYB is an active component of FYN kinase (12). Since FYN kinase is known to modulate
the epithelial-mesenchymal transition (15) and stimulate proliferation of OSCC
cells (16), FYB can modulate the
progression from dysplasia to invasive OSCC via FYN kinase
activity. FYB is expressed in T cells, myeloid cells and platelets,
in which it regulates receptor-mediated integrin activation and
adhesion (17). FYB enhances
programmed cell death receptor-1 expression in cluster of
differentiation 8-positive T cells and reduces the cytotoxic T
lymphocyte cytotoxicity (18).
Although the manner in which FYB products from OSCC are able to
affect T lymphocytes is yet to be investigated, FYB may promote
tumor progression by reducing antitumor immunity in OSCC
patients.
SLC16A13, encoding a monocarboxylic acid
transporter, has been identified as a novel candidate gene for type
2 diabetes, with a possible role in triacylglycerol metabolism
(19). However, to the best of our
knowledge, no previous study has reported this gene as being
associated with tumor etiology. Notably, the present study
demonstrated that the expression of SLC16A13 mRNA was
significantly higher in OSCCs than it was in LPs. Furthermore, the
expression of SLC16A13 was significantly higher in highly
invasive OSCCs classified as being of higher YK grades (YK3-4D)
than in those of lower grades (YK1 and 2). The comparison between
OSCC groups using the YK classification almost completely matches
the comparison made between grades 3–4 and 1–2 for the mode of
invasion described by Jacobson et al (8). Hence, the expression of SLC16A13
mRNA in highly invasive OSCCs could also be validated by Jacobson's
classification. Since mode of invasion described by Yamamoto et
al (5) and Jacobson et al
(8) is largely associated with the
incidence of lymph node metastasis (5–7,8), the expression of SLC16A13 mRNA
could confer important predictive values for metastases and
recurrence risks for OSCC patients.
In the present study, the expression of KRT7
mRNA was not significantly higher in OSCCs than that in LPs; its
expression is known to be activated in several malignancies,
including gastric cancer (20),
cervical low-grade squamous intraepithelial lesions (21), lung cancer (22), urothelial carcinoma (23), esophageal carcinoma (24) and other squamous cell carcinomas
(25).
In conclusion, the present study identified marker
genes using mouse OSCC sub-clones originating from regional lymph
node metastasis. Using the same approach, it may be possible to
establish variable malignant phenotypes from other distal
metastases (e.g., of the lung) of primary mouse OSCC cells. Such an
approach could provide further information on the molecular basis
of progressive OSCCs.
Acknowledgements
The present study was supported in part by the
Grants-in-Aid for Scientific Research from the Japan Society for
the Promotion of Science (grant no. 26463055).
References
|
1
|
Gupta S, Kong W, Peng Y, Miao Q and
Mackillop WJ: Temporal trends in the incidence and survival of
cancers of the upper aerodigestive tract in Ontario and the United
States. Int J Cancer. 125:2159–2165. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Leemans CR, Braakhuis BJ and Brakenhoff
RH: The molecular biology of head and neck cancer. Nat Rev Cancer.
11:9–22. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ohkura S, Kondoh N, Hada A, Arai M,
Yamazaki Y, Sindoh M, Takahashi M, Matsumoto I and Yamamoto M:
Differential expression of the keratin-4, −13, −14, −17 and
transglutaminase 3 genes during the development of oral squamous
cell carcinoma from leukoplakia. Oral Oncol. 41:607–613. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kondoh N, Ohkura S, Arai M, Hada A,
Ishikawa T, Yamazaki Y, Shindoh M, Takahashi M, Kitagawa Y,
Matsubara O and Yamamoto M: Gene expression signatures that can
discriminate oral leukoplakia subtypes and squamous cell carcinoma.
Oral Oncol. 43:455–462. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yamamoto E, Kohama G, Sunakawa H, Iwai M
and Hiratsuka H: Mode of invasion, bleomycin sensitivity, and
clinical course in squamous cell carcinoma of the oral cavity.
Cancer. 51:2175–2180. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kaihara T, Kusaka T, Kawamata H, Oda Y,
Fujii S, Morita K, Imura J and Fujimori T: Decreased expression of
E-cadherin and Yamamoto-Kohama's mode of invasion highly correlates
with lymph node metastasis in esophageal squamous cell carcinoma.
Pathobiology. 69:172–178. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nakayama A, Ogawa A, Fukuta Y and Kudo K:
Relation between lymphatic vessel diameter and clinicopathologic
parameters in squamous cell carcinomas of the oral region. Cancer.
86:200–206. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jacobson PA, Enoroth CM, Killander D,
Moberger G and Mårtensson B: Histologic classification and grading
of malignancy in carcinomaof the larynx. Acta Radiol Ther Phys
Biol. 12:1–8. 1973. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kondoh N, Ishikawa T, Ohkura S, Arai M,
Hada A, Yamazaki Y, Kitagawa Y, Shindoh M, Takahashi M, Ando T, et
al: Gene expression signatures that classify the mode of invasion
of primary oral squamous cell carcinomas. Mol Carcinog. 47:744–756.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
RIan Freshney: Culture of animal cells: A
manual of basic technique and specialized applications. 6th.
Hoboken, N.J.: Wiley-Blackwell; pp. 208–211. 2010
|
|
11
|
Klopp AH, Zhang Y, Solley T,
Amaya-Manzanares F, Marini F, Andreeff M, Debeb B, Woodward W,
Schmandt R, Broaddus R, et al: Omental adipose tissue-derived
stromal cells promote vascularization and growth of endometrial
tumors. Clin Cancer Res. 18:771–782. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
da Silva AJ, Li Z, de Vera C, Canto E,
Findell P and Rudd CE: Cloning of a novel T-cell protein FYB that
binds FYN and SH2-domain-containing leukocyte protein 76 and
modulates interleukin 2 production. Proc Natl Acad Sci USA. 94:pp.
7493–7498. 1997; View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Workman P, Aboagye EO, Balkwill F, Balmain
A, Bruder G, Chaplin DJ, Double JA, Everitt J, Farningham DA,
Glennie MJ, et al: Guidelines for the welfare and use of animals in
cancer research. Br J Cancer. 102:1555–1577. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Brierley JD, Gospodarowicz MK and
Wittekind C: TNM classification of malignant tumours. 8th. New
York: Wiley-Blackwell; pp. 2722016
|
|
15
|
Lewin B, Siu A, Baker C, Dang D, Schnitt
R, Eisapooran P and Ramos DM: Expression of Fyn kinase modulates
EMT in oral cancer cells. Anticancer Res. 30:2591–2596.
2010.PubMed/NCBI
|
|
16
|
Li X, Yang Y, Hu Y, Dang D, Regezi J,
Schmidt BL, Atakilit A, Chen B, Ellis D and Ramos DM:
Alphavbeta6-Fyn signaling promotes oral cancer progression. J Biol
Chem. 278:41646–41653. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Engelmann S, Togni M, Thielitz A,
Reichardt P, Kliche S, Reinhold D, Schraven B and Reinhold A: T
cell-independent modulation of experimental autoimmune
encephalomyelitis in ADAP-deficient mice. J Immunol. 191:4950–4959.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li C, Li W, Xiao J, Jiao S, Teng F, Xue S,
Zhang C, Sheng C, Leng Q, Rudd CE, et al: ADAP and SKAP55
deficiency suppresses PD-1 expression in CD8+ cytotoxic T
lymphocytes for enhanced anti-tumor immunotherapy. EMBO Mol Med.
7:754–769. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
SIGMA Type 2 Diabetes Consortium, ;
Williams AL, Jacobs SB, Moreno-Macías H, Huerta-Chagoya A,
Churchhouse C, Márquez-Luna C, García-Ortíz H, Gómez-Vázquez MJ,
Burtt NP, et al: Sequence variants in SLC16A11 are a common risk
factor for type 2 diabetes in Mexico. Nature. 506:97–101.
2014.PubMed/NCBI
|
|
20
|
Huang B, Song JH, Cheng Y, Abraham JM,
Ibrahim S, Sun Z, Ke X and Meltzer SJ: Long non-coding antisense
RNA KRT7-AS is activated in gastric cancers and supports cancer
cell progression by increasing KRT7 expression. Oncogene.
35:4927–4936. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Paquette C, Mills AM and Stoler MH:
Predictive value of cytokeratin 7 immunohistochemistry in cervical
low-grade squamous intraepithelial lesion as a marker for risk of
progression to a high-grade lesion. Am J Surg Pathol. 40:236–243.
2016.PubMed/NCBI
|
|
22
|
Si LL, Lv L, Zhou WH and Hu WD:
Establishment and identification of human primary lung cancer cell
culture in vitro. Int J Clin Exp Pathol. 8:6540–6546.
2015.PubMed/NCBI
|
|
23
|
Chatterjee D, Das A and Radotra BD:
Invasive micropapillary carcinoma of urinary bladder: A
clinicopathological study. Indian J Pathol Microbiol. 58:2–6. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sano M, Aoyagi K, Takahashi H, Kawamura T,
Mabuchi T, Igaki H, Tachimori Y, Kato H, Ochiai A, Honda H, et al:
Forkhead box A1 transcriptional pathway in KRT7-expressing
esophageal squamous cell carcinomas with extensive lymph node
metastasis. Int J Oncol. 36:321–330. 2010.PubMed/NCBI
|
|
25
|
Regauer S, Beham A and Mannweiler S: CK7
expression in carcinomas of the Waldeyer's ring area. Hum Pathol.
31:1096–1101. 2000. View Article : Google Scholar : PubMed/NCBI
|