Introduction
Apoptosis, or programmed cell death, is an ordered
and orchestrated cellular process that occurs in physiological and
pathological conditions (1,2). Evasion of apoptosis serves a vital
function in carcinogenesis and numerous novel treatment strategies
that may restore the apoptotic signaling pathways towards
homeostasis may be feasible for eliminating cancer cells (3,4). Reactive
oxygen species (ROS), as signals and modulators for cellular redox
changes, are key mediators in the regulation of cell apoptosis
(5,6).
Modulation of the redox state may potentially trigger apoptotic
signaling pathways by cellular FLICE-like inhibitory protein
degradation, apoptosis signal-regulating kinase 1 activation,
cytochrome c release and interaction of caspase 9/Apoptotic
protease-activating factor 1 (7).
Previous studies (8–10) concerning the extrinsic and intrinsic
pathways of apoptosis have focused on mitochondria, which are the
major source of ROS generation and serve a central function in
apoptosis regulation (11). It has
been suggested that instead of elevating overall cellular ROS,
regulation of mitochondrial ROS during apoptosis may be a promising
therapeutic strategy in the treatment of various types of cancer
(7).
As the most abundant cellular redox buffer,
glutathione (GSH) is distributed in at least 9 different
subcellular compartments, and protects cells against ROS through
the oxidation of GSH to glutathione disulfide (GSSG). The reduced
to oxidized glutathione ratio [GSH/GSSG] may be measured to reflect
cellular redox changes (12–14). Alternatively, various redox-sensitive
fluorescent dyes, such as dihydrodichlorofluorescein
(H2DCF) have been frequently used to monitor ROS levels
in a number of studies focusing on natural drug-induced apoptosis
in cancer (15,16). However, conventional approaches lack
either spatiotemporal resolution or specificity resolution
(17). This is a critical knowledge
gap that has adversely limited the ability to comprehensively probe
into the underlying regulatory mechanisms of mitochondrial ROS in
natural drug-induced apoptosis in cancer, and has also impeded the
development of novel redox-based therapeutic strategies targeting
cancer. Previously, genetically-encoded redox-sensitive green
fluorescent protein (roGFP) probes have been developed with a
suitable range of redox potentials to measure the spatial and
temporal intracellular oxidation status by ratiometric analysis,
which is compartmental, real-time, reversible and measures dynamic
variation trends compared with other methods for assessing
intracellular redox status (13,18–20).
Previously, human glutaredoxin1 (Grx1) was fused to the roGFP2
biosensor to facilitate specific real-time equilibration between
the sensor protein and the glutathione redox couple, consequently
allowing dynamic live imaging of the intracellular glutathione
redox potential in different cellular compartments, with
unprecedented sensitivity and temporal resolution (14,21).
Therefore, the ratiometric Grx1-roGFP2 biosensor targeted to
mitochondria may be extremely useful in quantifying mitochondrial
redox changes associated with natural drug-induced apoptosis in
cancer.
Oridonin, a natural and safe ent-kaurene diterpenoid
compound isolated from the plant Rabdosia rubescens, has
been demonstrated to induce ROS to mediate apoptosis in multiple
types of cancer, including human laryngeal cancer cells (22), human cervical carcinoma HeLa cells
(23), hepatocellular carcinoma cells
(24,25), human epidermoid carcinoma A431 cells
(26,27), human histiocytic lymphoma U937 cells
(28), esophageal cancer KYSE-150
cells (29) and rheumatoid arthritis
fibroblast-like synoviocytes (30).
Furthermore, the combined use of AG1478 and oridonin augments the
production of ROS and apoptosis (22,26);
Therefore, targeting epidermal growth factor receptor combined with
oridonin may be a potentially effective anti-neoplastic therapy.
Notably, neither study (31,32) demonstrated that oridonin-induced
apoptosis may be conferred by compartmental redox changes in live
cells. In the present study, using live-cell imaging, it was
demonstrated that the mitochondrial-targeted
glutaredoxin1-redox-sensitive green fluorescent protein 2
(mtGrx1-roGFP2) probe expressed in HepG2 cells may be used as a
real-time reporter of dynamic redox changes caused by oridonin. The
possible regulatory mechanisms of mitochondrial redox signaling
contributing to oridonin-induced apoptosis in HepG2 cells were also
assessed.
Materials and methods
Reagents and antibodies
Unless stated otherwise, reagents were purchased
from Thermo Fisher Scientific, Inc. (Waltham, MA, USA). Oridonin
(C20H28O6, MW=364.43; Chemical
Abstracts Service registry number, 28957-04-2; lot, H1616026) was
purchased from Shanghai Aladdin Bio-Chem Technology Co., Ltd.
(Shanghai, China). The oridonin was dissolved in ≥99.7% dimethyl
sulphoxide (DMSO; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
with 150 mM as original stock solution. Subsequent to mixing by
vortex (1,500 rpm for 10 sec), the solution was then filtered
through a 0.22-µm filter, which was then stored at 4°C prior to
use. The solution was diluted in serum-free Dulbecco's modified
Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) medium
prior to use. The maximum final concentration of DMSO in medium was
maintained below 0.1% (v/v) and did not exert any detectable effect
on cell growth or cell death (23,33). SS31,
a small aromatic-cationic mitochondria-targeted peptide
(H-D-Arg-Dmt-Lys-Phe-NH2), may be selectively
concentrated in the inner mitochondrial membrane and reduce
mitochondrial ROS generation (34–36). It
was provided by Stealth BioTherapeutics, Inc., (Newton, MA,
USA).
Primary antibodies against β-actin (catalog no.
8457P) and poly adenosine 5′-diphosphate ribose polymerase (PARP;
catalog no. 9532T) were purchased from Cell Signaling Technology,
Inc. (Danvers, MA, USA). A goat anti-rabbit immunoglobulin G
horseradish peroxidase (HRP)-conjugated secondary antibody (catalog
no. HS101-01) used for western blotting were purchased from Beijing
Transgen Biotech Co., Ltd. (Beijing, China).
Gene constructs
The mtGrx1-roGFP2-containing vector (200 ng/µl)
(21) was obtained from Professor
Dick (German Cancer Research Center, Heidelberg, Germany). The
mtGrx1-roGFP2 open reading frame was polymerase chain reaction
Deutsches Krebsforschungszentrum, Im Neuenheimer Feld-amplified
with the following primers: mt-forward,
5′-CGAGCTCAAGCTTCGAATTCGCCACCATGGCCTCCACTCGTGTCCTC-3′; and
mt-reverse, 5′-ACCCGGTAGAATTATCTAGATTACTTGTACAGCTCGTCCATGC-3′ by
polymerase chain reaction (PCR), which was performed in a 20 µl
total reaction volume using the vector as DNA template and
LongAmp® TaqDNA polymerase (New England BioLabs,
Inc., Ipswich, MA, USA). The PCR reaction was run as follows: 94°C
for 3 min as an initial denaturation step; amplification for 35
cycles with denaturation at 94°C for 30 sec, annealing at 58°C for
45 sec, and extension at 65°C for 90 sec; and final extension at
65°C for 10 min. The mtGrx1-roGFP2 open reading frame was cloned
into the pLenti-CMV-PGK-puro vector (Invitrogen; Thermo Fisher
Scientific, Inc.) using EcoR I and Xba I (20,000
U/ml; New England BioLabs, Inc.) to generate
pLenti-CMV-mtGrx1-roGFP2-PGK-puro vectors at 16°C for 12 h.
Restriction digestion was conducted with EcoR I and
Xba I (20,000 units/ml; New England BioLabs, Inc.) at 37°C
for 3 h, and then DNA sequencing was performed by Openlab Co., Ltd.
(Beijing, China) to verify the construction of a recombinant
vector. The vector, along with the helper plasmids [pLP/VSVg, pLP1
and pLP2, (Invitrogen; Thermo Fisher Scientific, Inc.)], were used
to prepare virus stocks for transduction experiments.
Stable cell line generation
The human hepatoblastoma HepG2 cell line (37,38),
obtained from the American Type Culture Collection (Manassas, VA,
USA), was cultured in DMEM containing 10% (v/v) fetal bovine serum
(Gibco; Thermo Fisher Scientific, Inc.) and antibiotics (100 U/ml
penicillin and 100 µg/ml streptomycin) at 37°C in an atmosphere
containing 5% CO2. HepG2 cells stably expressing the
mtGrx1-roGFP2 biosensor were generated by lentiviral transduction
(Thermo Fisher Scientific, Inc.) of the plasmids into interim 293T
cells (American Type Culture Collection) for 12–24 h at 37°C and
subsequent selection with 2 µg/ml puromycin (39). Colonies were screened for
mtGrx1-roGFP2 expression by western blotting and Olympus IX71
reverse fluorescence microscopy (magnification, ×100; Olympus
Corporation, Tokyo, Japan) at an excitation wavelength of 488 nm
with an emission filter of 507 nm. For 12 h prior to treatment, the
culture medium was altered to DMEM medium with 2% FBS to decrease
the serum effect.
Cell viability assay
The viability of cells was measured with
TransDetect™ Cell Counting Kit (CCK, catalog no. FC101-01;
Beijing Transgen Biotech Co., Ltd., Beijing, China) according to
the manufacturer's protocol (Beijing Transgen Biotech Co., Ltd.)
with minor modifications. Briefly, 100 µl HepG2-mtGrx1-roGFP2 cell
suspension was dispensed into 96-well plate (5×103
cells/well) and the plate was pre-incubated at 37°C for 24 h,
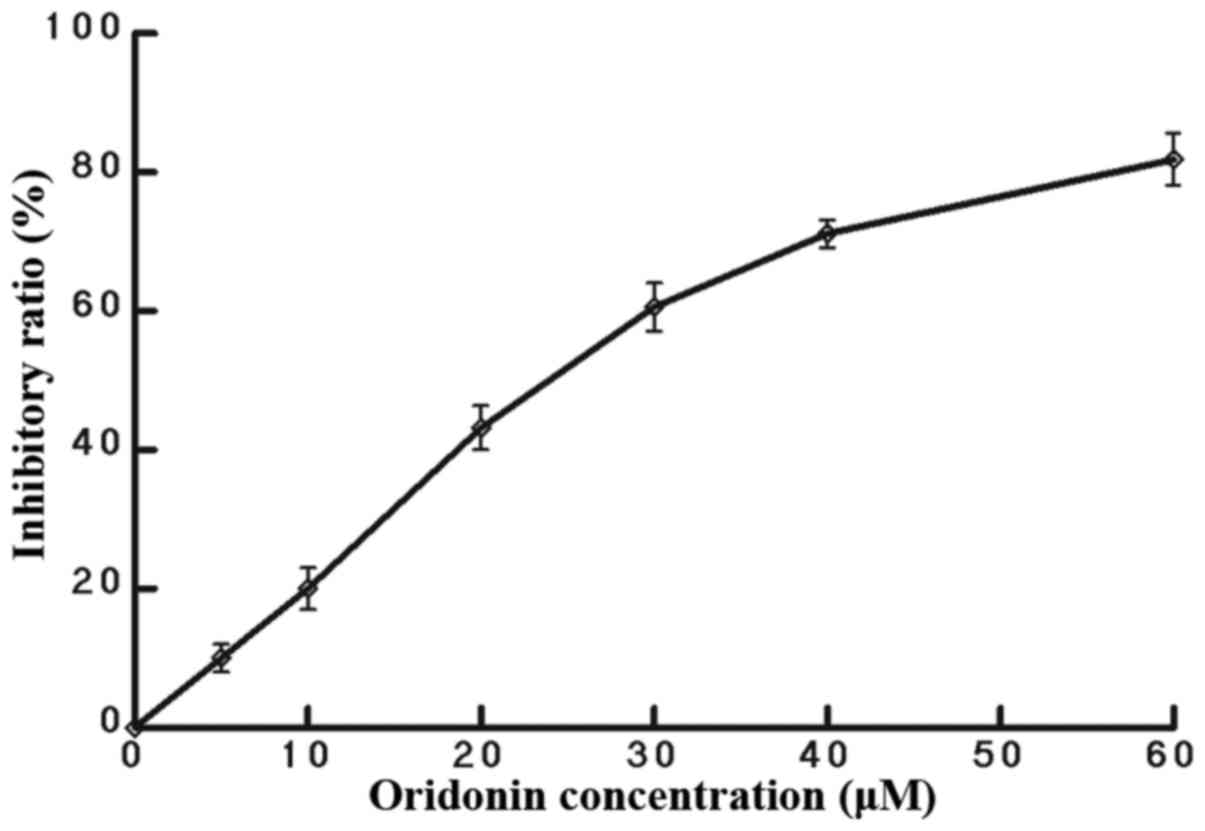
followed by treatment with various concentrations of oridonin (0,
5, 10, 20, 30 or 60 µM) for 8 h. Next, 10 µl (100 µl/ml) of CCK
working solution was added to each well of the plate and then
incubated at 37°C for 2 h. The absorbance of each well at 450 nm
was measured under a multimode plate reader (PerkinElmer, Inc.,
Waltham, MA, USA). The results representing the average of 5
parallel samples were expressed as the relative percentage of cell
growth inhibition.
Fluorescence microscopy
MtGrx1-roGFP2-expressing HepG2 cells placed in a
Nest-35 confocal dish, where the temperature was maintained at 37°C
by a stage top incubator (Tokai Hit Elmer, Inc., Waltham, MA, USA),
were treated with H2O2 (1 mM) or
dithiotheritol (DTT; 2 mM) and/or oridonin (25 µM) or SS31 (100 nM)
and imaged using a Nikon A1R confocal laser scanning system (Nikon
Corporation, Tokyo, Japan) equipped with a 0.95 numerical aperture
(water immersion) objective lens at magnification ×40. For
excitation of 405-nm (channel 1) and 488-nm (channel 2),
fluorescence was measured with a bandpass filter of 500–554 nm.
Images were captured and exported to ImageJ software (version
1.50i; National Institutes of Health, Bethesda, MD, USA).
Subsequent to setting thresholds to avoid low signal ratio
artifacts, false-color ratio images were obtained by dividing the
405-nm image by the 488-nm image on a pixel per pixel basis, as
previously described (21).
To confirm the mitochondrial localization of the
mtGrx1-roGFP2 probe in stable transfectants, the HepG2 cells
expressing mtGrx1-roGFP2 were incubated with a pre-warmed (37°C)
rosamine-based MitoTracker probe (100 nM; Thermo Fisher Scientific,
Inc.) for 20 min at 37°C in an atmosphere of 5% CO2
according to the manufacturer's protocol (Thermo Fisher Scientific,
Inc.). Cells were visualized using Olympus FV1000 confocal laser
scanning microscopy equipped with a magnification, ×40 objective
(Olympus Corporation) at 579 nm excitation and 599 nm emission
wavelengths.
Analysis of apoptotic cells by flow
cytometry
The percentage of apoptotic of cells exposed to
oridonin (25 µM) with or without SS31 (100 nM) for 8 h were
determined using a commercially available Annexin V-phycoerythrin
(PE) apoptosis detection kit according to the manufacturer's
protocol (BD Biosciences, Franklin Lakes, NJ, USA) with minor
modifications. Briefly, cells were collected and washed twice in
ice cold PBS and then resuspended in 100 µl 1X binding buffer at a
density of 1×105 cells/ml, and then incubated with 5 µl
Annexin V-PE and 5 µl 7-aminoactinomycin (7-AAD) in the dark for 15
min at room temperature. Finally, 400 µl of 1X binding buffer was
added to each tube. Samples were analyzed using a BD FACSCalibur
(BD Biosciences) and BD CellQuest Pro software (version 5.1; BD
Biosciences) and evaluated based on the percentage of apoptotic
cells that were Annexin V-PE-positive and 7-AAD-positive.
Observation of morphological
changes
Apoptotic nuclear morphology was assessed using
Hoechst 33342 (Beijing Solarbio Science & Technology Co., Ltd.,
Beijing, China) in PBS buffer, as described previously (33). HepG2-mtGrx1-roGFP2 cells were grown in
culture at 37°C for 12 h. Following 12 h of incubation at 37°C, the
cells were perfused with oridonin (25 µM) in the presence or
absence of SS31 (100 nM) at 37°C for 8 h. For morphological
observation, cells were incubated with Hoechst 33342 (1 mg/ml) for
25 min at 37°C, washed and then examined using an Olympus FV1000
confocal laser scanning microscopy (Olympus Corporation) equipped
with a ×40 objective at 350 nm excitation and 461 nm emission
wavelengths.
Western blot analysis
MtGrx1-roGFP2-expressing HepG2 cells were treated
with oridonin (25 µM) with or without SS31 (100 nM) at 37°C. After
8 h, cells were washed twice with ice-cold PBS and solubilized in
lysis buffer containing 20 mM Tris (pH 7.5), 135 mM NaCl, 2 mM
EDTA, 2 mM DTT, 25 mM β-glycerophosphate, 2 mM sodium
pyrophosphate, 10% glycerol, 1% Triton X-100, 1 mM sodium
orthovanadate (Sigma-Aldrich; Merck KGaA), 10 mM Na fluoride (NaF;
Sigma-Aldrich; Merck KGaA), 10 µg/ml aprotinin, 10 µg/ml leupeptin
and 1 mM phenylmethane sulfonyl (PMSF; Sigma-Aldrich; Merck KGaA,)
for 30 min on ice. Lysates were centrifuged (15,000 × g) at 4°C for
15 min. Soluble protein were denatured in SDS in a boiling water
bath for 5 min, and quantified using Easy II Protein Quantitative
Kit (BCA; catalog no. DQ111-01; Beijing Transgen Biotech Co., Ltd).
A total of 10 µl protein was electrophoresed on an 8–12% gel using
SDS-PAGE, and transferred to a nitrocellulose membrane. The
membrane was blocked with 5% skim milk for 1 h at room temperature,
then incubated overnight with the indicated primary antibodies
against PARP (1:1,000 dilution; catalog no. 9532T) at 4°C, prior to
incubation with goat anti-rabbit IgG HRP-conjugated secondary
antibody (1:10,000 dilution; catalog no. HS101-01) for 2 h at room
temperature. Detection was performed using an EasySee Western Blot
Kit (catalog no. DW101-01; Beijing Transgen Biotech Co., Ltd.).
Statistical analysis
All experimental data obtained from cultured cells
are expressed as the mean ± standard deviation. Statistical
analysis was performed using SPSS (version 19.0; IBM Corp., Armonk,
NY, USA). The data were analyzed using one-way analysis of
variance, followed by Bonferroni's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Mitochondrial-targeted Grx1-roGFP2
biosensor functions in HepG2 cells
Previously, it was demonstrated that ROS induced by
oridonin is capable of restoring apoptotic signaling pathways in
HepG2 cells (32); however, whether
mitochondrial ROS contribute to oridonin-induced apoptosis remains
unknown. To monitor mitochondrial ROS in HepG2 cells during
oridonin-induced apoptosis, stably transfected HepG2 cells were
created with a Grx1-roGFP2 biosensor targeted to mitochondria. As
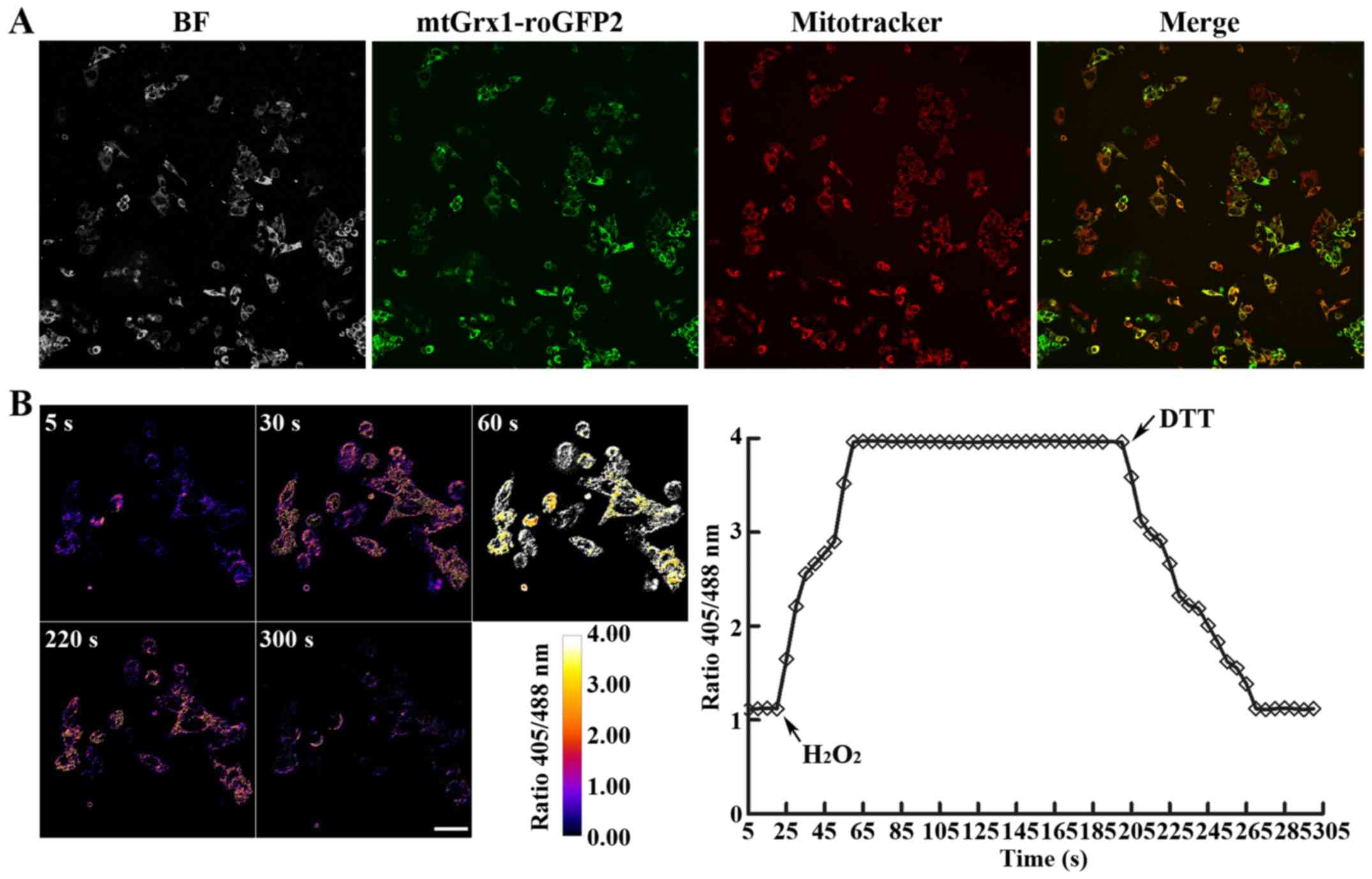
demonstrated in Fig. 1A, the
expressed Grx1-roGFP2 co-localized with the mitochondrial-selective
probe following confocal laser scanning microscopy, indicating that
Grx1-roGFP2 was correctly targeted to mitochondria in the
transgenic cells.
To determine whether Grx1-roGFP2 was responsive to
mitochondrial redox changes and was able to be ratiometrically
measured, HepG2 cells were treated with 1 mM
H2O2, followed 180 sec later by exposure to 2
mM DTT. As indicated in Fig. 1B, the
addition of H2O2 to the growth medium led to
an immediate and marked response, with the 405/488 nm fluorescence
ratios of Grx1-roGFP2 in the mitochondria of HepG2 cells ranging
from 1.21 to 3.86, which was maintained for 135 sec until injection
of DTT induced its reduction to 1.23. Taken together, these results
revealed that Grx1-roGFP2 responded, as reflected in the altered
405/488 nm excitation fluorescence ratio, to mitochondrial redox
changes triggered by H2O2 and DTT as
authoritative oxidants and reductants, therefore providing a
real-time detection system for dynamic and reversible changes to
mitochondrial redox status induced by anticancer drugs.
Oridonin induces mitochondrial redox
changes in HepG2 cells in a time-dependent manner
The primary aim of the present study was to
establish a roGFP-based redox-sensing system to monitor redox
changes in mitochondria, and to delineate the molecular mechanisms
by which mitochondrial ROS control oridonin-induced apoptosis in
HepG2 cells. To address this, Grx1-roGFP2 molecules targeting
mitochondria, as aforementioned, were used to measure the
perturbation of ROS levels reflected by the 405/488 nm excitation
fluorescence ratio, as distinguished from the fluorescent intensity
of ROS-specific probe H2DCF, which has traditionally
been used as the most applicable tool. However, H2DCF
exhibits additional disadvantages, such as being destructive,
integrated, irreversible, point-in-time and providing only a static
redox status (17,21,40).
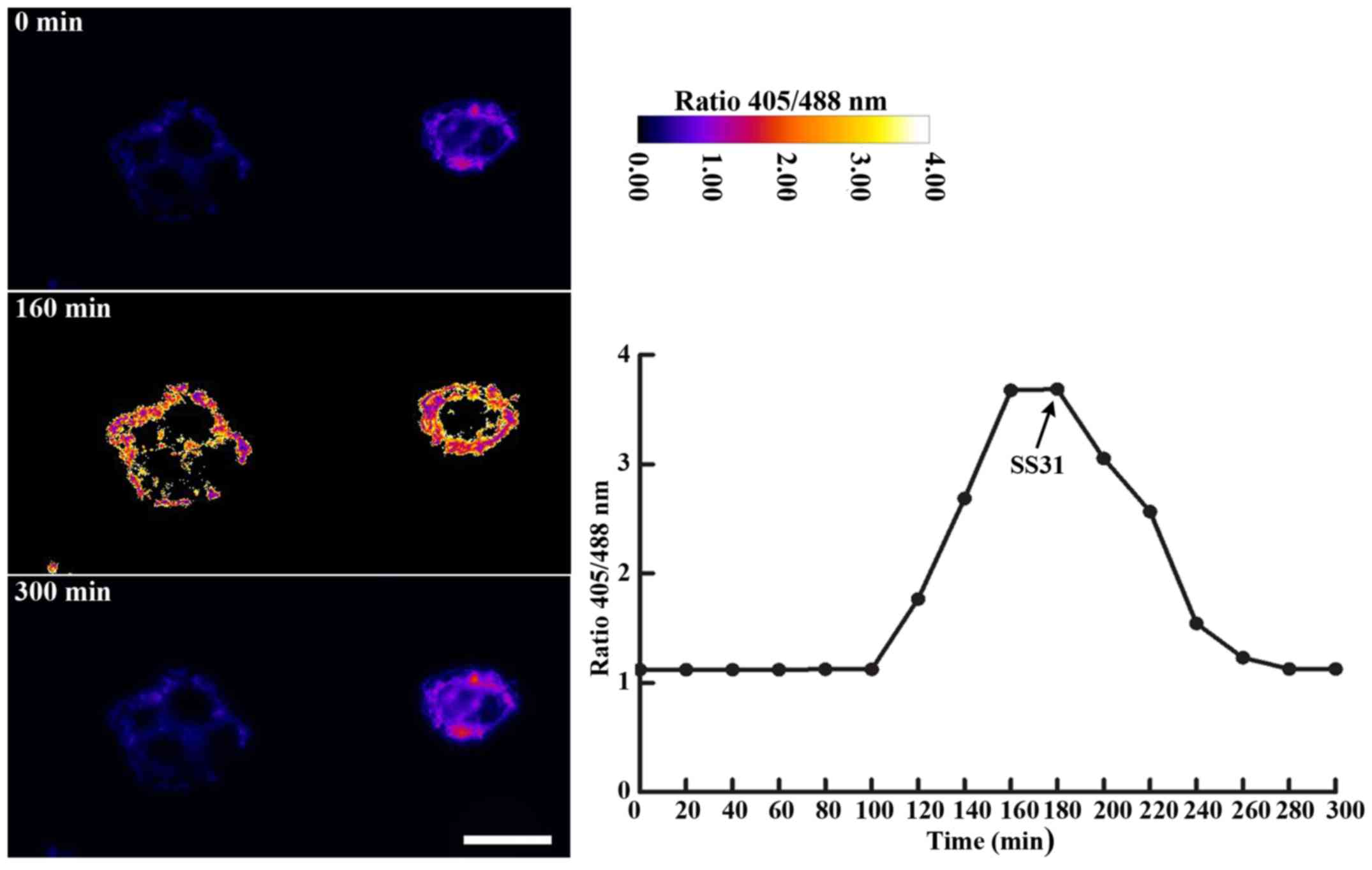
Representative fluorescence microscope images of mitochondrial
Grx1-roGFP2 with corresponding ratiometric analysis are presented
in Fig. 2. In the time course
experiments, stimulation of HepG2 cells with oridonin at a
concentration of 25 µM, responsible for a ~50% inhibitory ratio at
8 h (Fig. 3), resulted in a 3-fold
increase in progressive oxidation of the sensor, which plateaued
after 160 min. As anticipated, SS31 prevented the oridonin-induced
oxidation of mtGrx1-roGFP2, with a decline from 3.65 to 1.22 in the
405/488 nm excitation fluorescence ratio. In general, using
mitochondria-localized Grx1-roGFP2, the present study has provided
evidence that oridonin causes a disturbance of mitochondrial redox
poise in a manner that is reversible and may be captured in real
time.
Mitochondrial ROS mediate
oridonin-induced apoptosis in HepG2 cells
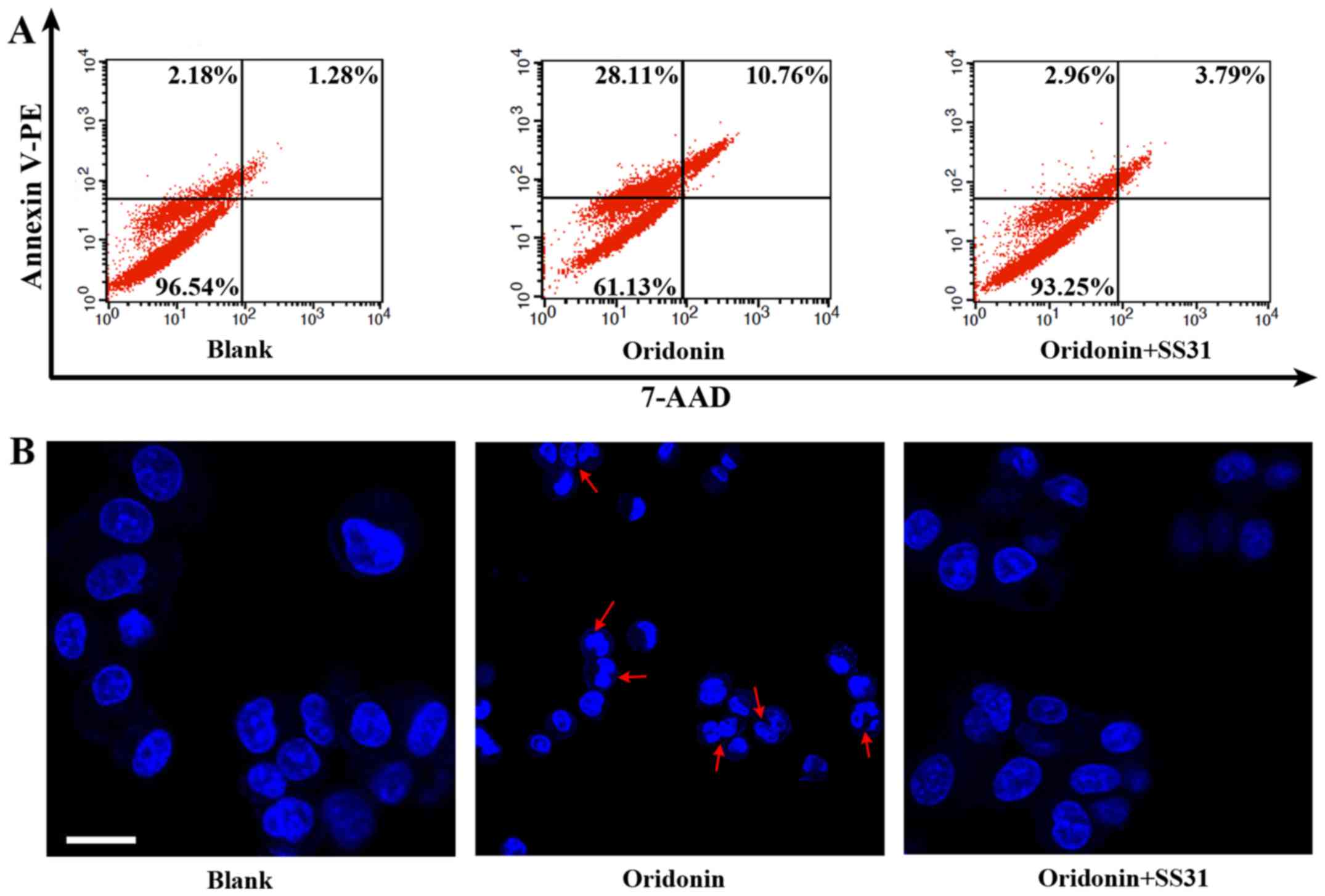
To additionally explore whether mitochondrial ROS
act as mediators in oridonin-induced apoptosis, HepG2 cells were
co-incubated with SS31 (100 nM) and oridonin (25 µM) for 8 h at
37°C. Compared with untreated cells, a marked increase by almost
34.41% in cell apoptosis, as indicated by flow cytometry using an
Annexin V-PE Apoptosis Detection kit (Fig. 4A), was observed in response to
oridonin treatment. Furthermore, inhibition of apoptosis was
induced by SS31, accompanied by a decrease in cell growth
inhibition ratio from 38.87 to 5.75%. Consistent with the apoptotic
percentage, apoptotic morphology was detected by Hoechst 33342
staining. As demonstrated in Fig. 4B,
nuclear condensation and fragmentation were identified in the
HepG2-mtGrx1-roGFP2 cells treated with oridonin, whereas SS31
treatment resulted in the integrity of cell morphology, similar to
untreated HepG2 cells. Collectively, these data implicate ROS in
the mitochondria as important contributors to oridonin-induced cell
apoptosis.
Mitochondrial ROS contribute to
oridonin-induced PARP activation in HepG2 cells
Hypothetically, oxidant stress-induced redox changes
may trigger the activation of PARP, which serves an indispensable
role in commonly described initiation pathways and redundant cell
death pathways (41–43). To elucidate the molecular mechanism
responsible for oridonin-induced apoptosis mediated by
mitochondrial redox changes in HepG2 cells, the expression levels
of PARP were determined by western blot analysis. As demonstrated
in Fig. 5, cleaved PARP protein
expression was markedly increased by oridonin at a concentration of
25 µM. However, the activation of PARP cleavage was inhibited by
SS31 administration. The results additionally confirm that redox
signaling in mitochondria serves as a modulator of oridonin-induced
apoptosis through the activation of PARP.
Discussion
Previous studies have indicated that oridonin
possesses anti-proliferative and apoptotic activities against a
variety of cancer cells (44,45). However, the underlying regulatory
mechanisms of mitochondrial ROS in oridonin-induced HepG2 apoptosis
remain largely unknown due to limitations of subcellular imaging
resolution. Previously, it has been argued that mitochondria are
more sensitive to redox perturbation compared with other
subcellular compartments, and studies have suggested that
mitochondria serve a potential role in sensing and signaling
cellular redox challenge in vital biological processes such as cell
death and abiotic stress response, based on the
mitochondrial-targeted redox-sensitive GFP (41,46). In
the present study, the mitochondrial-targeted Grx1-roGFP2
biosensor, which has been demonstrated as a valuable tool in
sensing compartmentalized redox status and consequently ROS
homeostasis/signaling in human cancer cell lines (47), was applied to monitor temporal changes
in the redox status of mitochondria in HepG2 cells in response to
either H2O2/DTT or oridonin/SS31 (Figs. 1B and 2), indicating that a redox-sensing system in
HepG2 cells was successfully established. Based on the regulation
of mitochondrial redox changes in oridonin-induced apoptosis
(Fig. 4), additional experiments were
conducted to elucidate the potential mechanisms of action of
mitochondrial redox changes in HepG2 cells, as demonstrated in
Fig. 5, which were partially
dependent on PARP cleavage. Taken together, the present study
provides evidence in support of mitochondrial ROS as a potential
mediator in the apoptotic activities of oridonin in HepG2 cells,
which provides insight on the molecular mechanisms by which
mitochondrial redox signaling regulates apoptosis in response to
oridonin in cancer therapy. Additional investigations are required
for the development of mitochondrial-specific oridonin as a novel
promising anticancer therapeutic strategy.
Consistent with previous studies (14,18,20,47),
the mtGrx1-roGFP2 redox biosensor exhibited excellent specificity
and sensitivity, as exemplified by the response to oridonin in
HepG2 cells. Future studies should focus on certain key protein in
the signaling pathways associated with apoptosis, including tumor
protein (p)53 (35,48) using mtGrx1-roGFP2 to determine whether
p53 participates in oridonin-induced apoptosis through the
generation of mitochondrial ROS. In principle, such redox-sensing
systems may be applied to dissect the regulatory mechanisms of
redox signaling, or for drug screening and toxicology (18,21). Redox
signaling mechanisms associated with HIV and HIV-Mycobacterium
tuberculosis co-infection were examined by performing oxidative
stress and antioxidant defense pathway-focused human gene
expression arrays (RT2 Profiler™ PCR arrays), using Grx1-roGFP2
(18). Compared with TNF-related
apoptosis-inducing ligand (TRAIL) as an acknowledged apoptosis
inducer (21), oridonin altered the
redox state of the mitochondria to the extent that the
mitochondrial sensor demonstrated highly rapid and marked changes
in oxidation (Fig. 2), suggesting
that the activity of oridonin as a redox-active molecule may be
superior to that of TRAIL, and potentially an excellent therapeutic
candidate for targeting cancer. However, it should be noted that
the activity of the mitochondrial sensor mtGrx1-roGFP2 has not been
compared or normalized between HeLa and HepG2 cell lines.
Additional studies are required to explore the issue as to which
subcellular compartment is more susceptible to redox changes
induced by oridonin.
In HepG2 cells, multiple mechanisms, including the
inhibition of telomerase, induction of endoplasmic reticulum stress
and regulation of p53, MAPK and mitochondrial signaling pathways,
have been implicated in the apoptotic activity of oridonin
(25,49,50).
Whether other mechanisms, such as PARP activation, are associated
with the anti-HepG2 activity of oridonin remains unknown. To the
best of our knowledge, for the first time, the present study
investigated the involvement of PARP activated by mitochondrial ROS
in contributing to oridonin-induced HepG2 apoptosis (Fig. 5), as previously demonstrated in
menadione-triggered cell death pathways (41). This provides further insight into the
molecular mechanisms of oridonin function in HepG2 cells.
Acknowledgements
The authors would like to thank Professor Dick
German Cancer Research Center, Heidelberg, Germany for kindly
providing the mtGrx1-roGFP2 plasmid. The present study was
supported by the Natural Science Fundamental Research Program of
Henan Provincial Education Department (grant no. 16A180046) and
Zhengzhou Science and Technology Development Plan (grant no.
20150350).
References
|
1
|
Mohan H: Textbook of pathology. 5th.
Jaypee Brothers Medical Publishers; New Delhi: pp. 21–60. 2010,
View Article : Google Scholar
|
|
2
|
Merkle CJ: Cellular adaptation, injury and
deathPathophysiology: Concepts of altered health states. Porth CM
and Matfin G: 8th. Wolters Kluwer/Lippincott Williams and Wilkins;
Philadelphia, PA: pp. 94–111. 2009
|
|
3
|
Ghobrial IM, Witzig TE and Adjei AA:
Targeting apoptosis pathways in cancer therapy. CA Cancer J Clin.
55:178–194. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wong RS: Apoptosis in cancer: From
pathogenesis to treatment. J Exp Clin Cancer Res. 30:872011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Parri M and Chiarugi P: Redox molecular
machines involved in tumor progression. Antioxid Redox Signal.
19:1828–1845. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tochhawng L, Deng S, Pervaiz S and Yap CT:
Redox regulation of cancer cell migration and invasion.
Mitochondrion. 13:246–253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang L, Wang K, Lei Y, Li Q, Nice EC and
Huang C: Redox signaling: Potential arbitrator of autophagy and
apoptosis in therapeutic response. Free Radic Biol Med. 89:452–465.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liao W, Yu Z, Lin Z, Lei Z, Ning Z,
Regenstein JM, Yang J and Ren J: Biofunctionalization of Selenium
nanoparticle with dictyophora indusiata polysaccharide and its
antiproliferative activity through death-receptor and
mitochondria-mediated apoptotic pathways. Sci Rep. 5:186292015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xue M, Ge Y, Yu C, Zheng Z, He X and Zhao
J: Apoptosis is induced by docosahexaenoic acid in breast cancer
cells via death receptor and mitochondria-mediated pathways. Mol
Med Rep. 16:978–982. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang YQ, Xiao CX, Lin BY, Shi Y, Liu YP,
Liu JJ, Guleng B and Ren JL: Silencing of pokemon enhances
caspase-dependent apoptosis via fas- and mitochondria-mediated
pathways in hepatocellular carcinoma cells. PLoS One. 8:e689812013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kroemer G: Mitochondrial control of
apoptosis: An overview. Biochem Soc Symp. 66:1–15. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mari M, Morales A, Colell A, Garcia-Ruiz C
and Fernandez-Checa JC: Mitochondrial glutathione, a key survival
antioxidant. Antioxid Redox Signal. 11:2685–2700. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Morgan B, Sobotta MC and Dick TP:
Measuring E(GSH) and H2O2 with roGFP2-based
redox probes. Free Radic Biol Med. 51:1943–1951. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kolossov VL, Hanafin WP, Beaudoin JN, Bica
DE, DiLiberto SJ, Kenis PJ and Gaskins HR: Inhibition of
glutathione synthesis distinctly alters mitochondrial and cytosolic
redox poise. Exp Biol Med (Maywood). 239:394–403. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chan CK, Supriady H, Goh BH and Kadir HA:
Elephantopus scaber induces apoptosis through ROS-dependent
mitochondrial signaling pathway in HCT116 human colorectal
carcinoma cells. J Ethnopharmacol. 168:291–304. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shen S, Zhang Y, Zhang R and Gong X:
Sarsasapogenin induces apoptosis via the reactive oxygen
species-mediated mitochondrial pathway and ER stress pathway in
HeLa cells. Biochem Biophys Res Commun. 15:519–524. 2013.
View Article : Google Scholar
|
|
17
|
Ezerina D, Morgan B and Dick TP: Imaging
dynamic redox processes with genetically encoded probes. J Mol Cell
Cardiol. 73:43–49. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bhaskar A, Munshi M, Khan SZ, Fatima S,
Arya R, Jameel S and Singh A: Measuring glutathione redox potential
of HIV-1-infected macrophages. J Biol Chem. 290:1020–1038. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
van Lith M, Tiwari S, Pediani J, Milligan
G and Bulleid NJ: Real-time monitoring of redox changes in the
mammalian endoplasmic reticulum. J Cell Sci. 124:2349–2356. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hanson GT, Aggeler R, Oglesbee D, Cannon
M, Capaldi RA, Tsien RY and Remington SJ: Investigating
mitochondrial redox potential with redox-sensitive green
fluorescent protein indicators. J Biol Chem. 279:13044–13053. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gutscher M, Pauleau AL, Marty L, Brach T,
Wabnitz GH, Samstag Y, Meyer AJ and Dick TP: Real-time imaging of
the intracellular glutathione redox potential. Nat Methods.
5:553–559. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kang N, Zhang JH, Qiu F, Tashiro S,
Onodera S and Ikejima T: Inhibition of EGFR signaling augments
oridonin-induced apoptosis in human laryngeal cancer cells via
enhancing oxidative stress coincident with activation of both the
intrinsic and extrinsic apoptotic pathways. Cancer Lett.
294:147–158. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang YH, Wu YL, Tashiro S, Onodera S and
Ikejima T: Reactive oxygen species contribute to oridonin-induced
apoptosis and autophagy in human cervical carcinoma HeLa cells.
Acta Pharmacol Sin. 32:1266–1275. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen G, Wang K, Yang BY, Tang B, Chen JX
and Hua ZC: Synergistic antitumor activity of oridonin and arsenic
trioxide on hepatocellular carcinoma cells. Int J Oncol.
40:139–147. 2012.PubMed/NCBI
|
|
25
|
Wang H, Ye Y and Yu ZL: Proteomic and
functional analyses demonstrate the involvement of oxidative stress
in the anticancer activities of oridonin in HepG2 cells. Oncol Rep.
31:2165–2172. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu Y, Fan SM, Ye YC, Tashiro S, Onodera S
and Ikejima T: The tyrphostin AG1478 augments oridonin-induced A431
cell apoptosis by blockage of JNK MAPK and enhancement of oxidative
stress. Free Radic Res. 46:1393–1405. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu Y, Fan SM, Song JK, Tashiro S, Onodera
S and Ikejima T: Hydroxyl radical (OH) played a pivotal role in
oridonin-induced apoptosis and autophagy in human epidermoid
carcinoma A431 cells. Biol Pharm Bull. 35:2148–2159. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zang L, He H, Xu Q, Yu Y, Zheng N, Liu W,
Hayashi T, Tashiro S, Onodera S and Ikejima T: Reactive oxygen
species H2O2 and •OH, but not
O2•(−) promote oridonin-induced phagocytosis of
apoptotic cells by human histocytic lymphoma U937 cells. Int
Immunopharmacol. 15:414–423. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pi J, Cai H, Jin H, Yang F, Jiang J, Wu A,
Zhu H, Liu J, Su X, Yang P and Cai J: Qualitative and quantitative
analysis of ROS-mediated oridonin-induced oesophageal cancer
KYSE-150 cell apoptosis by atomic force microscopy. PLoS One.
10:e01409352015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shang CH, Zhang QQ and Zhou JH: Oridonin
inhibits cell proliferation and induces apoptosis in rheumatoid
arthritis fibroblast-like Synoviocytes. Inflammation. 39:873–880.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu ZZ, Fu WB, Jin Z, Guo P, Wang WF and Li
JM: Reactive oxygen species mediate oridonin-induced apoptosis
through DNA damage response and activation of JNK pathway in
diffuse large B cell lymphoma. Leuk Lymphoma. 57:888–898. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang J, Wu L, Tashiro S, Onodera S and
Ikejima T: Reactive oxygen species mediate oridonin-induced HepG2
apoptosis through p53, MAPK and mitochondrial signaling pathways. J
Pharmacol Sci. 107:370–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bao R, Shu Y, Wu X, Weng H, Ding Q, Cao Y,
Li M, Mu J, Wu W, Ding Q, et al: Oridonin induces apoptosis and
cell cycle arrest of gallbladder cancer cells via the mitochondrial
pathway. BMC Cancer. 14:2172014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li J, Chen X, Xiao W, Ma W, Li T, Huang J,
Liu X, Liang X, Tang S and Luo Y: Mitochondria-targeted antioxidant
peptide SS31 attenuates high glucose-induced injury on human
retinal endothelial cells. Biochem Biophys Res Commun. 404:349–356.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao K, Zhao GM, Wu D, Soong Y, Birk AV,
Schiller PW and Szeto HH: Cell-permeable peptide antioxidants
targeted to inner mitochondrial membrane inhibit mitochondrial
swelling, oxidative cell death and reperfusion injury. J Biol Chem.
279:34682–34690. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao K, Luo G, Giannelli S and Szeto HH:
Mitochondria-targeted peptide prevents mitochondrial depolarization
and apoptosis induced by tert-butyl hydroperoxide in neuronal cell
lines. Biochem Pharmacol. 70:1796–1806. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
López-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009. View Article : Google Scholar
|
|
38
|
Capes-Davis A, Theodosopoulos G, Atkin I,
Drexler HG, Kohara A, MacLeod RA, Masters JR, Nakamura Y, Reid YA,
Reddel RR and Freshney RI: Check your cultures! A list of
cross-contaminated or misidentified cell lines. Int J Cancer.
127:1–8. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zufferey R, Nagy D, Mandel RJ, Naldini L
and Trono D: Multiply attenuated lentiviral vector achieves
efficient gene delivery in vivo. Nat Biotechnol. 15:871–875. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rota C, Chignell CF and Mason RP: Evidence
for free radical formation during the oxidation of
2′-7′-dichlorofluorescin to the fluorescent dye
2′-7′-dichlorofluorescein by horseradish peroxidase: Possible
implications for oxidative stress measurements. Free Radic Biol
Med. 27:873–881. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Loor G, Kondapalli J, Schriewer JM,
Chandel NS, Vanden Hoek TL and Schumacker PT: Menadione triggers
cell death through ROS-dependent mechanisms involving PARP
activation without requiring apoptosis. Free Radic Biol Med.
49:1925–1936. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ye W, Zhong Z, Zhu S, Zheng S, Xiao J,
Song S, Yu H, Wu Q, Lin Z and Chen J: Advanced oxidation protein
products induce chondrocyte death through a redox-dependent, poly
(ADP-ribose) polymerase-1-mediated pathway. Apoptosis. 22:86–97.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lu P, Kamboj A, Gibson SB and Anderson CM:
Poly(ADP-ribose) polymerase-1 causes mitochondrial damage and
neuron death mediated by Bnip3. J Neurosci. 34:15975–15987. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li CY, Wang EQ, Cheng Y and Bao JK:
Oridonin: An active diterpenoid targeting cell cycle arrest,
apoptotic and autophagic pathways for cancer therapeutics. Int J
Biochem Cell Biol. 43:701–704. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ikezoe T, Chen SS, Tong XJ, Heber D,
Taguchi H and Koeffler HP: Oridonin induces growth inhibition and
apoptosis of a variety of human cancer cells. Int J Oncol.
23:1187–1193. 2003.PubMed/NCBI
|
|
46
|
Schwarzlander M, Fricker MD and Sweetlove
LJ: Monitoring the in vivo redox state of plant mitochondria:
effect of respiratory inhibitors, abiotic stress and assessment of
recovery from oxidative challenge. Biochim Biophys Acta.
1787:468–475. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Albrecht SC, Sobotta MC, Bausewein D,
Aller I, Hell R, Dick TP and Meyer AJ: Redesign of genetically
encoded biosensors for monitoring mitochondrial redox status in a
broad range of model eukaryotes. J Biomol Screen. 19:379–386. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Johnson TM, Yu ZX, Ferrans VJ, Lowenstein
RA and Finkel T: Reactive oxygen species are downstream mediators
of p53-dependent apoptosis. Proc Natl Acad Sci USA. 93:pp.
11848–11852. 1996; View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang H, Ye Y, Chui JH, Zhu GY, Li YW, Fong
DW and Yu ZL: Oridonin induces G2/M cell cycle arrest and apoptosis
through MAPK and p53 signaling pathways in HepG2 cells. Oncol Rep.
24:647–651. 2010.PubMed/NCBI
|
|
50
|
Wang H, Ye Y, Chu JH, Zhu GY, Fong WF and
Yu ZL: Proteomic and functional analyses reveal the potential
involvement of endoplasmic reticulum stress and alpha-CP1 in the
anticancer activities of oridonin in HepG2 cells. Integr Cancer
Ther. 10:160–167. 2011. View Article : Google Scholar : PubMed/NCBI
|