Introduction
Cancer has become an important factor threatening
human health. Statistics show that hepatocellular carcinoma (HCC)
is one of the world's high incidence of malignant tumors, and in
China, it is ranked second in the highly lethal cancer (1). There are many factors leading to HCC,
but their nature mostly lead to the abnormal expression of the
coding gene or non-coding gene (2–4), and then
cause abnormal proliferation of cells and escape from the body
immune system monitoring.
In recent years, STAT3 and AKT have become the two
most important protooncogene. In many malignant tumors, abnormal
expressions of phosphorylated STAT3 (p-STAT3) and AKT (p-AKT) are
found. Signal transducer and activator of transcription (STATs) is
a cytoplasmic protein family that is first found in the
interferon-induced gene transcription study. It plays a key role in
the signal transduction of cytokine (CK) (5). Up to now, the STATs family found in
mammalian cells has STAT1, STAT2, STAT3, STAT4, STAT5 and STAT6.
STAT3 was originally found as an acute phase response factors
(APRF). At present, STAT3 has been found to be closely associated
with tumor inflammatory response, the transformation, survival,
proliferation, invasion and metastasis of tumor cells. Studies have
shown that the hyperactivity of STAT3 in HCC cells can promote the
proliferation, invasion and metastasis of tumor cells and inhibit
the apoptosis of tumor cells, then enhance the malignant biological
behavior of tumor (6). Li et
al (7) found that RNA
interference on STAT3 significantly inhibited cell proliferation
and reduced tumor volume in mice. In addition, the activation of
STAT3 can activate the expression of many downstream genes, such as
Bcl-2, Bcl-xL, myeloid cell leukemia-1 (Mcl-1), X-linked inhibitor
of apoptosis protein (XIAP), etc (8),
and these genes have an effect on the mechanism of multiple tumors.
Yang et al (9). found that
evodiamine could inhibit cell proliferation and induce apoptosis by
inhibiting STAT3 activity and down-regulating STAT3-mediated gene
expression in HCC.
Abnormal cell signal transduction is an important
factor leading to cell carcinogenesis, the signal transduction
pathway will transmit the signals of abnormal growth, proliferation
and differentiation to cells and lead to cancer. AKT is an
important protein kinase in the signal transduction pathway, which
is the downstream target protein of PI3K and the core of PI3K/AKT
signal transduction pathway (10).
AKT has three subtypes, which are AKT1, AKT2 and AKT3. AKT2 is one
of the important subtypes of AKT, which not only has the universal
characteristics of AKT, but also has its own unique biological
function. AKT2 is currently considered to be a key gene of the
PI3K/AKT2 signal transduction pathway, which mainly mediates the
adhesion, movement, invasion and metastasis of PI3K-dependent cells
in vivo. Researchers have transfected the full-length
wild-type AKT2 cDNA into human breast and ovarian cancer cells, and
the results show that transfected cells can increase tumor cell
invasion and metastasis by up-regulating β-integrin (11). However, the relationship of STAT3 and
AKT2 is unknown. In this paper, the correlation of STAT3 and AKT in
HCC cells was investigated.
Materials and methods
Ethics
In the present study, all methods were subject to
approval by the Institutional Animal Care and Use Committee of
People Hospital of Henan Zhengzhou.
Cell cultures
HCC cells (SMMC7721 cells and QGY-7703 cells) were
cultured in RPMI-1640 medium (FCS, ICN Biomedical Japan Co., Tokyo,
Japan) containing 10% fetal bovine serum, 100 µg/ml streptomycin
and 100 U/ml penicillin and incubated in an incubator (37°C, 5%
CO2). The cells used in the experiment were in the
logarithmic growth phase.
Cell transfection
According to the manufacturer's instructions,
si-Ctrl, si-STAT3 and si-AKT2 were transfected to the cells using
Lipofectamine 2000 (Invitrogen), respectively. Before transfection,
HCC cells were seeded to achieve 80–90% fusions. After 8 h of
transfection, the supernatant was removed and washed with 1xPBS for
three times, and RPMI-1640 medium was incubated with the cells for
related detection. The transfection was permanent.
RT-PCR
The total RNA was extracted with FASTAGEN-RNAfast200
kit (Invitrogen), and the reverse transcription conditions were as
follows: The reaction was carried out at 37°C for 15 min and at
85°C for 5 sec. The amplification reaction conditions were as
follows: Pre-denaturation at 94°C for 5 min, denaturation at 94°C
for 30 sec, annealing at 50°C for 30 sec, extension at 30°C for 30
sec, 30 cycles, and then extension at 72°C for 5 min. The PCR
products were electrophoresed on 3% agarose gel. Bio-Rad image
analysis system was used to take pictures, and Quantity One
software was used to analyze the data (Bio-Rad, Berkeley, CA, USA).
The relative expression quantity of the target gene was expressed
by the gray scale ratio between the target band and β-actin
stripe.
Western blot analysis
Cells were collected, and the total protein was
extracted from the cells with RIPA lysis buffer. BCA protein
quantitative kits (Invitrogen) were used to measure protein sample
concentration. 50 µg total protein were separated by SDS-PAGE and
transferred to PVDF membranes, and then incubated with STAT3,
p-STAT3, AKT2, p-AKT2, mTOR, p-mTOR, Cyclin D1, Bcl-xl and β-actin
antibody (Guangzhou RiboBio Co., Ltd., Guangzhou, China) overnight
at 4°C after the addition of 50 mg/l skimmed milk powder for 1.5 h.
Finally, the membranes were washed for 3 times and incubated with
horseradish peroxidase (HRP)-conjugated secondary antibody
(1:1,000) at room temperature for 2 h, and developed by ECI.
MTT assay
SMMC7721 cells and QGY-7703 cells were divided into
four groups (si-Ctrl group, si-STAT3 group, si-STAT3+pcDNA group,
si-STAT3+pcDNA-AKT2 group) and inoculated into 96-well plates
(2×105 cells/well). On the second day of inoculation, 6
wells were set in each group, and 20 µl of MTT solution (5 mg/ml)
were added to each well. After incubation at 37°C for 4 h, the
supernatant was removed and 150 µl of dimethyl sulfoxide (DMSO) was
added to each well, and then the absorbance of each well was
measured by a microplate reader (wavelength 492 nm) within 1 h
after mixing.
Migration and invasion assays
Transwell chambers (8 mm pore size; Corning
Incorporated, Corning, NY, USA) were used to determine the ability
of migration and invasion in HCC cells. The experimental grouping
method was the same as section 2.5. For migration assay, cells were
digested with trypsin and made into 2×105 cells/ml of
cell suspension. 200 µl of cell suspension were added to each well
of the upper chamber of the Transwell, and 500 µl of 1640 medium
containing serum was added to the lower chamber. After 48 h of
incubation, the cells on the upper surface were removed by a cotton
swab, and the cells on the lower surface of the membrane were fixed
with 90% alcohol for 30 min. Then, the cells were stained with
crystal violet for 20 min and rinsed with water. Cells were counted
under a microscope in 5 random fields and the results were
calculated as a percentage.
The invasion assay, 50 µl Matrigel was added to the
upper surface of the polycarbonate film of the transwell chamber to
solidify the gel. 500 µl of 1640 medium containing serum was added
to the lower chamber. After 48 h of incubation, the cells were
treated as described above.
Chromatin immunoprecipitation
assay
According to the manufacturer's instructions, the
Chromatin Immunoprecipitation (ChIP) assay was carried out with
ChIP assay kits (EMD Millipore, Billerica, MA, USA). Briefly, the
IL-6 stimulated (400 U/ml) and unstimulated SMMC7721 cells were
fixed with 1% formaldehyde at room temperature for 10 min and
neutralized with glycine for 5 min, followed by removing the
supernatant and washing with lxPBS for twice. Then, the cells were
lysed and collected in 1.5 ml EP tube (RNase-free). The RNA
fragment was disrupted by ultrasound for 5 min, and supernatant was
collected after centrifugation. Then, si-STAT3 and IgG antibody was
added into supernatant and incubated overnight at 4°C. After that,
proteinA/G was added to complexes and incubated at 4°C for 2 h.
After washing the pellets, elution buffer was added and the samples
were heated at 65°C for 1 h to remove cross-linking. Trizol method
was uesd to extract RNA and PCR products were electrophoresed on 3%
agarose gel, and the following steps were described as above.
Dual-Luciferase reporter gene
assay
SMMC7721 cells were inoculated into 24-well plates
(1×105 cells/well) and tested with Dual-Luciferase
Reporter Assay kit (DLR; Promega Corp., Madison, WI, USA) according
to the corresponding kit instructions. Briefly, 100 µl cell lysate
was added to each well and shaken slowly at room temperature for 15
min, and then adding 100 µl luciferase assay reagent II (LARII) to
cell lysate (20 µl). Firefly luciferase activity was detected by
fluorescence luminescence detector. Then, adding 100 µl renilla
luciferase detection reagent to determine renilla luciferase
activity. The ratio of Firefly and TK Renilla Luciferase activity
was used as the reporter gene activity value. TK Renilla
fluorescence value was used as an internal reference.
Animal experiment
Forty nude mice were randomly divided into si-Ctrl
group (control group), si-STAT3 group, si-STAT3+pcDNA group and
si-STAT3+pcDNA-AKT2 group, and each group had 10 mice. In si-STAT3
group, 0.1 ml (containing 1.0×107 cells)
SMMC-7721/si-STAT3 cell suspension (SMMC-7721 cells transfected
with STAT3-siRNA lentiviral vector) or QGY-7703/si-STAT3 cell
suspension (QGY-7703 cells transfected with STAT3-siRNA lentiviral
vector) was subcutaneously inoculated to the left armpit of nude
mouse. In si-STAT3+pcDNA-AKT2 group, 0.1 ml HCC cell suspension
(SMMC-7721 cells transfected with si-STAT3 lentiviral vector and
pcDNA-AKT2 or QGY-7703 cells transfected with si-STAT3 lentiviral
vector and pcDNA-AKT2) was subcutaneously inoculated in nude mouse.
The control group was inoculated with SMMC7721 cell suspension or
QGY-7703 cell suspension. The long diameter (a) and short diameter
(b) of the tumor was measured with a vernier caliper every week,
and the tumor volume (V) was calculated with the formula
V=0.5xaxb2.
Statistical analysis
SPSS 19.0 statistical software was used to analyze
the data. Student's t-test or one-way analysis of variance (ANOVA)
was used to compare quantitative variables. All data were expressed
as means ± standard deviation (SD). P<0.05 was considered
statistically significant.
Results
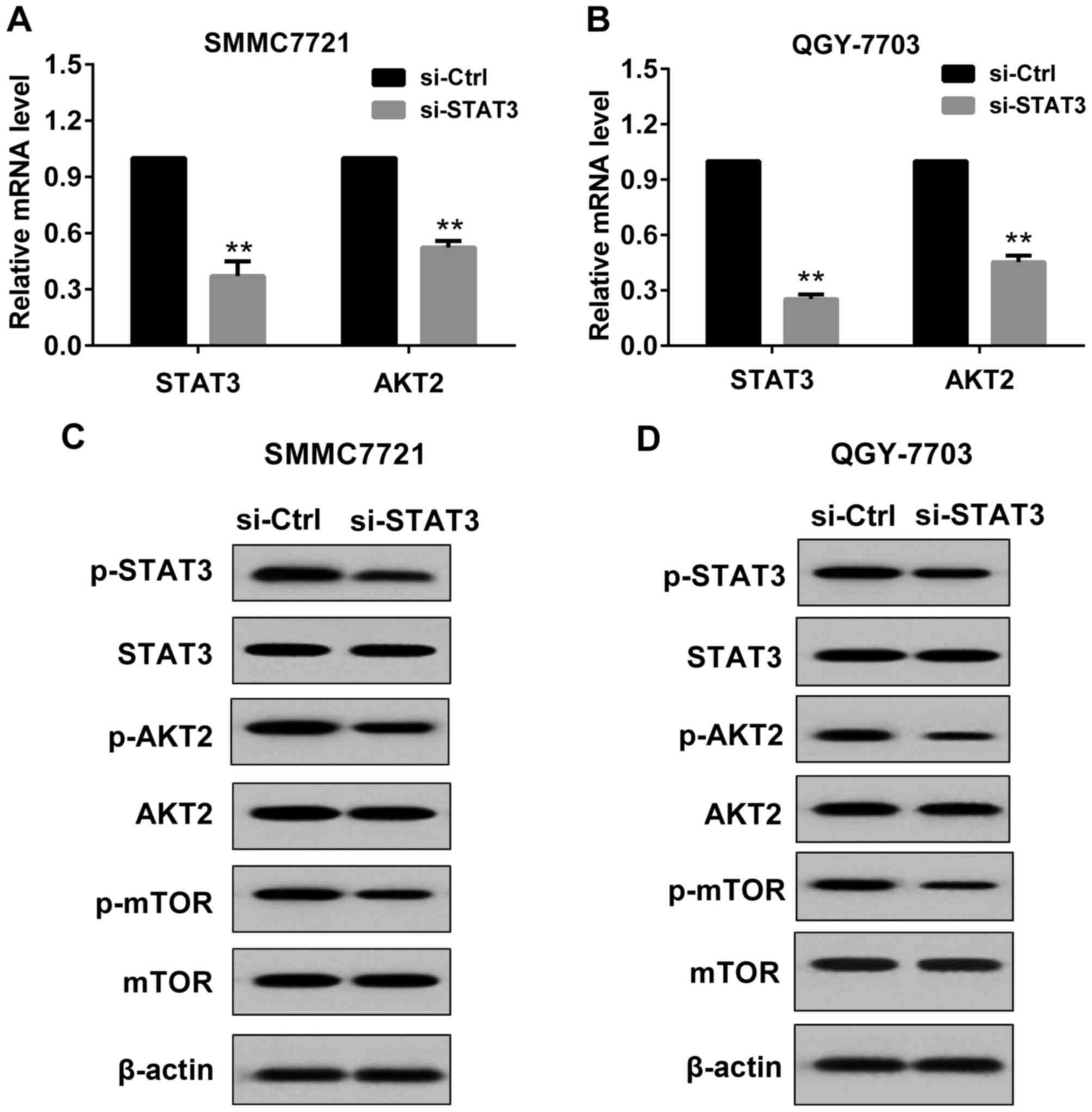
Effect of si-STAT3 on the expression
of STAT3 and AKT2 in HCC cells
In the present study, si-STAT3 or si-Control was
transfected to SMMC7721 cells and QGY7703 cells. The transfection
was permanent. As indicated in Fig.
1, after 48 h of transfection, STAT3 and AKT2 expression was
detected by RT-PCR and western blot respectively. Compared with the
si-Control, in SMMC7721 cells transfected with si-STAT3, the mRNA
level of STAT3 and AKT2 was significantly reduced (Fig. 1A), and the protein levels of
p-STAT3(Tyr705), p-AKT2(Ser473) and
p-mTOR(Ser2448) (downstream molecule of AKT2) were
markedly decreased (Fig. 1C). These
results indicated that transfection with si-STAT3 was effective in
SMMC7721 cells. Similar results were observed in QGY7703 cells
(Fig. 1B and D).
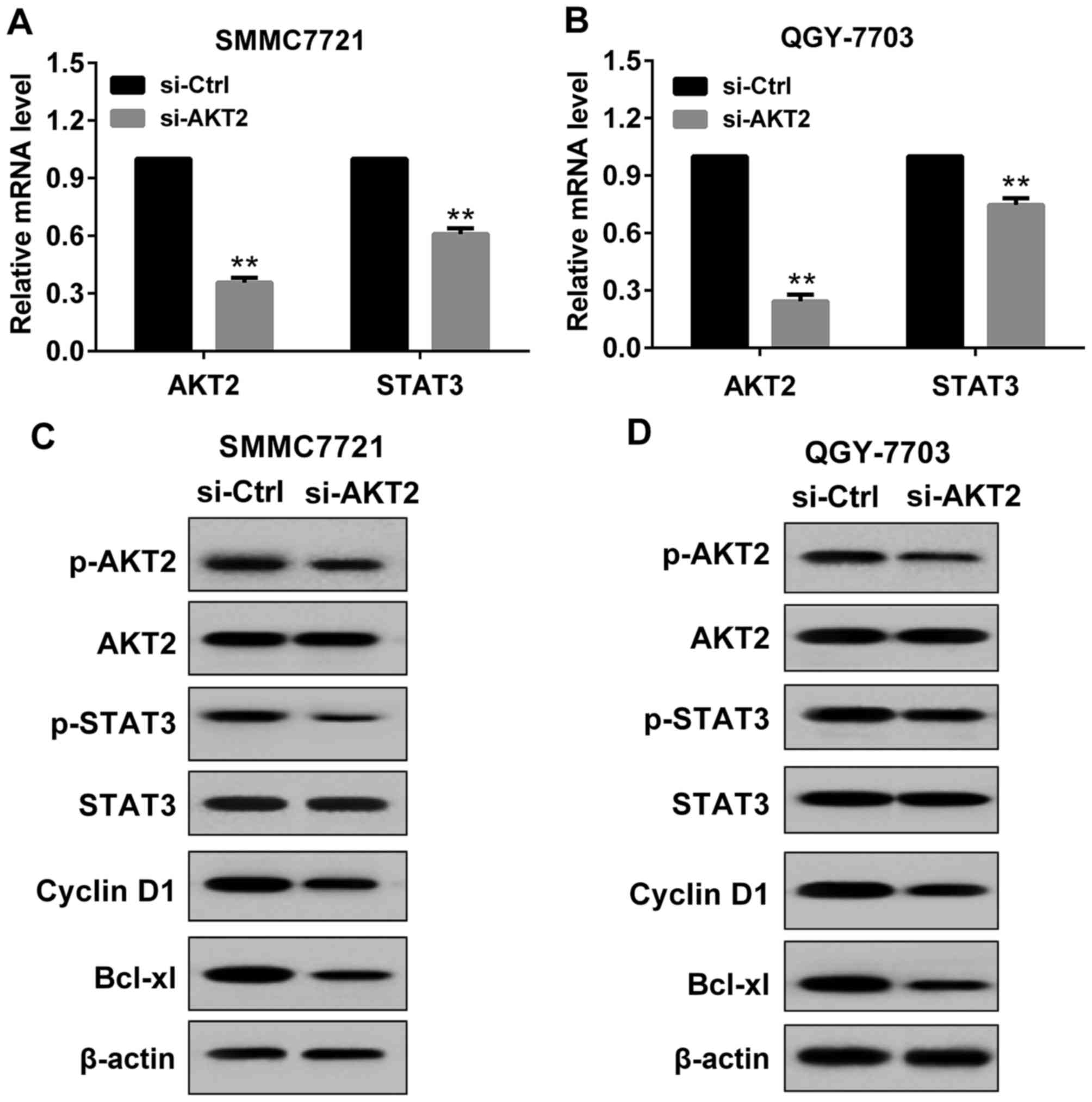
Effect of si-AKT2 on the expression of
AKT2 and STAT3 in HCC cells
In the present study, si-AKT2 or si-Control was
transfected to SMMC7721 cells and QGY7703 cells. The transfection
was permanent. As indicated in Fig.
2, after 48 h of transfection, AKT2 and STAT3 expression was
detected by RT-PCR and western blot respectively. Compared with the
si-Control, in SMMC7721 cells transfected with si-AKT2, the mRNA
level of AKT2 and STAT3 was significantly decreased (Fig. 2A), and the protein levels of p-AKT2,
p-STAT3 and its downstream molecules (Cyclin D1 and Bcl-xl) were
obviously reduced (Fig. 2C). These
results suggested that transfection with si-AKT2 was effective in
SMMC7721 cells. Similar results were obtained in QGY7703 cells
(Fig. 2B and D).
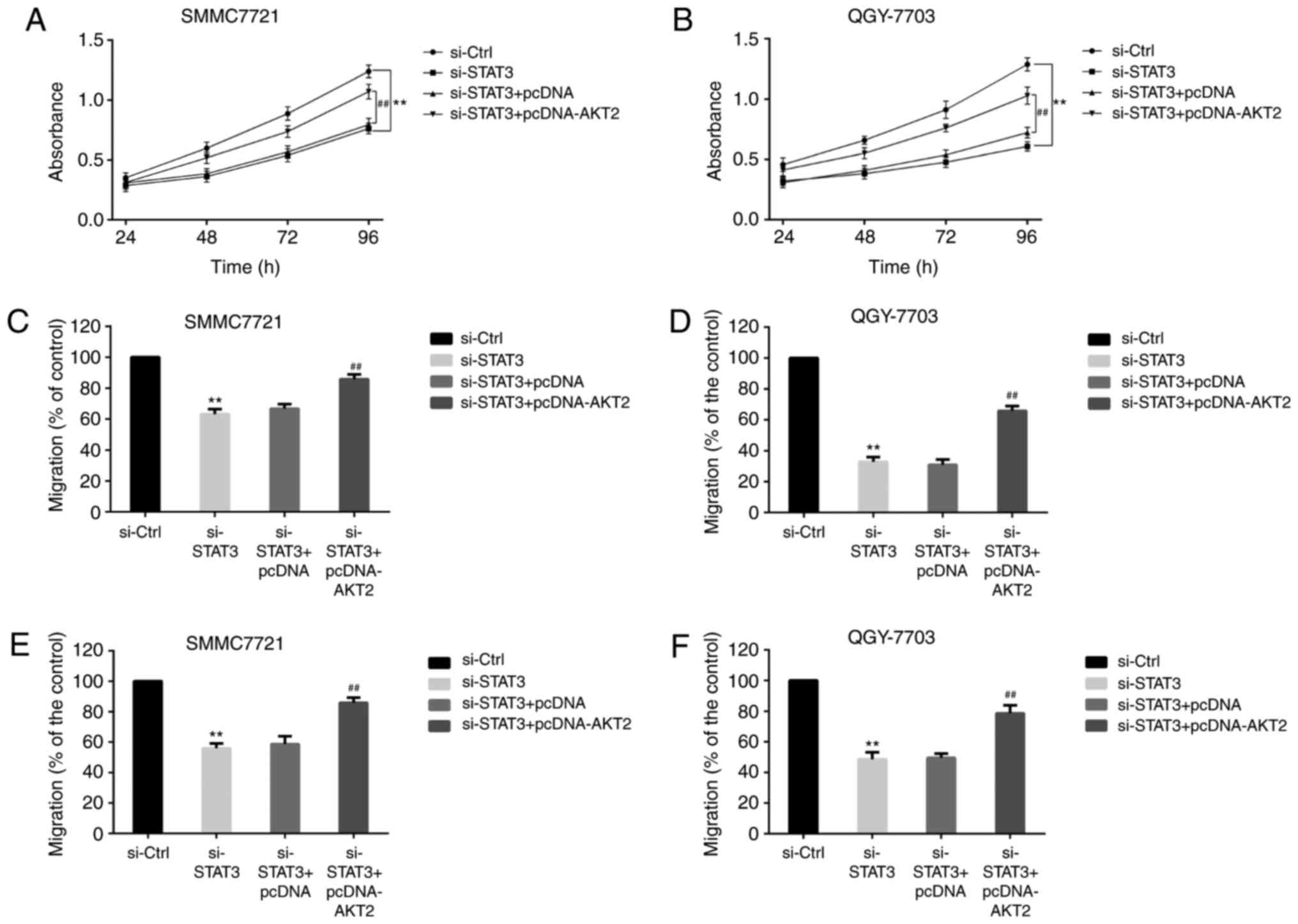
Effect of si-STAT3 and AKT2 on the
proliferation, migration and invasion of HCC cells
HCC cells were divided into si-Control group,
si-STAT3 group, si-STAT3+pcDNA group and si-STAT3+pcDNA-AKT2 group.
MTT analysis (Fig. 3) showed that the
absorbance of SMMC-7721 cells (Fig.
3A) and QGY-7703 cells (Fig. 3B)
was lower in si-STAT3 group than in si-Control group. And compared
with si-STAT3+pcDNA group, the absorbance of HCC cells (Fig. 3A and B) was increased in
si-STAT3+pcDNA-AKT2 group. The results illustrated that si-STAT3
could reduce the proliferation of HCC cells, and AKT2 could change
the phenomenon.
Transwell assays demonstrated that the migration of
SMMC-7721 cells (Fig. 3C) and
QGY-7703 cells (Fig. 3D) was reduced
in si-STAT3 group when compared with si-Control group. In contrast,
the migration of HCC cells (Fig. 3C and
D) was increased in si-STAT3+pcDNA-AKT2 group when compared
with si-STAT3+pcDNA group. Similarly, compared with si-Control
group, the invasion of SMMC-7721 cells (Fig. 3E) and QGY-7703 cells (Fig. 3F) was decreased in si-STAT3 group. And
compared with si-STAT3+pcDNA group, the invasion of HCC cells
(Fig. 3E and F) was increased in
si-STAT3+pcDNA-AKT2 group. These data indicated that STAT3
regulated the proliferation, migration and invasion of HCC by
AKT2.
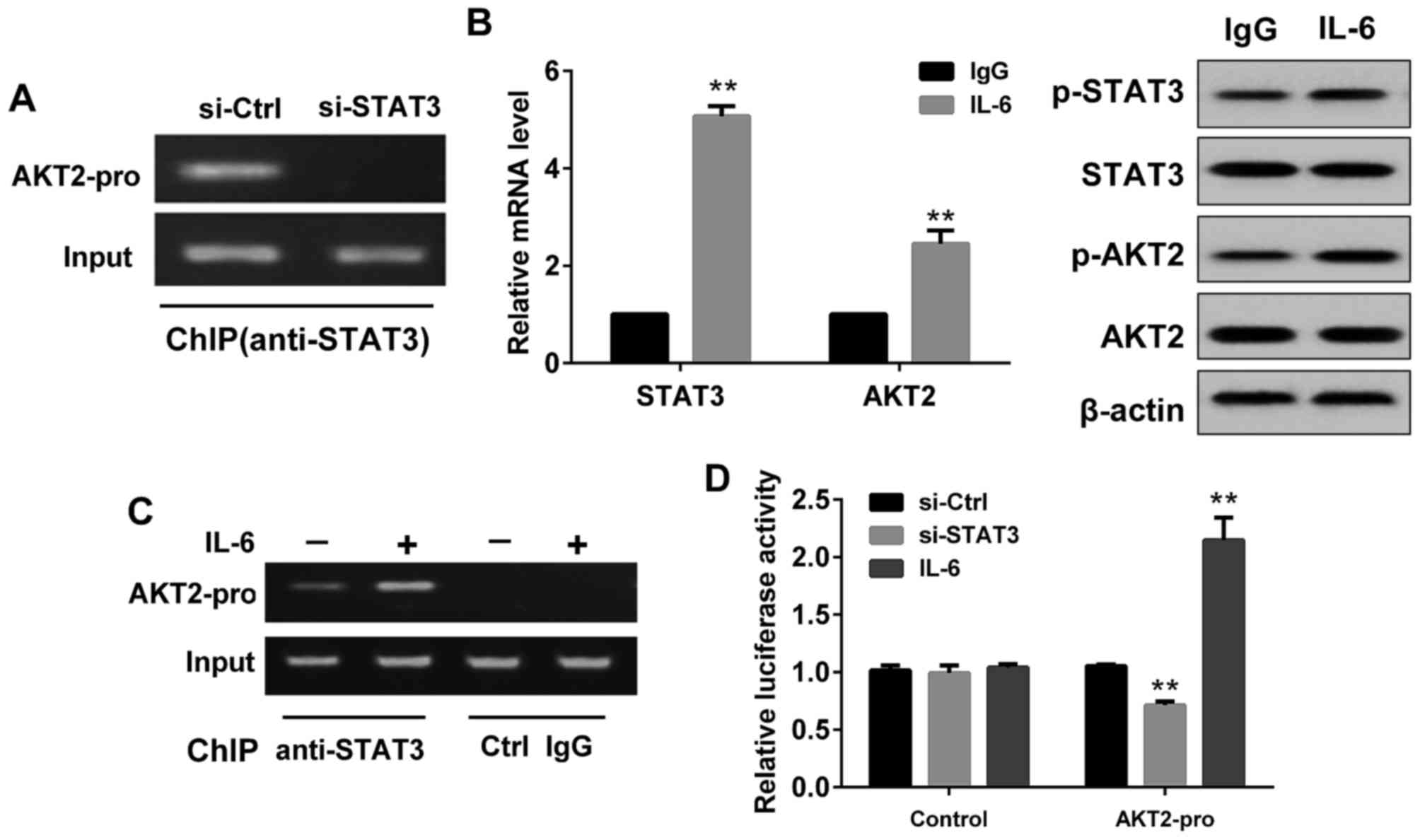
Effect of STAT3 on AKT2
expression
As demonstrated in Fig.
4, the ChIP assay showed that STAT3 could bind to AKT2 promoter
in SMMC-7721 cells, while no binding was detected in cells
transfected with si-STAT3 (Fig. 4A).
Compared with the control group (IgG stimulated SMMC7721 cells),
the mRNA levels of STAT3 and AKT2 in IL-6 (a known inducer of
STAT3) stimulated SMMC7721 cells were increased (Fig. 4B), and the protein levels of p-STAT3
and p-AKT2 were also enhanced (Fig.
4B). Moreover, ChIP assay showed STAT3 could better combine
with AKT2 promoter after IL-6 stimulation (Fig. 4C). DLR assay showed that the
fluorescence activity of the luciferase reporter vector containing
the AKT2 promoter sequence in SMMC7721 cells transfected with
si-STAT3 was significantly lower than that of the no-load without
the AKT2 promoter sequence (Fig. 4D).
However, in SMMC7721 cells treated with IL-6, the fluorescence
activity of the luciferase reporter vector containing the AKT2
promoter sequence was significantly higher than that of the no-load
without the AKT2 promoter sequence (Fig.
4D).
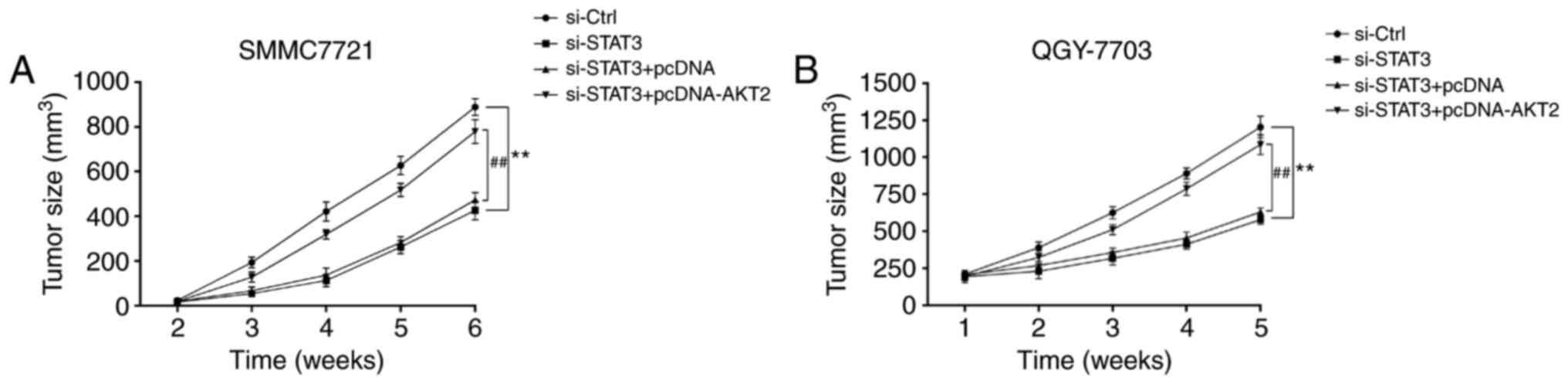
si-STAT3 inhibits tumor growth of
hepatoma xenografts in nude mice
As indicated in Fig.
5, from the second week, nude mice were subcutaneously
inoculated with SMMC-7721 cells and QGY7703 cells, the volume of
tumor in si-STAT3 group was significantly smaller than that in
si-Control group (Fig. 5A and B).
However, the tumor volume in si-STAT3+pcDNA-AKT2 group was markedly
higher than that in si-STAT3+pcDNA group (Fig. 5A and B).
Discussion
HCC is the fourth most common malignant tumor in the
world. Although surgical treatment and adjuvant chemotherapy are
continually improving, the survival rate of patients with HCC is
always very low. At present, due to the development of
chemotherapy, molecular targeted therapy and gene therapy, these
technologies are expected to bring new hope to patients with
HCC.
Studies have shown that abnormal activation of STAT3
is often associated with the occurrence of tumors (12), which can inhibit tumor cell apoptosis
and enhance the tolerance of tumor cells to chemotherapeutic drugs
(13). At the same time, p-STAT3
cannot only activate the expression of genes related to
proliferation, apoptosis and invasion, but also inhibit the
expression of genes associated with anti-tumor responses (14–17). It
had been reported that RNA interference STAT3 could significantly
inhibit cell proliferation and reduce the tumor volume of
tumor-bearing mice (7). In addition,
Li et al (18) found that
blocking the expression of STAT3 by interfering w (16) ith RNA interference (RNAi)
significantly inhibited the expression of Cyclin D1 and Bcl-2 in
cancer cells and promoted the apoptosis of tumor cells. Kunigal
et al (19) reported that
blocking the expression of STAT3 by RNAi could up-regulate the
expression of Fas protein, down-regulate Bcl-xL protein, and
promote the apoptosis of tumor cells. In this study, we had similar
findings. We discovered that silencing STAT3 by RNAi could reduce
mRNA level of AKT2 in HCC cells, down-regulate the expression of
p-AKT2 and its downstream molecules, and inhibit the proliferation,
migration and invasion of HCC cells. Moreover, we also found that
silencing AKT2 by RNAi could attenuate the expression of p-STAT3
and its downstream molecules in HCC cells. These results indicated
that STAT3 and AKT2 could interact with each other. However, the
mechanism between them would need further study.
AKT2 is defined as an oncogene (20,21), which
is closely related to the occurrence and development of HCC
(22). In the present study, we found
AKT2 could reverse the inhibitory effect of si-STAT3 in HCC cells.
Therefore, we speculated that STAT3 might promote the
proliferation, migration and invasion of HCC cells by regulating
AKT2. Further research confirmed this hypothesis. ChIP experiment
demonstrated that STAT3 in HCC cells could bind to AKT2 promoter,
and the combination was more closely after IL-6 (a known inducer of
STAT3) stimulation, while no binding was found in HCC cells
transfected with STAT3. Dual-Luciferase reporter gene assay
indicated that STAT3 could promote transcription of AKT2 by binding
to the AKT2 promoter, and then increase AKT2 expression. Thus, the
research demonstrated for the first time that STAT3 regulated AKT2
expression in HCC cells by a direct mechanism. Furthermore, nude
mice experiment also confirmed this finding.
In this study, we chose SMMC7721 and QGY-7703 cell
lines as research objects because the two hepatoma cell lines could
grow more rapidly and stably, and have a high the tumor formation
rate in immunodeficient mice. However, the two hepatoma cell lines
exhibited some differences in the intensity of their responses to
the treatments. We believed that it was related to the origin of
the two hepatoma cell lines. SMMC7721 cell line comes from a male
liver tissue, and QGY-7703 cell line comes from a female liver
tissue. Additionally, it inevitably has some improper operation
during the experimental, which is also the cause of the
difference.
In conclusion, our results revealed that STAT3 might
promote the occurrence and development of hepatocellular carcinoma
by regulating AKT2. And the result shed a new light on therapeutic
target for HCC.
Acknowledgements
This research was supported by Basic and frontier
technology research project of Henan Provincial Department of
science and technology in 2014 (NO.142300410073).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huang J, Wang Y, Guo Y and Sun S:
Down-regulated microRNA-152 induces aberrant DNA methylation in
hepatitis B virus-related hepatocellular carcinoma by targeting DNA
methyltransferase 1. Hepatology. 52:60–70. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Feitelson MA, Sun B, Satiroglu Tufan NL,
Liu J, Pan J and Lian Z: Genetic mechanisms of
hepatocarcinogenesis. Oncogene. 21:2593–2604. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hou J, Zhou Y, Zheng Y, Fan J, Zhou W, Ng
IO, Sun H, Qin L, Qiu S, Lee JM, et al: Hepatic RIG-I predicts
survival and interferon-α therapeutic response in hepatocellular
carcinoma. Cancer Cell. 25:49–63. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Darnell JE Jr: STATs and gene regulation.
Science. 277:1630–1635. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yuen JW, Poon LS, Chan AS, Yu FW, Lo RK
and Wong YH: Activation of STAT3 by specific galpha subunits and
multiple gbetagamma dimers. Int J Biochem Cell Biol. 42:1052–1059.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li WC, Ye SL, Sun RX, Liu YK, Tang ZY, Kim
Y, Karras JG and Zhang H: Inhibition of growth and metastasis of
human hepatocellular carcinoma by antisense oligonucleotide
targeting signal transducer and activator of transcription 3. Clin
Cancer Res. 12:7140–7148. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Levy DE and Darnell JE Jr: Stats:
Transcriptional control and biological impact. Nat Rev Mol Cell
Biol. 3:651–662. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang J, Cai X, Lu W, Hu C, Xu X, Yu Q and
Cao P: Evodiamine inhibits STAT3 signaling by inducing phosphatase
shatterproof 1 in hepatocellular carcinoma cells. Cancer Lett.
328:243–251. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pham NA, Tsao MS, Cao P and Hedley DW:
Dissociation of gemcitabine sensitivity and protein kinase B
signaling in pancreatic ductal adenocarcinoma models. Pancreas.
35:e16–e26. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Arboleda MJ, Lyons JF, Kabbinavar FF, Bray
MR, Snow BE, Ayala R, Danino M, Karlan BY and Slamon DJ:
Overexpression of AKT2/protein kinase Bbeta leads to up-regulation
of beta1 integrins, increased invasion and metastasis of human
breast and ovarian cancer cells. Cancer Res. 63:196–206.
2003.PubMed/NCBI
|
|
12
|
Lee H, Herrmann A, Deng JH, Kujawski M,
Niu G, Li Z, Forman S, Jove R, Pardoll DM and Yu H: Persistently
activated Stat3 maintains constitutive NF-κB activity in tumors.
Cancer Cell. 15:283–293. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sansone P and Bromberg J: Targeting the
interleukin-6/Jak/stat pathway in human malignancies. J Clin Oncol.
30:1005–1014. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fukuda A, Wang SC, Morris JP IV, Folias
AE, Liou A, Kim GE, Akira S, Boucher KM, Firpo MA, Mulvihill SJ and
Hebrok M: Stat3 and MMP7 contribute to pancreatic ductal
adenocarcinoma initiation and progression. Cancer Cell. 19:441–455.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kortylewski M, Xin H, Kujawski M, Lee H,
Liu Y, Harris T, Drake C, Pardoll D and Yu H: Regulation of the
IL-23 and IL-12 balance by Stat3 signaling in the tumor
microenvironment. Cancer Cell. 15:114–123. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lesina M, Kurkowski MU, Ludes K, Rose-John
S, Treiber M, Klöppel G, Yoshimura A, Reindl W, Sipos B, Akira S,
et al: Stat3/Socs3 activation by IL-6 transsignaling promotes
progression of pancreatic intraepithelial neoplasia and development
of pancreatic cancer. Cancer Cell. 19:456–469. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li N, Grivennikov SI and Karin M: The
unholy trinity: Inflammation, cytokines and STAT3 shape the cancer
microenvironment. Cancer Cell. 19:429–431. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li GH, Wei H, Lv SQ, Ji H and Wang DL:
Knockdown of STAT3 expression by RNAi suppresses growth and induces
apoptosis and differentiation in glioblastoma stem cells. Int J
Oncol. 37:103–110. 2010.PubMed/NCBI
|
|
19
|
Kunigal S, Lakka SS, Sodadasu PK, Estes N
and Rao JS: Stat3-siRNA induces fas-mediated apoptosis in vitro and
in vivo in breast cancer. Int J Oncol. 34:1209–1220.
2009.PubMed/NCBI
|
|
20
|
Rössig L, Jadidi AS, Urbich C, Badorff C,
Zeiher AM and Dimmeler S: Akt-dependent phosphorylation of p21Cip1
regulates PCNA binding and proliferation of endothelial cells. Mol
Cell Biol. 21:5644–5657. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shah A, Swain WA, Richardson D, Edwards J,
Stewart DJ, Richardson CM, Swinson DE, Patel D, Jones JL and
O'Byrne KJ: Phospho-akt expression is associated with a favorable
outcome in non-small cell lung cancer. Clin Cancer Res.
11:2930–2936. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Barderas R, Mendes M, Torres S, Bartolomé
RA, López-Lucendo M, Villar-Vázquez R, Peláez-García A, Fuente E,
Bonilla F and Casal JI: In-depth characterization of the secretome
of colorectal cancer metastatic cells identifies key proteins in
cell adhesion, migration and invasion. Mol Cell Proteomics.
12:1602–1620. 2013. View Article : Google Scholar : PubMed/NCBI
|