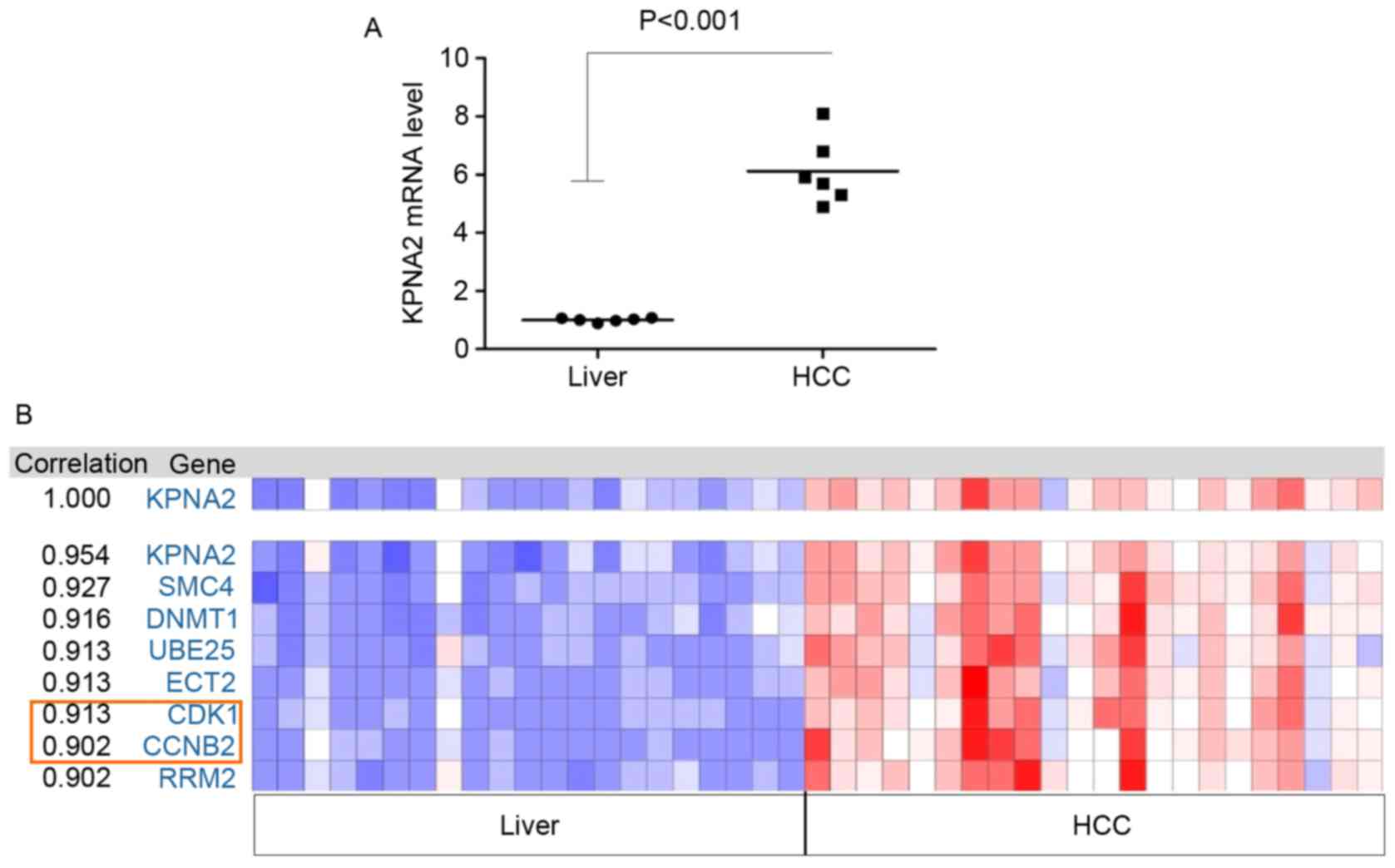
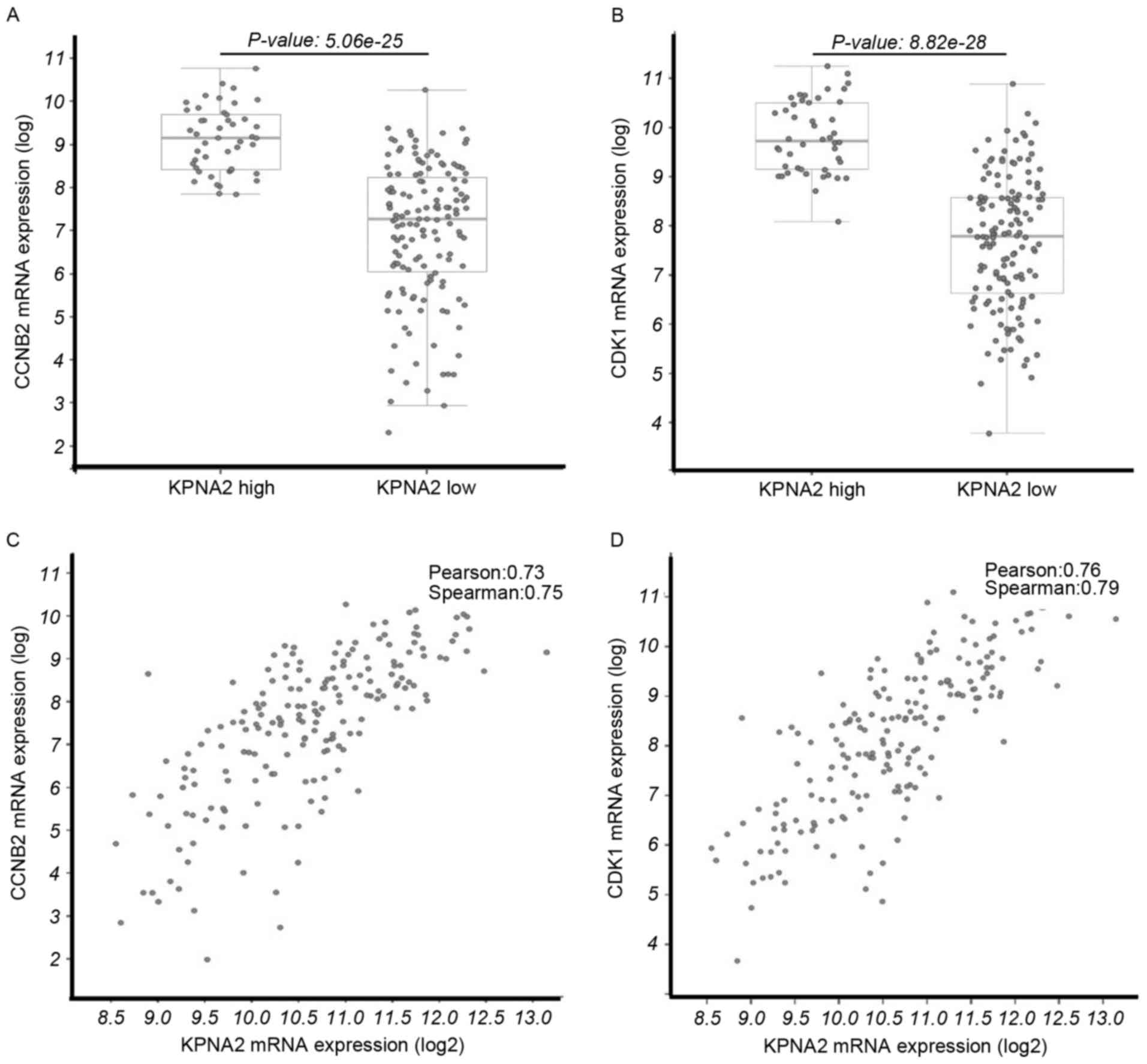
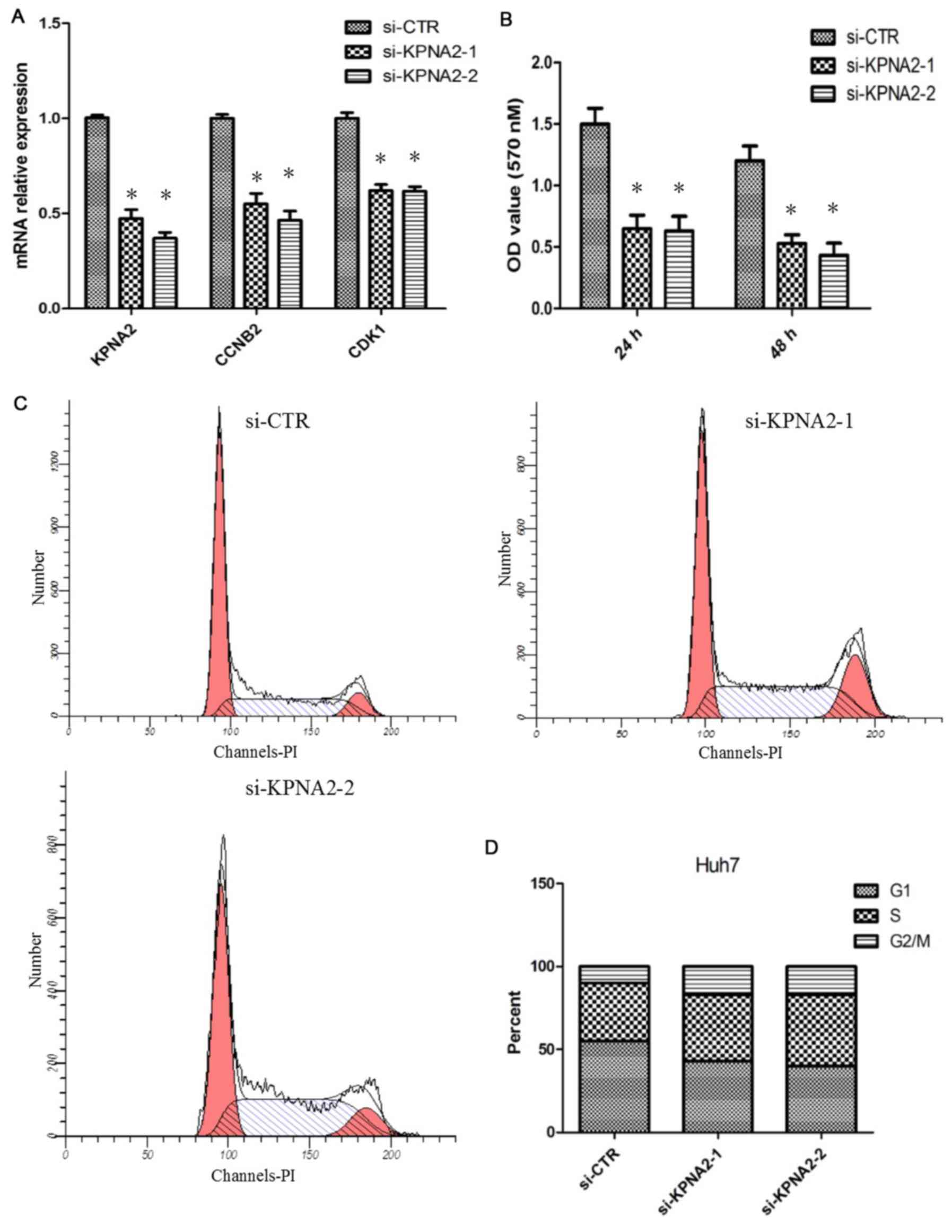
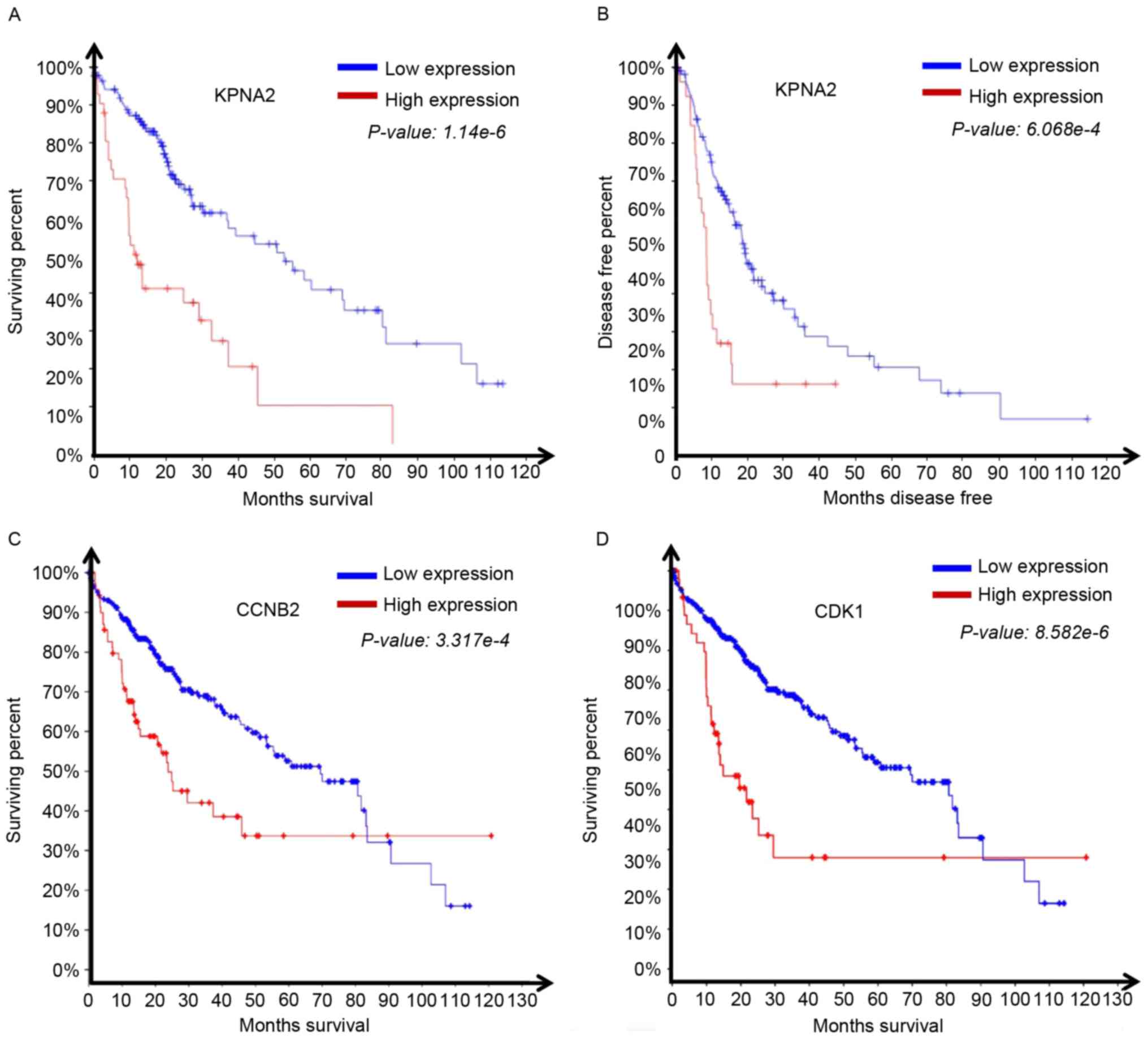
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Housman G, Byler S, Heerboth S, Lapinska
K, Longacre M, Snyder N and Sarkar S: Drug resistance in cancer: An
overview. Cancers (Basel). 6:1769–1792. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sukowati CH, Rosso N, Crocè LS and
Tiribelli C: Hepatic cancer stem cells and drug resistance:
Relevance in targeted therapies for hepatocellular carcinoma. World
J Hepatol. 2:114–126. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alshareeda AT, Negm OH, Green AR, Nolan
CC, Tighe P, Albarakati N, Sultana R, Madhusudan S, Ellis IO and
Rakha EA: KPNA2 is a nuclear export protein that contributes to
aberrant localisation of key proteins and poor prognosis of breast
cancer. Br J Cancer. 112:1929–1937. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang L, Wang HY, Li JD, Wang JH, Zhou Y,
Luo RZ, Yun JP, Zhang Y, Jia WH and Zheng M: KPNA2 promotes cell
proliferation and tumorigenicity in epithelial ovarian carcinoma
through upregulation of c-Myc and downregulation of FOXO3a. Cell
Death Dis. 4:e7452013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Teng SC, Wu KJ, Tseng SF, Wong CW and Kao
L: Importin KPNA2, NBS1, DNA repair and tumorigenesis. J Mol
Histol. 37:293–299. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Christiansen A and Dyrskjøt L: The
functional role of the novel biomarker karyopherin α 2 (KPNA2) in
cancer. Cancer Lett. 331:18–23. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim IS, Kim DH, Han SM, Chin MU, Nam HJ,
Cho HP, Choi SY, Song BJ, Kim ER, Bae YS and Moon YH: Truncated
form of importin alpha identified in breast cancer cell inhibits
nuclear import of p53. J Biol Chem. 275:23139–23145. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li XL, Jia LL, Shi MM, Li X, Li ZH, Li HF,
Wang EH and Jia XS: Downregulation of KPNA2 in non-small-cell lung
cancer is associated with Oct4 expression. J Transl Med.
11:2322013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Maass T, Sfakianakis I, Staib F, Krupp M,
Galle PR and Teufel A: Microarray-based gene expression analysis of
hepatocellular carcinoma. Curr Genomics. 11:261–268. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Malumbres M and Barbacid M: Cell cycle,
CDKs and cancer: A changing paradigm. Nat Rev Cancer. 9:153–166.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dorée M and Hunt T: From Cdc2 to Cdk1:
When did the cell cycle kinase join its cyclin partner? J Cell Sci.
115:2461–2464. 2002.PubMed/NCBI
|
|
17
|
Huang Y, Sramkoski RM and Jacobberger JW:
The kinetics of G2 and M transitions regulated by B cyclins. PLoS
One. 8:e808612013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kosugi S, Hasebe M, Tomita M and Yanagawa
H: Systematic identification of cell cycle-dependent yeast
nucleocytoplasmic shuttling proteins by prediction of composite
motifs. Proc Natl Acad Sci USA. 106:pp. 10171–10176. 2009;
View Article : Google Scholar : PubMed/NCBI
|