Introduction
Cases of liver cancer may be divided into two
categories, based on the primary tumor site: Primary liver cancer
(PLC) and metastatic cancer of the liver (liver metastases). Among
the various types of PLC, hepatocellular carcinoma (HCC) is the
predominant histological form, accounting for the majority of PLC
cases (1). An estimated 782,500
incident PLC cases and 745,500 mortalities occurred worldwide in
2012, with China alone reporting ~50% of the total number of cases
and mortalities (2). HCC development
is a multi-step process, and 80% of HCC develops in cirrhotic
livers (3). HCCs are clinically
heterogeneous and exhibit genetic alterations (4). At present, an early diagnosis of HCC,
without pathological verification, is achieved by analyzing serum
α-fetoprotein (AFP) levels combined with imaging techniques
(5). There is an urgent requirement
to develop novel molecular tools for assisting early HCC diagnosis,
prognosis and treatment stratification. Molecular profiling of gene
expression has improved the understanding of the mechanisms of HCC
development, allowing the identification of biomarkers for HCC
diagnosis and the stratification of patients with HCC for prognosis
and therapy (6). Takuji Yoshimura
et al (7) previously
identified that the gametocyte specific factor 1 (GTSF1) gene, a
member of the evolutionarily conserved UPF0224 family, is expressed
predominantly in male germ cells. It has been suggested that the
expression pattern of GTSF1 and its high conservation may serve an
important role in germ cell development (8). A meta-analysis of gene expression data
suggested that, as male GTSF1-knockout mice are sterile owing to a
large proportion of dead germ cells, aberrant overexpression of
GTSF1 may have a role in the apoptosis resistance of Mycosis
Fungoides (MF) (9). In the present
study, screening of gene expression in liver cancer samples, all
histologically HCC, revealed an association between GTSF1
expression and liver cancer cell proliferation and apoptosis. To
the best of our knowledge, there have been no prior studies
examining GTSF1 gene expression and its association with liver
cancer.
Materials and methods
Samples
A total of 24 patients with liver cancer and 32
normal liver controls were retrospectively included in the present
study at the Songjiang Hospital Affiliated to The Shanghai First
People's Hospital (Shanghai Jiaotong University, Shanghai, China)
from April 2009 to December 2014. Normal liver tissues were
collected from patients undergoing resection of hepatic
hemangiomas. Paired liver cancer and adjacent non-tumor liver
tissues (≥1-cm from the tumor edge) were obtained from patients
undergoing resection of liver cancer tumors. The
clinicopathological data of the patients were collected
retrospectively from the Songjiang Hospital Affiliated to The
Shanghai First People's hospital databases. No local or systemic
treatment had been administered to these patients prior to surgery.
The characteristics of patients with and without liver cancer are
summarized in Table I.
 | Table I.Patients characteristics with or
without HCC. |
Table I.
Patients characteristics with or
without HCC.
| Clinical
parameters | Liver cancer
(n=24) | Normal controls
(n=32) | P-value |
|---|
| Age (years),
mean | 63.1±9.9 | 51.6±12.7 | 0.654 |
| ± SD |
|
|
|
| Sex (%) |
|
|
|
|
Male | 18 (75.0) | 21 (65.6) | 0.642 |
|
Female | 6 (25.0) | 11 (34.4) | 0.591 |
| AFP level (%) |
|
|
|
|
Normal | 8 (33.3) | 30 (93.8) | 0.001a |
|
Abnormal | 16 (66.7) | 2 (6.2) | 0.001a |
| HBV infection
(%) |
|
|
|
|
Positive | 17 (70.8) | 3 (9.4) | 0.001a |
|
Negative | 7 (29.2) | 29 (90.6) | 0.001a |
Samples of the resected tumor specimens and the
normal liver controls were stored immediately in liquid nitrogen at
−196°C until analysis. Genomic DNA was obtained by digestion with
recombinant polymerase chain reaction (PCR)-grade proteinase K
(Roche Diagnostics, Basel, Switzerland). Liver samples were
pulverized in liquid nitrogen at −196°C for 10 sec prior to
incubation in tail-buffer (Roche Diagnostics, Basel, Switzerland).
In total, 750 µl Tris-EDTA buffer (Invitrogen; Thermo Fisher
Scientific, Inc.) with RNase A (20 µg/ml) were added to the
digested tails and incubated for 10 min at room temperature. The
samples were subsequently centrifuged at 4°C for 10 min at 10,000 ×
g. Supernatant containing the genomic DNA (600 µl) was transferred
into a 2-ml reaction tube and precipitated by adding 60 µl 3 M NaAc
and 1,200 µl 100% EtOH. Total RNA was isolated from each of the
frozen samples with an RNeasy® Mini kit (Qiagen Benelux
B. V., Venlo, The Netherlands), according to the manufacturer's
protocol. Ethical approval was granted by the Institutional Review
Board of the Songjiang Hospital Affiliated to The Shanghai First
People's Hospital, (Shanghai Jiaotong University), and written and
informed consent was obtained from all patients.
Serum AFP and hepatitis B virus (HBV)
detection
Serum samples were analyzed using an Architect AFP
assay (Abbott Pharmaceutical Co., Ltd., Lake Bluff, IL, USA) and
results were calculated using the conversion equation as follows:
Conversion factor, 0.83 kU/l=1 µg/l. The results for AFP levels
were provided by the Clinical Laboratory of Songjiang Hospital
Affiliated to The Shanghai First People's Hospital, Shanghai
Jiaotong University, Shanghai, China. In order to detect hepatitis
B virus (HBV), serological markers of HBV were quantified using an
enzyme immunoassay kit (Abbott Pharmaceutical Co., Ltd.). HBV DNA
was detected with a Cobas TaqMan HBV test version 2.0 (lower limit
of detection, 20 IU/ml; Roche Diagnostics).
RNA/DNA extraction and reverse
transcription (RT)-quantitative (q)PCR
Total RNA and genomic DNA from human tissue samples
cells were obtained from the pathology department (Songjiang
Hospital Affiliated to The Shanghai First People's Hospital,
Shanghai, China) and were extracted using a GeneJET RNA
Purification kit (cat. no. K0731; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. Concentrations were
quantified using a NanoDrop 1000 (Thermo Fisher Scientific, Inc.).
An RT reaction was performed using 1 µg total RNA with a High
Capacity cDNA Reverse Transcription kit (cat. no. 4368814, Applied
Biosystems; Thermo Fisher Scientific, Inc.). The mRNA levels of
GTSF1 were determined by qPCR using a SYBR® Green Master
Mix kit and an ABI 7500 Real-Time PCR System (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The primer sequences were as
follows: β-actin forward, 5′-AAGATGACCCAGATCATGTTTGAG-3′ and
reverse, 5′-GCAGCTCGTAGCTCTTCTCCAG-3′; and GTSF1 forward,
5′-CACAAGCATCCTGTCTCATGTG-3′ and reverse,
5′-CTACACTTCTGGTCTGGGATTAC-3′. β-actin was used as an internal
control, and relative quantification was conducted using the
comparative cycle threshold method (10); the method was used to analyze the
relative changes in GTSF1 expression from the qPCR experiments. All
PCR reactions were performed under the following conditions:
Initial denaturation at 95°C for 30 sec, followed by annealing for
30 sec at 55°C and extension of DNA for 1 min at 74°C. These steps
were repeated 25–30 times prior to final extension for 5 min at
74°C. The experiments were performed in triplicate.
Vector construction and tumorigenicity
assays in nude mice
To construct the GTSF1 expression vector
(pcDNA3.0-GTSF1), a gene fragment encompassing the full-length
GTSF1 sequence and its 5′- and 3′-flanking regions was amplified
and then cloned into the BamHI and EcoRI sites in
pcDNA3.0 (Invitrogen; Thermo Fisher Scientific, Inc.).
Amplification of DNA fragments corresponding to amino acids 1–167
of the GTSF1 sequence was performed via PCR with primers
5′-CACAAGCATCCTGTCTCATGTG-3′ and 5′-GGCAGGGTATCATCTTTCTATTC-3′. The
PCR was performed at 95°C for 5 min, then 94°C for 1 min, 65°C for
50 sec and 72°C for 40 sec, for 30 cycles. Extension was performed
at 72°C for 10 min and 4°C for 10 min, using Phusion PCR Master Mix
(Thermo Fisher Scientific, Inc.). GTSF1 DNA was cloned into the
pcDNA3.0 expression vector. Digestion products were purified using
a NucleoSpin® extract II kit (Macherey-Nagel, Hœrdt,
France) and visualized with ethidium bromide, prior to ligation
using T4 DNA Ligase (Invitrogen; Thermo Fisher Scientific, Inc.)
overnight at 16°C. The purified plasmid DNA was verified by DNA
sequencing using the Sanger sequencing method performed on 3730XL
sequencers (Data Collection v3.0 and Sequencing Analysis v5.2;
Thermo Fisher Scientific, Inc.). The sequence blast was processed
using Blast software (BLAST+, v2.0.0, https://blast.ncbi.nlm.nih.gov/Blast.cgi; Shanghai
Sangong Pharmaceutical Co., Ltd., Shanghai, China). All
experimental procedures were in accordance with the Guide for the
Care and Use of Laboratory Animals, Songjiang Hospital Affiliated
to The Shanghai First People's Hospital Ethical Guidelines for
Animal Experiments. HepG2 cell lines (Shanghai Chinese Academy of
Sciences, Shanghai, China) were transfected with pcDNA3.0-GTSF1
[named Ad-shNC (GTSF1-positive)] or pcDNA3.0 empty vector [named
Ad-shGTSF1 (GTSF1-negative control)] using
Lipofectamine™ 2000 Transfection Reagent (cat. no.
11668027; Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocols. Transfected
pcDNA3.0-GTSF1-transfected cells were incubated at 37°C for 1–2
days and these cells (2×106) were suspended in 100 µl
PBS prior to being injected subcutaneously into either side of the
posterior flank of each 5–6-week-old male BALB/c athymic nude
mouse. A total of 12 mice were divided into two groups (Shanghai
Chinese Academy of Sciences, Shanghai, China). A total of 6 mice
were injected with Ad-shNC (GTSF1-positive)-transfected HepG2 cells
(total of 2×106 targeted cells) and the other group was
injected with Ad-shGTSF1 (GTSF1-negative control)-transfected HepG2
cells. The mice were housed in polypropylene cages under standard
experimental conditions (20–22°C, 55% humidity, food and water
ad libitum, 12 h light/dark cycle) and checked at least once
a week until tumors became palpable. Mice were sacrificed using
20–30% CO2 gas flow. Tumor sizes did not exceed 20 mm
(2.0 cm) in any direction in an adult mouse and tumor growth was
observed for >8 weeks (pre-determined end-point). The tumor
dimensions were measured every 3 days using a digital caliper and
the tumor volume was calculated using the following formula: V=π/6
× (larger diameter) × (smaller diameter)2.
RNA oligoribonucleotides and cell
transfections
The small interfering RNA (siRNA) targeting human
GTSF1 mRNA (NM_144594.2; https://www.ncbi.nlm.nih.gov/nuccore/NM_144594.2;
accessed October 27, 2017; NC_000081.6; https://www.ncbi.nlm.nih.gov/nuccore/NC_000081.6;
accessed November 10, 2017) were designated as siRNA1
(5′-UUCUCCGAACGUGUCACGUdTdT-3′) and siRNA2
(5′-ACGUGACACGUUCGGAGAAdTdT-3′). The two siRNAs against GTSF1 were
designed using the Whitehead Institute Web Server (http://jura.wi.mit.edu/bioc/siRNAext/)
and were chemically synthesized by Shanghai GenePharma Co., Ltd.
(Shanghai, China) to target GTSF1 mRNA. The control RNA duplex (NC)
for siRNA1 and siRNA 2 was non-homologous to any human genome
sequences. For the in vivo tumorigenicity assay, all
pyrimidine nucleotides in the NC or siRNA1 and siRNA2 duplex were
substituted with their 2-O-methyl analogs to improve RNA stability.
The anti-GTSF1 mRNA with the following sequences: siRNA1 (228–250)
forward, 5′-GGCUACUUGUCCCUUCAAUDTDT-3′ and reverse,
5′-AUUGAAGGGACAAGUAGCCDTDT-3′; and siRNA2 (478–500) forward,
5′-CCUGCGAGCAACAUAGUUAdTdT-3′ and reverse,
5′-UAACUAUGUUGCUGCAGGdTdT-3′, were 2-O-methyl-modified
oligoribonucleotides designed as an inhibitor of GTSF1 mRNA. All
RNA oligoribonucleotides were purchased from GenePharma (Shanghai
GenePharma Co., Ltd., Shanghai, China). Reverse transfection of RNA
oligoribonucleotide(s) was performed using Lipofectamine
RNAiMAX® (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. The transfection
efficiency, examined by fluorescein amidite-conjugated siRNA and
fluorescence-activated cell sorting analysis, was ~75% in PLC/PRF/5
and Huh-7 cell lines (Shanghai Chinese Academy of Sciences). A
total of 50 nmol/l RNA duplex and 200 nmol/l miRNA inhibitor were
used for each transfection. The above siRNAs were transfected into
HCC PLC/PRF/5 and Huh-7 cell lines. In total, 3×103
cells from each cell line were seeded onto 96-well plates. The
siRNA oligomer was diluted in 50 µl Opti-MEM® I Reduced
Serum medium without serum (with a final RNA concentration of 40
nM), prior to being diluted 1 µl in 50 µl OptiMEM® I
Reduced Serum medium, mixed gently and incubated for 5 min at room
temperature. Following incubation, the diluted oligomer was
combined with the diluted Lipofectamine™ 2000, mixed
gently and incubate for a further 20 min at room temperature. The
cells were then incubated at 37°C in a 5% CO2 incubator
for 24–96 h until subsequent experimentation. Subsequently, 24 h
following the siRNA transfection of HCC PLC/PRF/5 and Huh-7 cells,
the cells were transfected with 200 ng plasmid in a 24-well plate
using Lipofectamine 2000® (Invitrogen; Thermo Fisher
Scientific, Inc.).
Cell growth and colony formation
assay
Cell growth was determined by using an MTS assay
(Promega Corporation, Madison, WI, USA). Briefly, siRNA-transfected
HCC PLC/PRF/5 and Huh-7 cells transfected with either empty vectors
or 3xFlag-tagged Gls2 expressing vectors (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) were cultured at 37°C in a 96-well plate
in complete Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo
Fisher Scientific, Inc.) at 37°C for 2 weeks. This MTS Cell
Proliferation assay is based on the reduction of MTS tetrazolium
compound by viable cells to generate a colored formazan product
that is soluble in cell culture media. The quantity of formazan dye
produced by viable cells can be quantified and measured at 450 nm
absorbance after 1 h of incubation at 37°C with CellTiter
96® Aqueous One Solution Reagent (Promega Corporation,
Madison, WI, USA, according to the manufacturer's protocol.
Following 24 h of transfection at 37°C, 3×103
siRNA-transfected HCC PLC/PRF/5 and Huh-7 cells were placed in a
fresh 6-well plate and maintained in DMEM containing 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.) for 2 weeks.
Colonies were fixed at room temperature for 30 min in 1% methanol
and stained at room temperature with 0.1% crystal violet in 20%
methanol for 15 min.
Statistical analysis
The numerical data are expressed as the mean ±
standard deviation. Differences in proportion were analyzed by the
χ2 test or an unpaired Student's t-test and one-way
analysis of variance (ANOVA) with a Student-Newman-Keuls (S-N-K)
post hoc test, as required. The odds ratios and 95% confidence
intervals were calculated along with Fisher's exact P-values, where
appropriate. All calculations were performed with SPSS software
version 19.0 (SPSS Inc., Chicago, IL, USA). All experiments were
repeated at least three separate times. P<0.05 was considered to
indicate a statistically significant difference.
Results
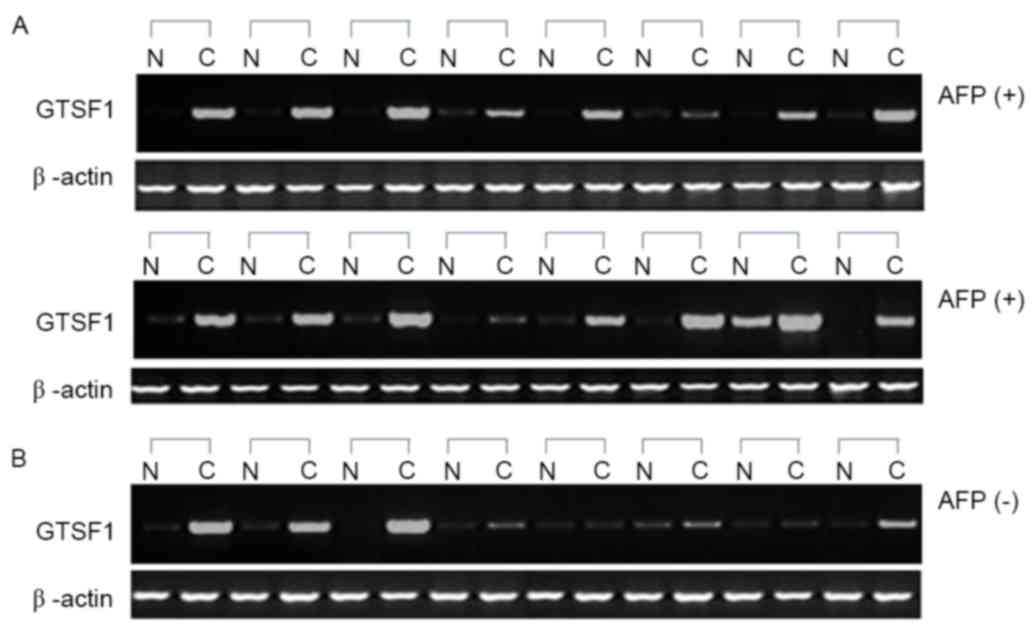
GTSF1 mRNA expression in human liver
cancer tissues and its association with AFP levels
The GTSF1 mRNA expression profiles of 32 normal
liver and 24 primary liver cancer samples, which were
histologically HCC, were analyzed. When comparing the 24 samples
from patients with liver cancer and their adjacent tissues, 22
exhibited significant differential expression of GTSF1 mRNA in
liver cancer tissues (Fig. 1). Only
1/32 healthy controls exhibited GTSF1 expression, with a complete
lack of expression being observed in the other 31 healthy subjects
and in the adjacent non-cancerous liver tissues of the 24 patients
with liver cancer. A higher frequency of GTSF1 mRNA expression was
observed in the samples from patients with HCC, compared with the
healthy control samples, as determined using an unpaired Student's
t-test (P<0.05). To examine the association between AFP levels
and GTSF1 mRNA expression, the χ2 test was used to
analyze the potential clinical implications of GTSF1 mRNA
expression. A total of 4/8 (50%) samples with negative AFP levels
exhibited GTSF1 mRNA expression, and 14/16 (87.5%) samples with
positive AFP levels exhibited GTSF1 mRNA expression (Fig. 1A and B). Although the ratio analysis
of the AFP-positive liver cancer samples (87.5%) was higher
compared with the AFP-negative liver cancer samples (50%), no
statistical significance between these two groups was observed
using a χ2 test (P=0.129). In addition, GTSF1 expression
was also compared in Hepatitis B virus (HBV)-infected HCC samples
and non-HBV-infected patient specimens to examine the association
between HBV infection and GTSF1 mRNA expression. A total of 5/7
(71.42%) samples that were HBV-negative exhibited GTSF1 mRNA
expression, and 13/17 (76.47%) samples that were HBV-positive
exhibited GTSF1 mRNA expression. No statistical significance
between these two groups was observed, using a χ2 test
(P=0.921).
Vector construction and GTSF1 mRNA
raises tumorigenicity in vivo
The significant overexpression of GTSF1 in liver
cancer samples prompted an examination of the potential biological
significance of GTSF1 in tumorigenesis. In order to confirm the
function of GTSF1 mRNA in liver cancer tumorigenicity, an Ad-shNC
vector, which was GTSF1-expressing (positive), was constructed
using GTSF1 mRNA gene sequencing. A negative control without the
GTSF1 vector sequence, termed Ad-shGTSF1 (GTSF1-negative control),
was also constructed. As an initial step, the capacity of colony
formation was evaluated in the HepG2 cells, which were transfected
with the Ad-shNC vector (GTSF1-positive) or the Ad-shGTSF1
(GTSF1-negative) control duplex. Ad-shNC vector- and
Ad-shGTSF1-transfected HepG2 cells (5×106 in 100 µl)
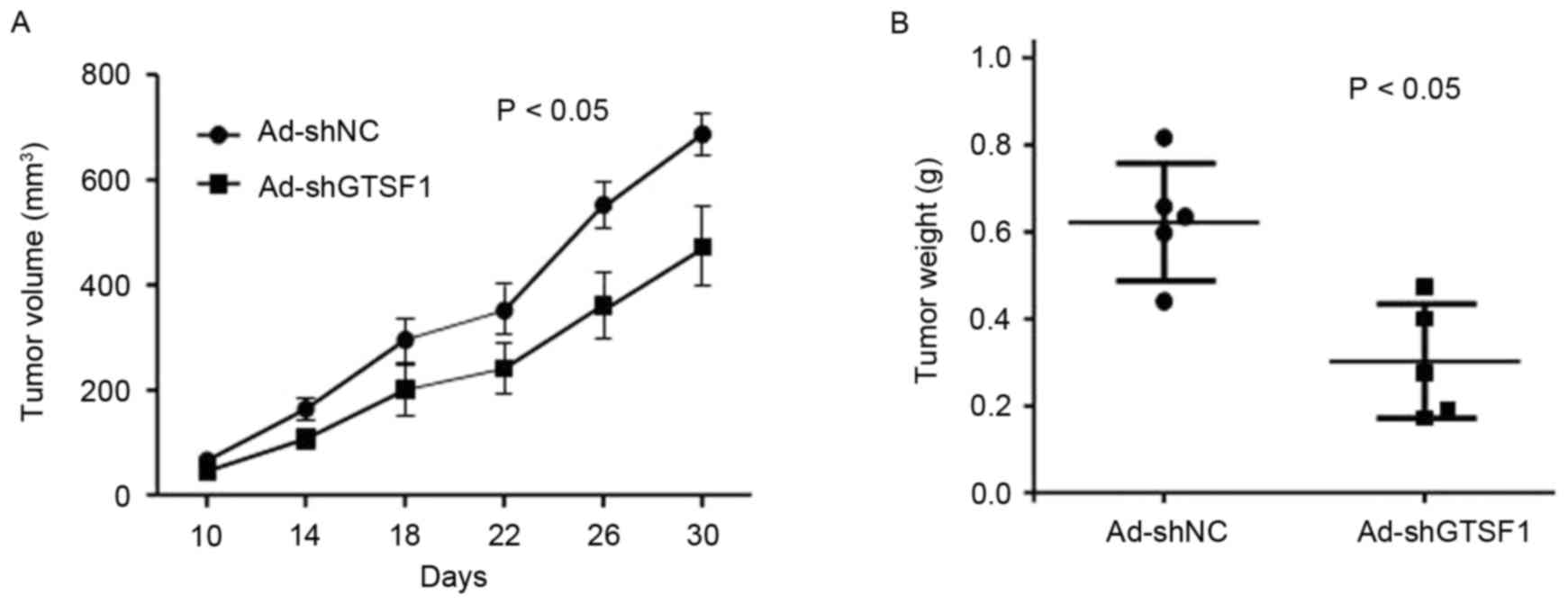
were injected subcutaneously into 6 nude mice. As demonstrated in
Fig. 2A, the tumor became visible at
10–30 days in the mice injected with Ad-shNCtransfected
(GTSF1-positive) HepG2 cells, and grew from 10–700 mm3
by the end of the observation period (30 days; mean size, 686±107
mm3 at the end of observation). By contrast, tumors
appeared at the injection sites of the mice treated with
Ad-shGTSF1-transfected (GTSF1-negative control) HepG2 cells, and
grew from 10–450 mm3 by the end of the observation
period (30 days; mean size, 448±92 mm3 at the end of
observation). A total of 30 days following injection, the sizes of
the tumors produced in the flanks of mice injected with GTSF1 was
increased compared with in those mice treated with Ad-shGTSF1
(GTSF1-negative control) (P<0.05). Consistently, the tumor
weight in mice following injection with Ad-shNC-transfected
(GTSF1-positive) HepG2 cells grew from 430–810 µg (mean weight,
610±98 µg by the end of observation), whereas mice treated with the
Ad-shGTSF1-transfected (GTSF1-negative control) HepG2 cells
exhibited tumors weighing 190–450 µg (mean weight, 308±73 µg by the
end of observation) (Fig. 2B;
P<0.05). There were significant differences in the sizes and
weights of tumors between those mice injected with GTSF1 and those
without it, as determined by a Student's t-test.
Expression of GTSF1 mRNA is a
prerequisite for proliferation in hepatoma cell lines
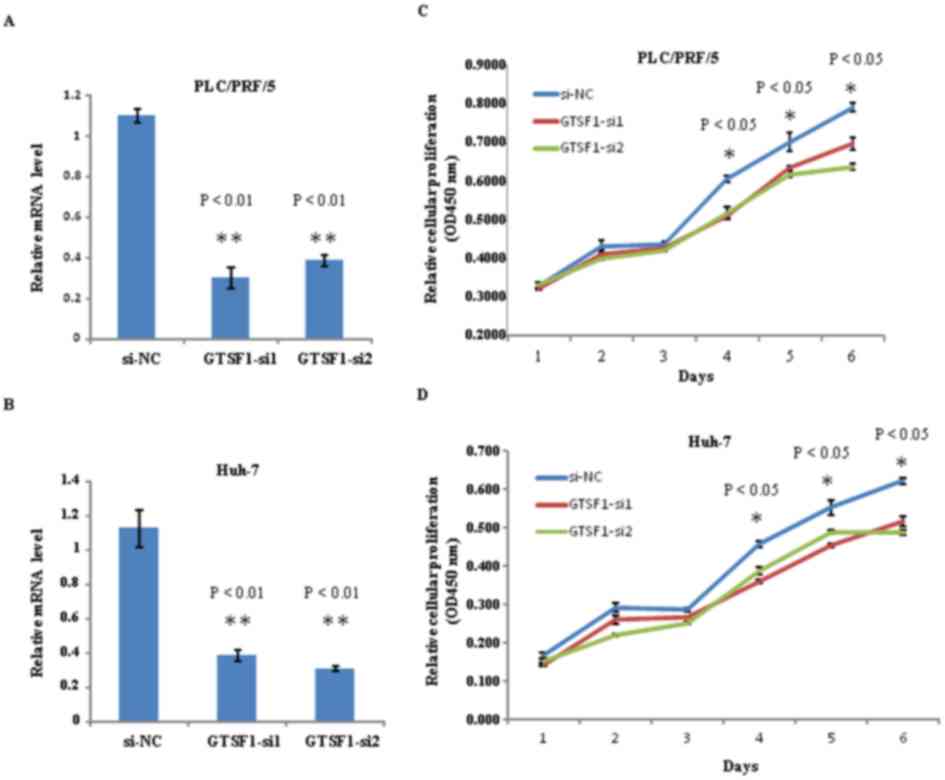
In order to investigate the function of GTSF1 mRNA,
two siRNA (GTSF1-siRNA1 and GTSF1-siRNA2) targeting GTSF1 were
designed to analyze the mRNA expression levels in PLC/PRF/5 and
Huh-7 cell lines. As an initial step in this protocol, the GTSF1
mRNA levels in PLC/PRF/5 and Huh-7 cells were examined, subsequent
to transfection with siRNA1-GTSF1, siRNA2-GTSF1 and GTSF1 mRNA
(si-NC). Using qPCR, reduced levels of GTSF1 mRNA were identified
in human liver cancer cells in the two groups with siRNA targeting
GTSF1, compared with the GTSF1 mRNA (si-NC). As indicated in
Fig. 3A and B, a 7 day-long regimen
of GTSF1 mRNA si-NC transfection was used to achieve a sustained
increased level of GTSF1 mRNA expression in the PLC/PRF/5 and Huh-7
cell lines, without two siRNAs, and analyzed using a Student's
t-test (P<0.01). To investigate the potential role of GTSF1 in
tumor proliferation, colony formation assays were used to measure
the proliferation of PLC/PRF/5 and Huh-7 cells transfected with
siRNA1-GTSF1 in duplicate, siRNA2-GTSF1 in duplicate, GTSF1 mRNA
(si-NC) or with no transfection. Notably, siRNA1-GTSF1 or siRNA2-
GTSF1-transfected cells exhibited fewer and smaller colonies
compared with GTSF1 mRNA-transfected and non-transfected cells from
days 4–6, as determined using a Student's t-test (P<0.05), as
demonstrated in Fig. 3C and D. This
result suggests that the GTSF1 gene serves an important role in the
proliferation of liver cancer cells. Notably, these results were
confirmed by one-way ANOVA followed by an S-N-K post-hoc test.
Consistent with the outcomes of the Student's t-tests, there were
significant differences between GTSF1 mRNA expression levels and
cell proliferation when comparing the GTSF1-transfected cells with
the siRNA1 or siRNA2- GTSF1-transfected cells (P<0.05); no
significant difference was present between the siRNA1- and the
siRNA2-GTSF1-transfected cells (P>0.05), as evaluated using
ANOVA and S-N-K analysis. These data indicate a
growth-proliferation role for GTSF1 mRNA expression, and suggest
that it is a prerequisite for the proliferation of hepatoma cell
lines.
Discussion
Hepatocarcinogenesis is a complex and multistep
process that involves the accumulation of genetic and epigenetic
alterations in regulatory genes. The identification of
cancer-associated molecules may lead to the development of novel
molecular targets for treatment, and of biomarkers for predicting
prognosis (11). The various
etiological factors of HCC, including HBV infection and hepatitis C
virus infection, may affect different signaling pathways (12). Therefore, multiple genetic and
epigenetic factors may affect HCC development (13). It has previously been reported in
several studies that the GTSF1 gene participates in DNA methylation
and retrotransposon activation in germ cells, particularly in cell
proliferation, and that it is present in MF tumor samples,
suggesting that it may serve a role in the apoptosis resistance of
MF (7,9,14).
Therefore, it may be concluded that the GTSF1 gene may serve a role
as a proliferation factor during various physiological processes,
including growth, development, differentiation and reproduction in
animals and plants. In terms of liver cancer, there have been
numerous studies regarding the molecular markers associated with
the development of liver cancer, and gene signatures with
diagnostic and prognostic potential have been identified by the
gene expression profiling of tumor tissues (15–17). In
the present study, it was identified that GTSF1 mRNA expression in
liver cancer tissue samples was increased compared with its
expression in adjacent non-tumor liver tissues and healthy control
liver tissues. Due to a statistically significant difference
between the patients with liver cancer and the healthy controls
(P<0.05), GTSF1 expression levels may be a potential biomarker
for liver cancer diagnosis. AFP is a glycoprotein known to be
expressed in HCC and is secreted into the blood of ~70% of patients
with liver cancer. Therefore, serum AFP levels are useful for the
early detection and differential diagnosis of HCC (18). For patients who undergo curative
hepatectomy for localized HCC, AFP levels are also useful for the
detection of recurrence, and are associated with prognosis
(19–21). Based on the data of the present study,
GTSF1 mRNA was expressed in 87.5% of AFP-positive samples, but only
in 50% of AFP-negative samples. Although there was no statistically
significant difference between these two groups (χ2
test, P=0.129), the proportion of samples that were GTSF1-positive
was almost equivalent to the number that were AFP-positive.
Notably, GTSF1 expression was identified in AFP-negative liver
cancer tissue samples, and may be a potential biomarker for
diagnosis in patients with AFP-negative liver cancer. In addition,
there was no statistical significance between GTSF1 expression in
the non-HBV-infected liver cancer samples and the HBV-infected
liver cancer specimens observed. It may be hypothesized that GTSF1
expression in liver cancer samples is irrelevant to HBV infection.
Notably, GTSF1 expression was significantly upregulated in the
majority of liver cancer tissues examined, but was not expressed in
the adjacent non-tumor normal liver tissue. These results suggest
that increased GTSF1 expression is a frequent event in human liver
cancer tissues and may be involved in hepatocarcinogenesis.
During tumor progression, the number, type,
distribution and expression levels of tumor markers in patients
with liver cancer exhibit variations that are closely associated
with the occurrence, development, metastasis, treatment response
and prognosis of tumors and patients (22). Few data are available concerning the
molecular mechanisms by which mRNAs modulate the process of
tumorigenesis and the behavior of cancer cells. To explore the
roles of GTSF1 in the liver in vivo, transfected HepG2 cells
were generated and injected to a mouse model using a clone of a
constructed GTSF1 gene plasmid. In agreement with previous
observations that GTSF1 was frequently upregulated in hepatoma cell
lines, and that GTSF1 may increase colony formation in
vitro, it was demonstrated that GTSF1 promoted tumor growth
in vivo. All these data emphasize a fundamental role of
GTSF1 in tumorigenesis, particularly in the development of liver
cancer. The present study revealed that GTSF1 significantly
increased tumorigenicity in Ad-shNC-transfected (GTSF1-positive)
HepG2 cells in a nude mouse xenograft model, whereas the absence of
GTSF1 inhibited increases in the size and weight of the tumors.
Therefore, GTSF1 expression may serve a critical role in the
proliferation of HCC tumor cells in vivo. Notably, the HepG2
cell line, originally thought to be an HCC cell line, was
identified as a hepatoblastoma cell line (23). This is important, as differences exist
between HepG2 cells and native human hepatocytes, including in the
drug-processing proteins in liver tissues and the genetic changes
that occur in HCC; these changes can affect protein concentrations
and the copy number (24–26). Conversely, genetic changes involved in
the development of liver cancer, the cancer protein secretomes and
the biological significance of human leucocyte antigen expression
in HCC tumors closely resemble those of the HepG2 cells observed in
previous studies (27–29).
Compared with previous studies, the results of the
present study indicated that GTSF1-transfected HepG2 cells injected
into mice produced tumors larger in size (mean, 686±107
mm3) and weight (mean, 610±98 µg) compared with the
tumors observed in the mice injected with GTSF1-negative HepG2
cells (mean size, 448±92 mm3; mean weight, 308±73 µg;
Fig. 2A and B). Therefore, the tumors
were bigger and heavier in the model that used GTSF1-transfected
HepG2 cells, compared with in those cells transfected with the
negative control, which confirmed that the introduction of GTSF1
significantly increases tumorigenicity in vivo. Although the
misidentified hepatoblastoma HepG2 cell line was used in the
present study, this is unlikely to have affected the ability of
GTSF1 to initiate tumor growth in vivo when using HepG2
cells as the vector. Due to the limitations of using the HepG2 cell
line, these results may indicate that GTSF1 significantly promoted
in vivo tumorigenicity in malignant liver tumors in general,
rather than in HCC specifically. The mechanism for this apparent
GTSF1 gene proliferation function in HCC in vivo should be
elucidated using verified HCC cell lines in the future.
To validate the effects of the GTSF1 gene on cell
proliferation and growth, siRNA protocols were used in the present
study. As it is possible to generate siRNAs and miRNAs targeted
against any cellular RNA, these molecules may be used to
downregulate the expression of almost any disease-causing gene
(30–32). The in vitro data obtained
during the present study indicated that GTSF1 knockdown by siRNA1
or siRNA2 significantly reduced the proliferation of PLC/PRF/5 and
Huh-7 cells. The cell proliferation, as evaluated by colony
formation, of liver cancer cell lines without GTSF1 siRNA was
significantly decreased in HCC cells. In addition, the levels of
GTSF1 expression were significantly decreased following GTSF1
knockdown in PLC/PRF/5 and Huh-7 cells. Therefore, it was
hypothesized that the result of interfering with the expression of
GTSF1 in PLC and HepG2 cells may be due to GTSF1 overexpression in
the tissues of patients with liver cancer, and that this result may
be associated with the observations concerning HCC tumor size and
weight from the mouse model. These results suggest that the
overexpression of GTSF1 is involved in regulating liver cancer
proliferation. The use of siRNA/miRNA is considered to have great
therapeutic potential, as it is possible to generate
siRNA/miRNA-based silencing of any gene implicated in HCC (33). Notably, GTSF1 mRNA expression was
significantly upregulated in the majority of the cancer cell lines
and cancer tissues examined, and that GTSF1 not only increased
colony formation in vitro but also tumorigenicity in
vivo.
In summary, the present study revealed the GTSF1
mRNA expression profile in liver cancer, and examined the potential
role of GTSF1 in tumorigenesis. The data suggest an important role
for the GTSF1 gene in the molecular etiology of
hepatocarcinogenesis, and suggest the potential application of
GTSF1 mRNA expression in liver cancer diagnosis and therapy. Due to
the limitations associated with the small number of cases involved,
the absence of data on hepatoblastoma tissues derived from
patients, and the patient cohort all originated from a single
region in the present study, the function of GTSF1 mRNA and protein
expression in liver cancer requires additional study.
Acknowledgements
The present study was supported by the Science of
Shanghai Songjiang District Fund in China (grant no. 13SJGGYY28),
and by the Key project of Shanghai Songjiang District Planning and
Growth Committee (grant no. 2012-III).
References
|
1
|
Balogh J, Victor D III, Asham EH,
Burroughs SG, Boktour M, Saharia A, Li X, Ghobrial RM and Monsour
HP Jr.: Hepatocellular carcinoma: A review. J Hepatocell Carcinoma.
3:41–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Caldwell S and Park SH: The epidemiology
of hepatocellular cancer: From the perspectives of public health
problem to tumor biology. J Gastroenterol. 44 Suppl 19:S96–S101.
2009. View Article : Google Scholar
|
|
4
|
Ferenci P, Fried M, Labrecque D, Bruix J,
Sherman M, Omata M, Heathcote J, Piratsivuth T, Kew M, Otegbayo JA,
et al: Hepatocellular carcinoma (HCC): A global perspective. J Clin
Gastroenterol. 44:239–245. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tsuchiya N, Sawada Y, Endo I, Saito K,
Uemura Y and Nakatsura T: Biomarkers for the early diagnosis of
hepatocellular carcinoma. World J Gastroenterol. 21:10573–10583.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zucman-Rossi J, Villanueva A, Nault JC and
Llovet JM: Genetic landscape and biomarkers of hepatocellular
carcinoma. Gastroenterology. 149:1226–1239.e4. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yoshimura T, Toyoda S, Kuramochi-Miyagawa
S, Miyazaki T, Miyazaki S, Tashiro F, Yamato E, Nakano T and
Miyazaki J: Gtsf1/Cue110, a gene encoding a protein with two copies
of a CHHC Zn-finger motif, is involved in spermatogenesis and
retrotransposon suppression in murine testes. Dev Biol.
335:216–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Krotz SP, Ballow DJ, Choi Y and Rajkovic
A: Expression and localization of the novel and highly conserved
gametocyte-specific factor 1 during oogenesis and spermatogenesis.
Fertil Steril. 91(5 Suppl): S2020–S2024. 2009. View Article : Google Scholar
|
|
9
|
van Kester MS, Borg MK, Zoutman WH,
Out-Luiting JJ, Jansen PM, Dreef EJ, Vermeer MH, van Doorn R,
Willemze R and Tensen CP: A meta-analysis of gene expression data
identifies a molecular signature characteristic for tumor-stage
mycosis fungoides. J Invest Dermatol. 132:2050–2059. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shimizu D, Inokawa Y, Sonohara F, Inaoka K
and Nomoto S: Search for useful biomarkers in hepatocellular
carcinoma, tumor factors and background liver factors (Review).
Oncol Rep. 37:2527–2542. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shao YY, Hsu CH and Cheng AL: Predictive
biomarkers of antiangiogenic therapy for advanced hepatocellular
carcinoma: Where are we? Liver Cancer. 2:93–107. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Llovet JM and Bruix J: Molecular targeted
therapies in hepatocellular carcinoma. Hepatology. 48:1312–1327.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yoshimura T, Miyazaki T, Toyoda S,
Miyazaki S, Tashiro F, Yamato E and Miyazaki J: Gene expression
pattern of Cue110: A member of the uncharacterized UPF0224 gene
family preferentially expressed in germ cells. Gene Expr Patterns.
8:27–35. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tanabe KK, Lemoine A, Finkelstein DM,
Kawasaki H, Fujii T, Chung RT, Lauwers GY, Kulu Y, Muzikansky A,
Kuruppu D, et al: Epidermal growth factor gene functional
polymorphism and the risk of hepatocellular carcinoma in patients
with cirrhosis. JAMA. 299:53–60. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li M, Zhao H, Zhang X, Wood LD, Anders RA,
Choti MA, Pawlik TM, Daniel HD, Kannangai R, Offerhaus GJ, et al:
Inactivating mutations of the chromatin remodeling gene ARID2 in
hepatocellular carcinoma. Nat Genet. 43:828–829. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jin F, Xiong WJ, Jing JC, Feng Z, Qu LS
and Shen XZ: Evaluation of the association studies of single
nucleotide polymorphisms and hepatocellular carcinoma: A systematic
review. J Cancer Res Clin Oncol. 137:1095–1104. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Snowberger N, Chinnakotla S, Lepe RM,
Peattie J, Goldstein R, Klintmalm GB and Davis GL: Alpha
fetoprotein, ultrasound, computerized tomography and magnetic
resonance imaging for detection of hepatocellular carcinoma in
patients with advanced cirrhosis. Aliment Pharmacol Ther.
26:1187–1194. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Grąt M, Kornasiewicz O, Lewandowski Z,
Hołówko W, Grąt K, Kobryń K, Patkowski W, Zieniewicz K and Krawczyk
M: Combination of morphologic criteria and α-fetoprotein in
selection of patients with hepatocellular carcinoma for liver
transplantation minimizes the problem of posttransplant tumor
recurrence. World J Surg. 38:2698–2707. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Raoul JL, Park JW, Kang YK, Finn RS, Kim
JS, Yeo W, Polite BN, Chao Y, Walters I, Baudelet C and Lencioni R:
Using modified RECIST and alpha-fetoprotein levels to assess
treatment benefit in hepatocellular carcinoma. Liver Cancer.
3:439–450. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Furihata T, Sawada T, Kita J, Iso Y, Kato
M, Rokkaku K, Shimoda M and Kubota K: Serum alpha-fetoprotein level
per tumor volume reflects prognosis in patients with hepatocellular
carcinoma after curative hepatectomy. Hepatogastroenterology.
55:1705–1709. 2008.PubMed/NCBI
|
|
22
|
Zhao YJ, Ju Q and Li GC: Tumor markers for
hepatocellular carcinoma. Mol Clin Oncol. 1:593–598. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
López-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009. View Article : Google Scholar
|
|
24
|
Hart SN, Li Y, Nakamoto K, Subileau EA,
Steen D and Zhong XB: A comparison of whole genome gene expression
profiles of HepaRG cells and HepG2 cells to primary human
hepatocytes and human liver tissues. Drug Metab Dispos. 38:988–994.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wiśniewski JR, Vildhede A, Norén A and
Artursson P: In-depth quantitative analysis and comparison of the
human hepatocyte and hepatoma cell line HepG2 proteomes. J
Proteomics. 136:234–247. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Capes-Davis A, Theodosopoulos G, Atkin I,
Drexler HG, Kohara A, MacLeod RA, Masters JR, Nakamura Y, Reid YA,
Reddel RR and Freshney RI: Check your cultures! A list of
cross-contaminated or misidentified cell lines. Int J Cancer.
127:1–8. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cevik D, Yildiz G and Ozturk M: Common
telomerase reverse transcriptase promoter mutations in
hepatocellular carcinomas from different geographical locations.
World J Gastroenterol. 21:311–317. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Srisomsap C, Sawangareetrakul P,
Subhasitanont P, Chokchaichamnankit D, Chiablaem K, Bhudhisawasdi
V, Wongkham S and Svasti J: Proteomic studies of cholangiocarcinoma
and hepatocellular carcinoma cell secretomes. J Biomed Biotechnol.
2010:4371432010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wadee AA, Paterson A, Coplan KA and Reddy
SG: HLA expression in hepatocellular carcinoma cell lines. Clin Exp
Immunol. 97:328–333. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dapas B, Farra R, Grassi M, Giansante C,
Fiotti N, Uxa L, Rainaldi G, Mercatanti A, Colombatti A, Spessotto
P, et al: Role of E2F1-cyclin E1-cyclin E2 circuit in human
coronary smooth muscle cell proliferation and therapeutic potential
of its downregulation by siRNAs. Mol Med. 15:297–306. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Farra R, Dapas B, Pozzato G, Giansante C,
Heidenreich O, Uxa L, Zennaro C, Guarnieri G and Grassi G: Serum
response factor depletion affects the proliferation of the
hepatocellular carcinoma cells HepG2 and JHH6. Biochimie.
92:455–463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Spänkuch B and Strebhardt K: Combinatorial
application of nucleic acid-based agents targeting protein kinases
for cancer treatment. Curr Pharm Des. 14:1098–1112. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Farra R, Grassi M, Grassi G and Dapas B:
Therapeutic potential of small interfering RNAs/micro interfering
RNA in hepatocellular carcinoma. World J Gastroenterol.
21:8994–9001. 2015. View Article : Google Scholar : PubMed/NCBI
|