Introduction
Breast cancer is the most frequently diagnosed
malignancy in women worldwide and poses a serious threat to women's
health (1). Breast cancer is the
second-leading cause of cancer-associated mortality in 2015,
accounting for ~11% of the total cancer mortalities in women
(2,3).
Although substantial progress has been made in surgical management
and chemotherapy treatment, the rate of relapse in patients with
advanced breast cancer remains high (4–6). It is
therefore essential to investigate further the molecular mechanisms
underlying breast cancer development.
Previous findings concerning microRNAs (miRNAs/miRs)
have substantially broadened knowledge concerning cancer
pathogenesis (7,8). MiRNAs are a class of short non-coding
RNA of ~20–22 nucleotides that can bind to complementary sequences
in the 3′ untranslated region (3′UTR) of mRNA to regulate gene
expression. Therefore, miRNAs are involved in a number of cellular
processes, including proliferation, apoptosis, development and
differentiation (9). Numerous studies
have demonstrated that miRNAs can perform oncogenic or tumor
suppressive roles in various types of cancers. Previous studies
have demonstrated the functional roles of miR-433 in several types
of cancer, including retinoblastoma (10), ovarian cancer (11), bladder cancer (11), oral squamous cell carcinoma (OSCC)
(12), gastric cancer (13), hepatocellular carcinoma (HCC)
(14) and lung cancer (15). The downstream targets of miR-433 have
been revealed to be strongly associated with cancer development,
including Notch1, cAMP responsive element binding protein 1
(CREB1), paired box protein Pax-6 (PAX6), histone deacetylase 6
(HDAC6), kirsten rat sarcoma viral oncogene homolog and hepatocyte
growth factor receptor (c-Met) (10,12,13,16).
MiR-433 expression was found to be downregulated in these cancer
types, and miR-433 was demonstrated to function as a tumor
suppressor by inhibiting cell proliferation, migration and
differentiation (10,12–14,16).
However, to the best of our knowledge, the role of miR-433 in
breast cancer development is largely unknown.
In the present study, miR-433 was downregulated in
breast cancer tissues and breast cancer cell lines. In vitro
functional assay indicated that miR-433 was able to inhibit cell
proliferation, reduce cell viability and also decrease apoptosis in
breast cancer cell lines. It was revealed that RAC-γ
serine/threonine-protein kinase (AKT3) is a direct target of
miR-433 in breast cancer cell lines. More importantly, analysis of
clinical specimens further indicated that the level of miR-433
expression was inversely correlated with AKT3 expression in breast
cancer tissues.
Materials and methods
Human clinical samples
Paired breast cancer and adjacent normal breast
cancer tissues (~5 cm away from cancerous tissues) were obtained
from 42 patients (mean age, 53 years old, age range: 27–68 years
old) undergoing surgery in Nanfang Hospital (Guangzhou, China). The
clinical samples were collected between July 2012 and December
2015. Informed consent was obtained from all patients enrolled in
the present study, and all clinical studies were approved by the
Ethics Committee of Nanfang Hospital. All the collected samples
were snap-frozen for further reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis.
Cell lines and cell culture
The human breast epithelial MCF-10A cell line, and
breast cancer cell lines BT-549, MCF-7, MDA-MB-453 and MDA-MB-231
were purchased from the Shanghai Institute of Cell Biology
(Shanghai, China). All cells were cultured in Dulbecco's modified
Eagle's medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher
Scientific, Inc.) under a humidified atmosphere of 5%
CO2 at 37°C.
RNA isolation and RT-qPCR
miRNA and mRNA were extracted from clinical samples
and cell lines using TRIzol reagent (Takara Biotechnology Co.,
Ltd., Dalian, China), according to the manufacturer's protocol.
MiRNAs were reverse transcribed into cDNA using the One Step
PrimeScript miRNA cDNA Synthesis kit (Takara Biotechnology Co.,
Ltd., Dailan, China), and mRNA was reverse transcribed into cDNA
using the PrimeScript RT Reagent kit (Takara Biotechnology Co.,
Ltd.). RT-qPCR was performed by using the SYBR Green PCR kit
(Takara Biotechnology Co., Ltd.) and the ABI 7500 FAST Real-Time
PCR system (Applied Biosystems, Thermo Fisher Scientific, Inc.).
The sequences of the primers used are as follows: miR-433 forward,
5′-TGCGGTACGGTGAGCTGTC-3′ and reverse, 5′-CCAGTGCAGGGTCCGAGGT-3′;
AKT3 forward, 5′-ATGAGCGATGTTACCATTGT-3′ and reverse,
5′-CAGTCTGTCTGCTACAGCCTGGATA-3′; U6 forward:
5′-CGCTTCGGCAGCACATATAC-3′, reverse: 5′-TTCACGAATTTGCGTGTCAT-3′;
GAPDH forward, 5′-GGTGAAGGTCGGAGTCAACG-3′, and reverse:
5′-CAAAGTTGTCATGGATGHACC-3′. The qPCR thermocycling conditions used
were 95°C for 30 sec; 40 cycles of 95°C for 5 sec and 60°C for 30
sec; 95°C for 15 sec, 60°C for 60 sec and 95°C for 15 sec. The
relative expression level of miR-433 and AKT3 mRNA were calculated
using 2−∆∆Cq method (17)
following normalization to U6 and GAPDH, respectively. All the
experiments were performed in triplicates.
Reagents and transfection
The miR-433 mimics, and miR-433 inhibitors
(anti-miR-433), as well as respective controls (miR-Ctrl and
anti-miR-Ctrl), and the small interfering RNA (siRNA) targeting
AKT3 (siAKT3), as well as its negative control (siNC), were
obtained from Shanghai GeneChem, Inc. (Shanghai, China). The pGCL
and pGCL-AKT3 plasmids were purchased from Shanghai GeneChem Co.,
Ltd. (Shanghai, China). In vitro transfection of these
oligonucleotides (50 nM) was performed using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in
accordance with the manufacturer's instructions. At 48 h
post-transfection, cells were processed for further
experimentation.
Cell proliferation assay
Breast cancer cells (BT-549 and MDA-MB-231) were
plated in 96-well plates overnight, and the cells were transfected
with 50 nM miR-433 mimics, or anti-miR-433, or their respective
controls (miR-Ctrl, anti-miR-Ctrl) using Lipofectamine 2000
reagent. At 48 h post-transfection, cells were further cultured in
DMEM supplemented with 10% FBS for 0, 24, 48 or 72 h prior to the
addition of MTT to each well. Following incubation with MTT for 4 h
at 37°C, MTT solution was removed, and dimethyl sulfoxide (DMSO)
was added to each well. Absorbance was then measured at 490 nm
using a spectrophotometer. For the AKT3 siRNA study and rescue
experiments, the cells were transfected with siAKT3 or the negative
control (siNC), miR-Ctrl with pGCL, miR-433 mimics + pGCL or
miR-433 mimcis + pGCL-AKT3. A total of 48 h after transfection, the
cells were further cultured for 48 h at 37°C and the MTT assay was
performed as aforementioned.
Cell viability assay
Breast cancer cells (BT-549 and MDA-MB-231) were
plated in 96-well plates overnight, and the cells were transfected
with miR-433 mimics, anti-miR-433, miR-Ctrl or anti-miR-Ctrl. A
total of 48 h after transfection, the cells were further cultured
in serum-free DMEM for 0, 24, 48 or 72 h prior to the addition of
MTT to each well. Following incubation with MTT for 4 h at 37°C,
the MTT solution was removed and DMSO was added to each well
followed by measuring the absorbance at 490 nm using a
spectrophotometer. For the AKT3 siRNA study and rescue experiment,
the cells were transfected with siAKT3 or the negative control
(siNC), miR-Ctrl + pGCL, miR-433 mimics + pGCL, or miR-433 mimcis +
pGCL-AKT3, and 48 h after transfection, the cells were further
cultured for 48 h at 37°C before MTT assay was conducted as
aforementioned.
Luciferase reporter assay
The predicted targets of miR-433 were analyzed by
using TargetScan (version 7.1), and AKT3 was found to one of the
targets of miR-433. The entire human AKT3 3′UTR, harboring the
miR-433 target sequence as well as the mutated (MUT) seed sequence,
were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China).
The AKT3 3′UTR reporter was generated by inserting the entire
wild-type (WT) 3′UTR or MUT 3′UTR of AKT3 mRNA into psiCHECK-2
vector (Promega Corporation, Madison, WI, USA). For the luciferase
reporter assay, the cells (BT-549 and MDA-MB-231) were
co-transfected with the AKT3 3′UTR reporter and the miR-433 mimics,
anti-miR-433 or their respective controls (miR-Ctrl, anti-miR-Ctrl)
using the Lipofectamine 2000 reagent, and 48 h following
transfection, firefly and Renilla luciferase activities were
measured by using the Dual Luciferase Assay kit (Promega
Corporation, Madison, WI, USA).
Cell apoptosis analysis
Breast cancer cells (BT-549 and MDA-MB-231) were
plated in 6-well plates overnight, and the cells were then
transfected with miR-433 mimics, anti-miR-433 or respective
controls (miR-Ctrl and anti-miR-Ctrl). At 48 h after transfection,
the cells were harvested and processed, and double staining with
annexin V-fluorescein isothiocyanate (FITC) and propidium iodide
was performed using the Annexin V-FITC Apoptosis Detection kit (BD
Biosciences, Franklin Lakes, NJ, USA) according to the
manufacturer's protocol. Cell apoptosis was analyzed with a FACScan
flow cytometer (BD Biosciences), and the cell apoptotic rates were
analyzed by the BD CellQuest Pro™ Software (Version 5.1; BD
Biosciences).
Western blot analysis
Proteins from breast cancer cells were extracted
using a modified Radioimmunoprecipitation Assay buffer
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) with proteinase
inhibitor cocktail (Complete mini; Roche Applied Science, Penzberg,
Germany). The concentration of protein was measured using the BCA
kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA) Protein lysates
(30 µg) were separated on 10 % SDS-PAGE and transferred to
polyvinylidene difluoride membranes. The membranes were blocked
with 5% skimmed milk for 1 h at room temperature, followed by
incubating with the following primary antibodies: Rabbit polyclonal
anti-B-cell lymphoma 2 (Bcl-2) antibody (1:1,500; cat. no. ab59348;
Abcam, Cambridge, USA), rabbit polyclonal anti-Bcl-associated X
(Bax) antibody (1:1,000; cat. no. ab53154; Abcam), rabbit
polyclonal anti-AKT3 antibody (1:1,000, cat. no. ab189643; Abcam)
and rabbit polyclonal anti-GAPDH antibody (1:2,000, cat. no.
ab9485; Abcam) overnight at 4°C. The membranes were subsequently
incubated with horseradish peroxidase conjugated secondary antibody
(1:2,000, cat. no. ab205718; Abcam) and visualized with enhanced
chemiluminescence reagent (GE Healthcare Life Sciences, Little
Chalfont, UK) according to the manufacturer's protocol.
Statistical analysis
All data are presented as the mean ± standard
deviation, and data analysis was performed using GraphPad Prism
(version 6.0; GraphPad Software, Inc., La Jolla, CA, USA).
Significant differences between groups for clinical samples were
analyzed by paired t-test. For in vitro functional assay,
significant differences were analyzed by one-way analysis of
variance followed by Dunnett's multiple comparison test or unpaired
t-test. The correlation between miR-433 levels and AKT3 mRNA levels
in breast cancer tissues was analyzed by Spearman's correlation
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
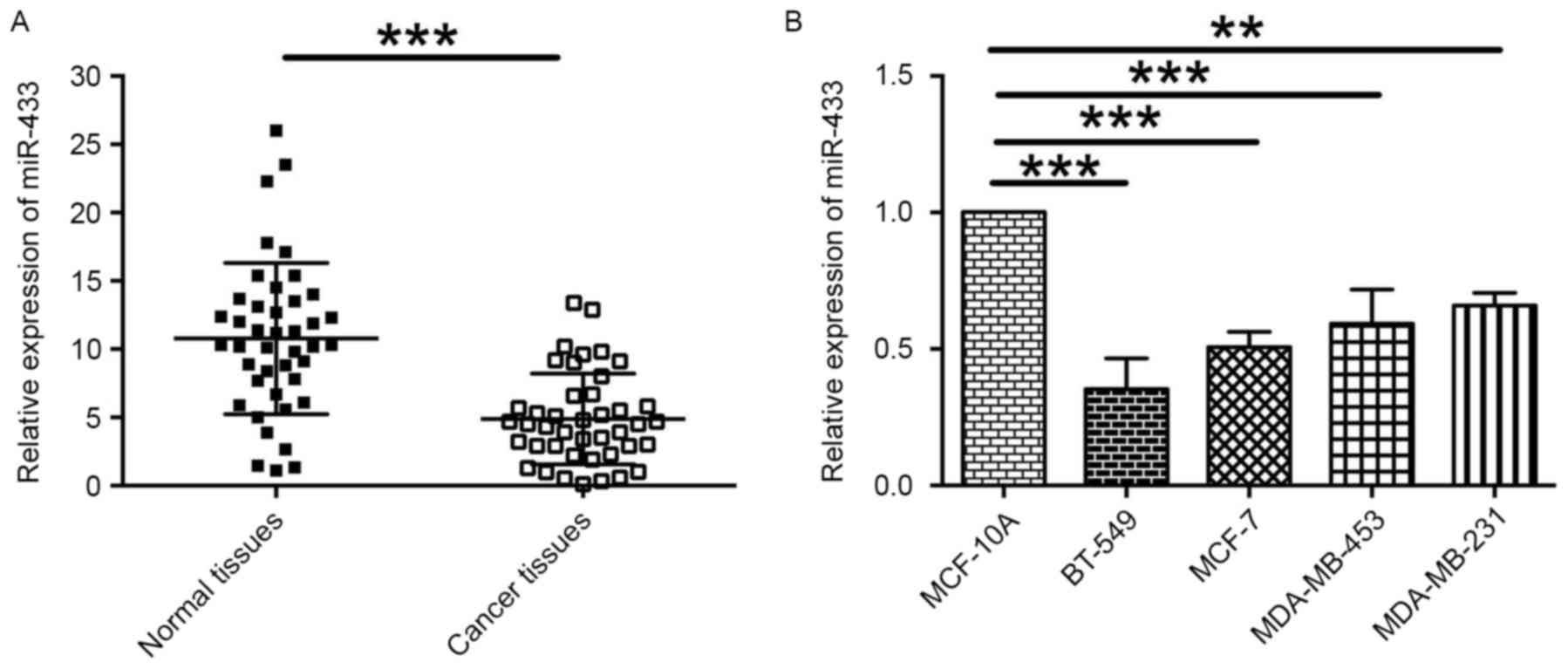
miR-433 expression is downregulated in
breast cancer tissues and breast cancer cell lines
To assess the role of miR-433 in breast cancer, the
levels of miR-433 expression in 42 paired breast cancer and normal
adjacent breast tissues were examined by RT-qPCR. The results
revealed that the level of miR-433 expression was significantly
lower in cancer tissues compared with normal adjacent breast
tissues (Fig. 1A). In addition, the
relative expression levels of miR-433 in different breast cancer
cells lines were also determined. It was indicated that the levels
of miR-433 expression in the breast cancer cell lines BT-549,
MCF-7, MDA-MB-453 and MDA-MB-231 was downregulated when compared
with the expression level of miR-433 in normal breast cell line,
MCF-10A (Fig. 1B). These data
indicate that miR-433 may be a tumor suppressor in breast
cancer.
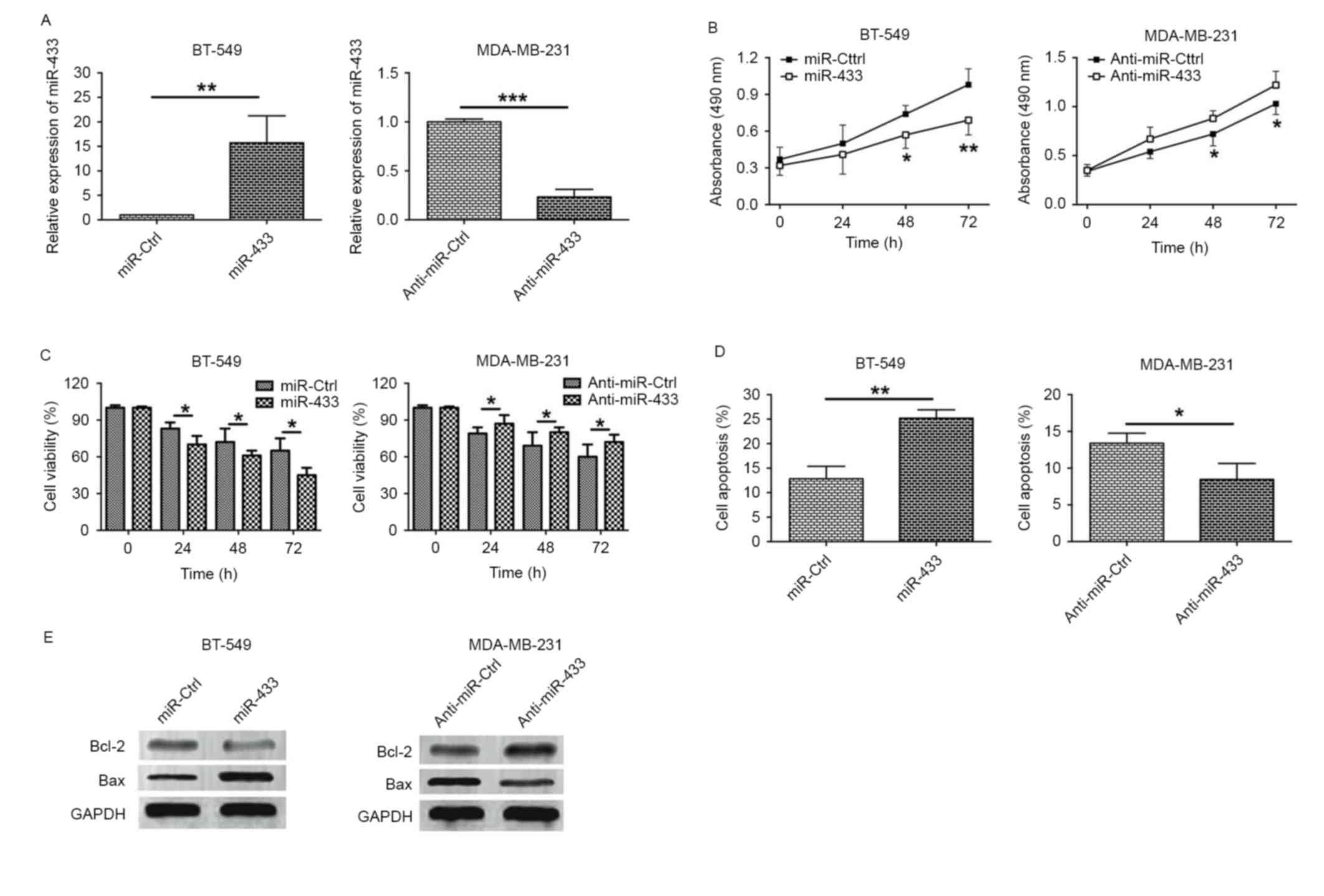
miR-433 inhibits proliferation in
breast cancer cell lines
To gain insights into the underlying mechanisms of
action of miR-433 in breast cancer, in vitro functional
assays including MTT and cell viability assays were performed.
RT-qPCR results revealed that the expression level of miR-433 was
increased >10 fold in BT-549 cells transfected with miR-433
mimics, while the expression level of miR-433 was significantly
decreased in MDA-MB-231 cells transfected with anti-miR-433
(Fig. 2A). The MTT assay revealed
that BT-549 cells transfected with miR-433 mimics exhibited lower
proliferative activity when compared with cells transfected with
miR-Ctrl. Additionally, the downregulation of miR-433 in MDA-MB-231
cells by transfection with anti-miR-433 was able to increase cell
proliferation (Fig. 2B). The
viability of BT-549 cells transfected with miR-Ctrl or miR-433
mimics and MDA-MB-231 cells transfected with anti-miR-Ctrl or
anti-miR-433 was also examined. As shown in Fig. 2C, transfection with miR-433 mimic in
BT-549 cells reduced cell viability when compared with miR-Ctrl
transfection, whereas MDA-MB-231 cells transfected with
anti-miR-433 exhibited a higher cell viability when compared with
cells transfected with anti-miR-Ctrl (Fig. 2C). These results indicate that miR-433
exerts a tumor suppressive function in breast cancer cells.
miR-433 induces apoptosis in breast
cancer cell lines
To examine the role of miR-433 on cell apoptosis,
flow cytometry and western blotting experiments were performed.
Flow cytometry results demonstrated that transfection of BT-549
cells with the miR-433 mimic significantly increased the proportion
of apoptotic cells when compared with cells transfected with
miR-Ctrl, whereas the proportion of apoptotic MDA-MB-231 cells
transfected with anti-miR-433 was significantly lower compared with
cells transfected with anti-miR-Ctrl (Fig. 2D). Expression levels of the
apoptosis-associated proteins, including Bcl-2 and Bax, were
examined. Overexpression of miR-433 in BT-549 cells decreased the
level of Bcl-2 protein expression and increased the level of Bax
protein expression, whereas downregulation of miR-433 in MDA-MB-231
cells upregulated Bcl-2 expression and downregulated Bax expression
(Fig. 2E).
AKT3 is a direct target of
miR-433
To predict the downstream targets of miR-433, the
predicted targets were analyzed by using TargetScan (version 7.1).
Of the predicted targets, AKT3 was selected for further experiments
owing to its known role in cancer development (18). To confirm that AKT3 was a target of
miR-433, AKT3 3′UTR luciferase reporter plasmids containing WT or
MUT potential binding sites for miR-433 were constructed and a
dual-luciferase reporter assay was conducted (Fig. 3A). The results of this assay revealed
that overexpression of miR-433 significantly decreased luciferase
activity in BT-549 cells transfected with plasmids containing the
WT 3′UTR of AKT3, but not in cells transfected with plasmids
containing the MUT 3′UTR of AKT3 (Fig.
3B). Downregulation of miR-433 by anti-miR-433 transfection
significantly increased luciferase activity in MDA-MB-231 cells
transfected with plasmids containing the WT 3′UTR of AKT3, but not
in cells transfected with plasmids containing the MUT 3′UTR of AKT3
(Fig. 3B). In addition, BT-549 cells
transfected with miR-433 mimics expressed lower levels of ATK3 mRNA
compared with cells transfected with miR-Ctrl (Fig. 3C), and MDA-MB-231 cells transfected
with anti-miR-433 exhibited increased AKT3 mRNA the expression
levels of AKT3 mRNA when compared to with cells transfected with
anti-miR-Ctrl (Fig. 3C). These
results suggest that AKT3 may be a direct target of miR-433.
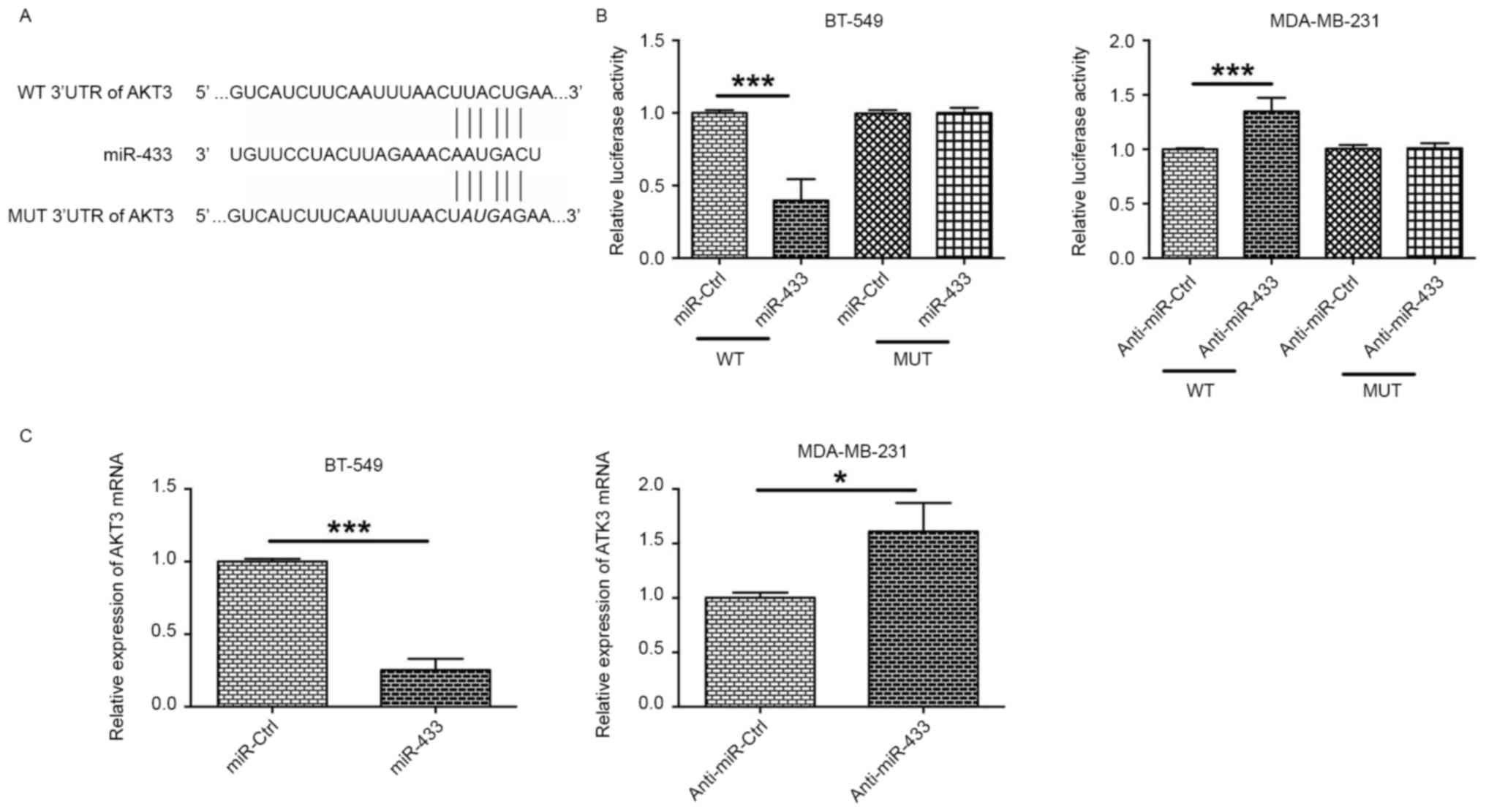 | Figure 3.AKT3 is a direct target of miR-433.
(A) Schematic representation of miR-433 putative binding sites in
the 3′UTR of AKT3 mRNA, and the mutations in the 3′UTR region of
AKT3. (B) In BT-549 cells, the relative luciferase activities were
measured following the co-transfection of the cells with miR-Ctrl
(or miR-433 mimics) and WT 3′UTR (or MUT 3′UTR). In MDA-MB-231, the
relative luciferase activities were measured following the
co-transfection of the cells with anti-miR-Ctrl (or anti-miR-433)
and WT 3′UTR (or MUT 3′UTR). (C) The relative expression of AKT3
mRNA in BT-549 cells transfected with miR-Ctrl or miR-433, and in
MDA-MB-231 cells transfected with anti-miR-Ctrl or anti-miR-433 was
examined by reverse transcription-quantitative polymerase chain
reaction. *P<0.05, ***P<0.001 (unpaired t-test; n=3). AKT3,
RAC-γ serine/threonine-protein kinase; miR, microRNA; 3′UTR, 3′
untranslated region; WT, wild-type; MUT, mutated; miR-Ctrl, control
scrambled microRNA. |
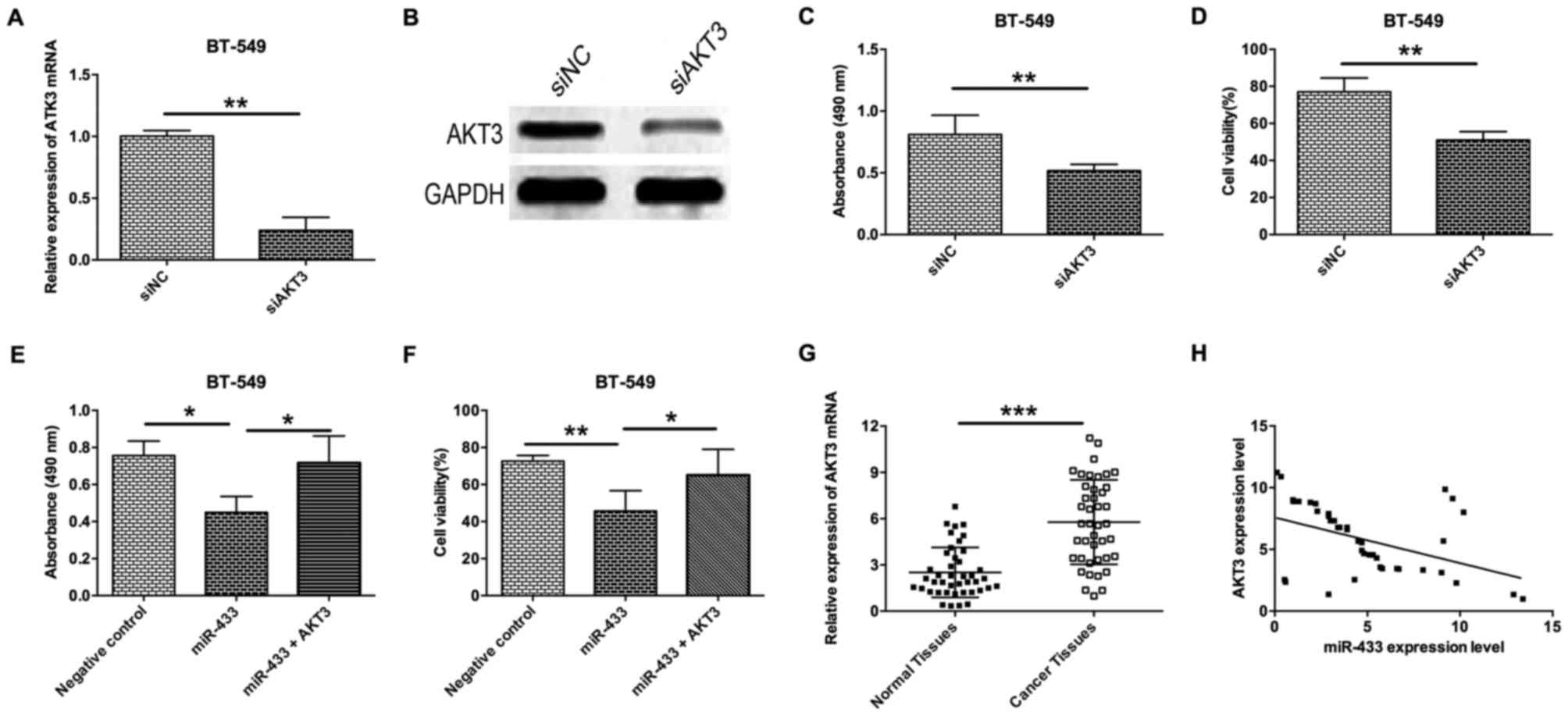
Inverse correlation between the
expression levels of miR-433 and AKT3
To further examine the role of AKT3 in breast cancer
cells, a siRNA experiment to determine the effect of AKT3-knockdown
on proliferation and cell viability of breast cancer cells was
performed. As shown in Fig. 4A and B,
AKT3-knockdown in BT-549 cells by transfection with siAKT3
significantly decreased the mRNA and protein expression levels
(Fig. 4A and B). Knockdown of AKT3 in
BT-549 cells was able to decrease cell proliferation and viability
(Fig. 4C and D). To further confirm
that ATK3 is a downstream target of miR-433, a rescue experiment
was also performed by transfecting BT-549 cells with miR-433 mimics
and AKT3-expressing vectors. The results revealed that
overexpression of AKT3 in miR-433-transfected BT-549 cells
significantly prevented the inhibitory effect of miR-433 on cell
proliferation and cell viability (Fig. 4E
and F). Analysis of clinical samples revealed that the level of
AKT3 mRNA expression in breast cancer tissues was significantly
higher compared with adjacent normal breast cancer tissues
(Fig. 4G), and Spearman's correlation
analysis demonstrated that miR-433 expression level was inversely
correlated with the level of AKT3 mRNA expression (Fig. 4H).
Discussion
Recently, substantial effort has been made to
improve diagnostic techniques and therapeutic approach for breast
cancer. However, the limited efficacy of novel therapeutics has
become a major obstacle owing to the inadequate understanding of
molecular mechanisms underlying breast cancer pathogenesis
(4–6).
The present study demonstrated the tumor suppressive role of
miR-433 in breast cancer cells. miR-433 expression was found to be
downregulated in breast cancer tissues and breast cancer cell
lines, and miR-433 overexpression was revealed to inhibit cell
proliferation and viability. Additionally, overexpression of
miR-433 was able to induce apoptosis possibly via targeting of AKT3
in breast cancer cells. Clinical sample analysis further confirmed
that miR-433 and AKT3 expression levels were inversely correlated
in breast cancer tissues.
The functional role of miR-433 has been determined
in various types of cancer. miR-433 has been demonstrated to be
downregulated in several types of cancer tissue, including
retinoblastoma, bladder cancer, OSCC, gastric cancer and HCC
(10,12–14,16).
Consistent with this, the results of the present study also
demonstrated the downregulation of miR-433 in breast cancer
tissues, which suggests that miR-433 may be tumor-suppressive in
breast cancer tissues. To gain further understanding of miR-433 in
breast cancer development, MTT assay was performed in the present
study to assess cell viability and proliferation, and it was
indicated that transfection with miR-433 was bale to inhibit cell
proliferation and reduce the viability of breast cancer cells.
These findings were in agreement with those of previous studies, as
miR-433 was shown to negatively regulate cell proliferation,
migration and invasion in retinoblastoma cells (10). Exogenous expression of miR-433 was
able to significantly inhibit cell proliferation, colony formation,
migration and invasion in bladder cancer cells (16). Restoring miR-433 expression in OSCC
cells dramatically suppressed cell growth, invasion and migration,
(12) and ectopic expression of
miR-433 inhibited cell proliferation, cell cycle progression, cell
migration and invasion in gastric cancer cells (19). Collectively, these results indicate
that miR-433 has a tumor-suppressive role in cancer cells in
vitro. In addition, overexpression of miR-433 induced cell
apoptosis and altered the expression levels of apoptosis mediators
(Bcl-2 and Bax). miR-433 induced cell cycle arrest and cell
apoptosis in retinoblastoma cells (10), and miR-433 accelerated apoptosis in
human dental pulp cells (20).
Therefore, the results of these previous studies indicate that the
inhibitory effect of miR-433 on breast cancer cell proliferation
and cell viability may involve the acceleration of cell
apoptosis.
AKT is a key mediator of the phosphoinositide
3-kinase (PI3K) pathway, and was found to regulate breast cancer
progression and metastasis by promoting migration and invasion
(21,22). Activation of the PI3K/AKT signaling
pathway is observed in >80% of breast cancer patients (23). AKT3 is one of the three isoforms of
AKT (AKT1, AKT2, and AKT3), and has been demonstrated to serve
notable roles in breast cancer development (24–29). AKT3
amplification has been suggested to represent a potentially
relevant oncogenic event in the subset of triple-negative breast
cancer (TNBC) (29). Downregulation
of AKT3 inhibited the growth of TNBC cell lines in 3D spheroid
cultures and in mouse xenograft models (24), whereas upregulation of AKT3 conferred
resistance to the AKT inhibitor MK2206 in breast cancer, and
suppression of AKT3 by miR-489 increases chemosensitivity in breast
cancer (28).
In the present study, it was found that AKT3 is a
direct target of miR-433, which was confirmed by luciferase
reporter assay. The knockdown of AKT3 was also able to
significantly inhibit proliferation and viability of breast cancer
cells. The level of AKT3 expression was inversely correlated with
miR-433 expression in breast cancer tissues. Collectively, these
data indicate that miR-433 was able to inhibit cell proliferation
and cell viability by modulating AKT3. However, caution must be
exercised as miR-433 may have downstream targets other than AKT3.
Previous studies have found that miR-433 targets Notch1, CREB1,
PAX6, HDAC6, KRAS and c-Met in other types of cancer (10,12,13,16).
Therefore, future studies must assess other targets of miR-433 in
breast cancer.
In summary, in the present study, it was revealed
that miR-433 was downregulated in breast cancer tissues and cell
lines. To the best of our knowledge, the present study is the first
to reveal that miR-433 may function as a tumor suppressor by
targeting AKT3 in breast cancer cells. Therefore miR-433 may be a
potential biomarker for breast cancer diagnosis and may serve as a
novel target for breast cancer therapy.
Acknowledgements
The present study was supported by a grant from the
Science and Technology Planning Project of Guangdong Province
(grant no. 2015B090904007).
References
|
1
|
Rudolph A, Chang-Claude J and Schmidt MK:
Gene-environment interaction and risk of breast cancer. Br J
Cancer. 114:125–133. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li C, Yang L, Zhang D and Jiang W:
Systematic review and meta-analysis suggest that dietary
cholesterol intake increases risk of breast cancer. Nutr Res.
36:627–635. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Srivastava S, Reid BJ, Ghosh S and Kramer
BS: Research needs for understanding the biology of overdiagnosis
in cancer screening. J Cell Physiol. 231:1870–1875. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Golubnitschaja O, Debald M, Yeghiazaryan
K, Kuhn W, Pešta M, Costigliola V and Grech G: Breast cancer
epidemic in the early twenty-first century: Evaluation of risk
factors, cumulative questionnaires and recommendations for
preventive measures. Tumour Biol. 37:12941–12957. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sonnenblick A, Pondé N and Piccart M:
Metastatic breast cancer: The Odyssey of personalization. Mol
Oncol. 10:1147–1159. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tzanninis IG, Kotteas EA,
Ntanasis-Stathopoulos I, Kontogianni P and Fotopoulos G: Management
and outcomes in metaplastic breast cancer. Clin Breast Cancer.
16:437–443. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hemmatzadeh M, Mohammadi H, Jadidi-Niaragh
F, Asghari F and Yousefi M: The role of oncomirs in the
pathogenesis and treatment of breast cancer. Biomed Pharmacother.
78:129–139. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pinweha P, Rattanapornsompong K,
Charoensawan V and Jitrapakdee S: MicroRNAs and oncogenic
transcriptional regulatory networks controlling metabolic
reprogramming in cancers. Comput Struct Biotechnol J. 14:223–233.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Leung AK: The Whereabouts of microRNA
Actions: Cytoplasm and beyond. Trends Cell Biol. 25:601–610. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li X, Yang L, Shuai T, Piao T and Wang R:
MiR-433 inhibits retinoblastoma malignancy by suppressing Notch1
and PAX6 expression. Biomed Pharmacother. 82:247–255. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Weiner-Gorzel K, Dempsey E, Milewska M,
McGoldrick A, Toh V, Walsh A, Lindsay S, Gubbins L, Cannon A,
Sharpe D, et al: Overexpression of the microRNA miR-433 promotes
resistance to paclitaxel through the induction of cellular
senescence in ovarian cancer cells. Cancer Med. 4:745–758. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang XC, Ma Y, Meng PS, Han JL, Yu HY and
Bi LJ: miR-433 inhibits oral squamous cell carcinoma (OSCC) cell
growth and metastasis by targeting HDAC6. Oral Oncol. 51:674–682.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo LH, Li H, Wang F, Yu J and He JS: The
tumor suppressor roles of miR-433 and miR-127 in gastric cancer.
Int J Mol Sci. 14:14171–14184. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tan Y, Ge G, Pan T, Wen D, Chen L, Yu X,
Zhou X and Gan J: A serum microRNA panel as potential biomarkers
for hepatocellular carcinoma related with hepatitis B virus. PLoS
One. 9:e1079862014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Del Vescovo V, Meier T, Inga A, Denti MA
and Borlak J: A cross-platform comparison of affymetrix and Agilent
microarrays reveals discordant miRNA expression in lung tumors of
c-Raf transgenic mice. PLoS One. 8:e788702013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu X, Zhu Y, Liang Z, Li S, Xu X, Wang X,
Wu J, Hu Z, Meng S, Liu B, et al: c-Met and CREB1 are involved in
miR-433-mediated inhibition of the epithelial-mesenchymal
transition in bladder cancer by regulating Akt/GSK-3β/Snail
signaling. Cell Death Dis. 7:e20882016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bellacosa A, Kumar CC, Di Cristofano A and
Testa JR: Activation of AKT kinases in cancer: Implications for
therapeutic targeting. Adv Cancer Res. 94:29–86. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Luo H, Zhang H, Zhang Z, Zhang X, Ning B,
Guo J, Nie N, Liu B and Wu X: Down-regulated miR-9 and miR-433 in
human gastric carcinoma. J Exp Clin Cancer Res. 28:822009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang K, Li L, Wu J, Qiu Q, Zhou F and Wu
H: The different expression profiles of microRNAs in elderly and
young human dental pulp and the role of miR-433 in human dental
pulp cells. Mech Ageing Dev 146–148. 1–11. 2015. View Article : Google Scholar
|
|
21
|
Dillon RL and Muller WJ: Distinct
biological roles for the akt family in mammary tumor progression.
Cancer Res. 70:4260–4264. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chin YR and Toker A: Function of Akt/PKB
signaling to cell motility, invasion and the tumor stroma in
cancer. Cell Signal. 21:470–476. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sultana A, Idress R, Naqvi ZA, Azam I,
Khan S, Siddiqui AA and Lalani EN: Expression of the androgen
receptor, pAkt, and pPTEN in breast cancer and their potential in
prognostication. Transl Oncol. May 12–2014.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chin YR, Yoshida T, Marusyk A, Beck AH,
Polyak K and Toker A: Targeting Akt3 signaling in triple-negative
breast cancer. Cancer Res. 74:964–973. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Grottke A, Ewald F, Lange T, Nörz D,
Herzberger C, Bach J, Grabinski N, Gräser L, Höppner F, Nashan B,
et al: Downregulation of AKT3 increases migration and metastasis in
triple negative breast cancer cells by upregulating S100A4. PLoS
One. 11:e01463702016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mosquera JM, Varma S, Pauli C, MacDonald
TY, Yashinskie JJ, Varga Z, Sboner A, Moch H, Rubin MA and Shin SJ:
MAGI3-AKT3 fusion in breast cancer amended. Nature. 520:E11–E12.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
O'Hurley G, Daly E, O'Grady A, Cummins R,
Quinn C, Flanagan L, Pierce A, Fan Y, Lynn MA, Rafferty M, et al:
Investigation of molecular alterations of AKT-3 in triple-negative
breast cancer. Histopathology. 64:660–670. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Stottrup C, Tsang T and Chin YR:
Upregulation of AKT3 confers resistance to the AKT inhibitor MK2206
in breast cancer. Mol Cancer Ther. 15:1964–1974. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Waugh MG: Amplification of chromosome 1q
genes encoding the phosphoinositide signalling enzymes PI4KB, AKT3,
PIP5K1A and PI3KC2B in breast cancer. J Cancer. 5:790–796. 2014.
View Article : Google Scholar : PubMed/NCBI
|