Introduction
Hepatocellular carcinoma (HCC) is estimated to be
the second most common cause of cancer-associated mortality
worldwide, with steadily increasing morbidity and mortality rates
(1,2).
Notably, approximately 50% of the liver cancer cases and
mortalities worldwide occur in China (2). Although surgical resection and liver
transplantation are regarded as sufficient treatment strategies to
extend the life expectancy of certain patients with HCC (3), the prognosis of these patients remains
unsatisfactory, since the disease is frequently diagnosed at an
advanced stage and is typically accompanied by severe liver
dysfunction. Therefore, there is an urgency to identify potential
biomarkers for the early detection and prognostic prediction of
HCC.
Mediator complex subunit 15 (MED15) is a subunit of
the MED multiprotein complex and functions as a crucial cofactor
for diverse signaling pathways, such as the sterol regulatory
element-binding protein (SREBP) and transforming growth factor-β
signaling pathways (4–7). It has been reported that MED15 promotes
transcriptional activation in eukaryotes and has been linked to
certain human diseases (7–9). For instance, MED15 regulates the
activity of the transcriptional activator SREBP and functions as a
transcriptional cofactor to control lipid homeostasis in
cardiovascular disease (5,7). In addition, previous studies have
demonstrated that dysregulation of MED15 expression promotes
tumorigenesis. Increased MED15 expression has been reported in
several human malignancies, including breast cancer, head and neck
squamous cell carcinoma (HNSCC), prostate cancer and testicular
germ cell tumors (6,10–14).
Furthermore, knockdown of MED15 significantly reduces the viability
of cancer cells, indicating that disrupting MED15 expression may
inhibit the progression of these types of cancer (10–12).
However, the expression level of MED15 in HCC and its prognostic
significance for clinical patients remain unknown.
In the present study, the aim was to investigate
MED15 mRNA and protein expression levels in HCC tissues using
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) and immunohistochemical analysis, respectively. The study
results revealed that MED15 levels were significantly upregulated
in HCC tissues as compared with those in the corresponding adjacent
non-tumor liver tissues. Additionally, analysis of The Cancer
Genome Atlas-Liver Hepatocellular Carcinoma (TCGA-LIHC) and
GSE14520 datasets revealed that MED15 expression levels were
associated with hypoxia-inducible factor 1α (HIF-1α) expression and
poor prognosis in patients with HCC. These findings suggest that
MED15 serves a role in liver tumorigenesis and may be a potential
therapeutic target for HCC treatment.
Materials and methods
Patients and tissue samples
The experiments of the present retrospective study
were reviewed and approved by the Ethics Committee of Nanfang
Hospital, Southern Medical University (Guangzhou, China). The
enrolled patients had not received any anticancer therapies prior
to surgery. All eligible patients provided informed consent prior
to the collection of HCC specimens and corresponding adjacent
non-tumor tissues. A total of 20 pairs of HCC tissues and matching
adjacent non-tumor tissues were obtained from patients undergoing
hepatectomy procedures between January 2014 and December 2015.
Each sample was divided into two parts, one part was
immediately stored in RNA keeper tissue stabilizer (Vazyme Biotech
Co., Ltd. Nanjing, China) and frozen at −80°C prior to RNA
extraction. The other was fixed overnight in 10% formaldehyde at
room temperature and paraffin-embedded using conventional
methods.
RNA extraction, cDNA synthesis and
RT-qPCR analysis
Total RNA was extracted from the HCC tissues and
paired adjacent non-tumor liver tissues using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), according to the manufacturer's protocol. RNAse-free DNase I
(Takara Biotechnology Co., Ltd., Dalian, China) was used to remove
genomic DNA contamination, while the quality and concentration of
total RNA were measured using a NanoDrop®
spectrophotometer (Thermo Fisher Scientific, Inc.). RNA was then
transcribed into cDNA using an RT kit (Takara Biotechnology Co.,
Ltd.) in a 20-µl reaction volume with 1 µg RNA. Subsequently,
amplification reactions were performed using a SYBR Green PCR kit
from Takara Biotechnology Co., Ltd., using the following primers:
MED15 sense, 5′-TGTCGTGTCTACGGCAACTC-3′, and anti-sense,
5′-CACTCTGCTGAGCCTGGAA-3′; β-actin sense,
5′-TCAAGATCATTGCTCCTCCTGA-3′, and anti-sense,
5′-CTCGTCATACTCCTGCTTGCTG-3′. β-actin was amplified as an internal
control. Gene-specific amplification was performed using a
LightCycler® 480 Instrument II (Roche Diagnostics,
Basel, Switzerland). Specific conditions for the qPCR reaction were
as follows: Preliminary denaturation at 95°C for 30 sec, followed
by 40 cycles of 95°C for 5 sec, and 60°C for 20 sec. A melting
curve analysis of the PCR products was used to assess the
specificity of amplification. Fold changes were calculated using
the relative quantification (2−ΔΔCq) method (15).
Hematoxylin and eosin staining
Human HCC tissues and matched adjacent non-tumor
liver tissues were fixed in 10% formaldehyde for 24 h, dehydrated
through an ethanol series for 2 h each, followed by two washes in
xylene (30 min each). After two immersions in paraffin (45 min
each), tissues were embedded and sectioned. Tissue sections were
incubated at 60°C for 1 h to remove the paraffin. Subsequently,
they were stained with hematoxylin for 7 min, de-stained in a
hydrochloric acid alcohol solution for 15 sec and then stained with
eosin for 3 min. Finally, sections were dehydrated through an
ethanol series for 1 min each, rinsed with xylene and coverslipped
with mounting medium. Staining results were observed and imaged
under a light microscope (Olympus BX 51; Olympus Corporation,
Tokyo, Japan).
Immunohistochemical assay
In total, 12 pairs of formalin-fixed and
paraffin-embedded HCC tissues were used for MED15
immunohistochemical staining. Briefly, paraffin-embedded tissues
were sliced into 4-µm sections and incubated at 60°C for 2 h.
Following deparaffinization, the sections were treated with 3%
hydrogen peroxide to inactivate the endogenous peroxidase, and
antigens were retrieved using a citrate buffer. Next, the sections
were incubated in PBS containing 5% normal goat serum (cat. no.
C-0005; BIOSS, Beijing, China) at room temperature for 1 h,
followed by incubation at 4°C overnight with a rabbit monoclonal
anti-human MED15 antibody (1:200 dilution; ab181158; Abcam,
Cambridge, UK) or with PBS alone, serving as the control. The
following day, sections were rinsed three times with PBS and then
incubated with a horseradish peroxidase-conjugated secondary
antibody (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA)
at room temperature for 1 h. Subsequent to rinsing with PBS, the
sections were stained with 3,3′-diaminobenzidine (Dako; Agilent
Technologies, Inc.) to induce a color reaction. The tissues were
then counterstained with hematoxylin for 1 min and de-stained with
acid-alcohol for 5 sec. Finally, the sections were dehydrated using
an ethanol series (1 min each), rinsed in xylene, dried and placed
on coverslips with a mounting medium. Immunohistochemical staining
results were assessed under a light microscope at ×200 and ×400
magnification. MED15 positive staining was mainly present in the
nuclei and manifested as a color varying between brown and light
yellow. Compared with the background, the staining intensity was
evaluated as follows: Light yellow, indicating weak expression;
brownish-yellow, indicating moderate expression; and brown,
indicating strong expression. MED15 protein expression in HCC was
further analyzed using clinical specimens from The Human Protein
Atlas (www.proteinatlas.org), which included
12 paired of HCC tissues and samples that can be filtered based on
the level of antibody staining via selecting one or several of the
following categories: high, medium, low and not detected.
TCGA-LIHC and GSE14520 datasets,
Oncomine gene expression array
In order to investigate the role of MED15 expression
in HCC, the current study analyzed the TCGA-LIHC dataset
(http://www.cbioportal.org/data_sets.jsp) that contains
331 HCC cases and the GSE14520 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14520)
that provides 246 HCC cases (hereafter defined as the TCGA-LIHC and
GSE14520 cohorts, respectively). These datasets comprise follow-up
information for patients and gene expression data. Patients with
available data on MED15 expression and follow-up information were
included into the present study. Correlations of MED15 expression
with the clinicopathological features, overall survival (OS) and
disease-free survival (DFS) of patients with HCC were analyzed.
Patients were organized according to their MED15 expression levels
from low to high expression, and then divided into two groups. The
first third of MED15 expression was defined as the MED15-low
expression group and the remainder patients were termed the
MED15-high expression group. The expression of MED15 in HCC was
further analyzed on Oncomine™ gene expression array
(https://www.oncomine.org/resource/login.html).
We used the OncoLnc online system (http://www.oncolnc.org/) to analyze the overall
survival of HCC patients from the TCGA-LIHC dataset. According to
the mRNA expression level of MED15 on the website, the first third
of MED15 expression was defined as the MED15-low expression group
and the remainder patients were termed the MED15-high expression
group. And the result of patients' overall survival was presented
on the website.
Gene set enrichment analysis
(GSEA)
The patients were divided into two groups according
to the MED15 mRNA expression level in the TCGA-LIHC dataset, which
were MED15 high- and low-expressed groups. Subsequently, GSEA
software (GSEA Desktop Application v3.0; Broad Institute, Inc.,
Cambridge, MA, USA) was used to analyze, annotate and interpret
enrichment results. Prior to running GSEA, four input files were
prepared; the GCT, GMT, CHIP and cls formatted files. The GMT file
was created in-house, and the GCT, CHIP and cls files were
downloaded from the TCGA dataset. Gene sets that were positively or
negatively correlated with the MED15 high- or low-expressed groups
were searched for.
Statistical analysis
Student's t-test was used to analyze the RT-qPCR
results, while Mann-Whitney U-test was used to evaluate the
association between MED15 expression and the clinicopathological
features of patients with HCC. OS and DFS time were calculated
using the Kaplan-Meier method and analyzed using log-rank tests.
Cox proportional hazards modeling was used for univariate and
multivariate analysis to investigate the effect of variables on
survival. Conventional clinicopathological variables and MED15
expression were enrolled into a Cox univariate regression analysis.
Statistically significant variables from the univariate regression
model were reanalyzed using the multivariate model. Statistical
analyses were conducted using SPSS version 17.0 software (SPSS,
Inc., Chicago, IL, USA). All statistical tests were two-sided, and
P<0.05 was considered to indicate a difference that was
statistically significant.
Results
MED15 mRNA expression is elevated in
HCC tissues
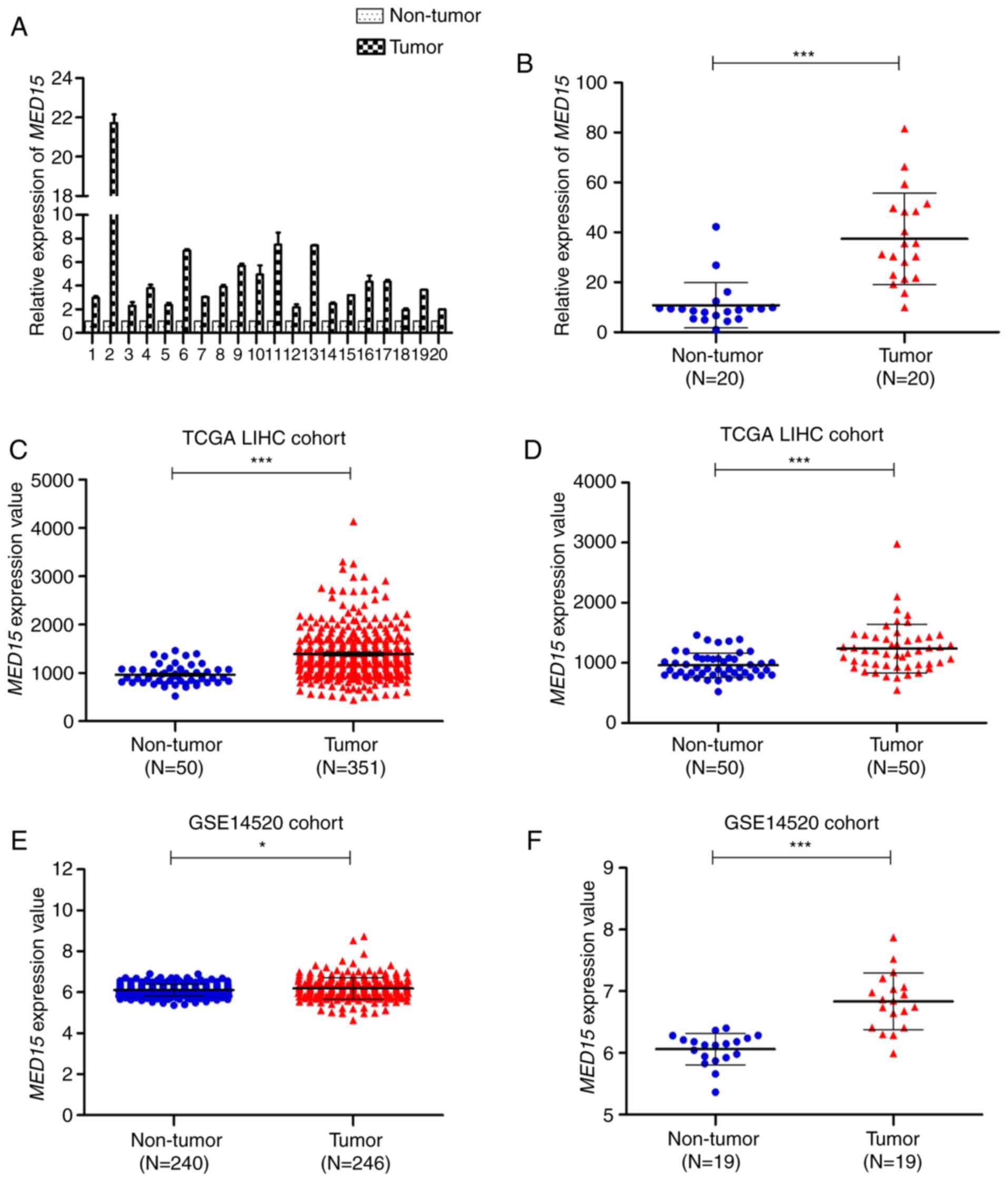
To investigate MED15 mRNA expression in HCC, 20
pairs of fresh HCC specimens and their corresponding adjacent
non-tumor tissues were used to measure the relative expression of
MED15 by RT-qPCR. It was observed that MED15 mRNA was notably
increased in HCC tissues compared with that in non-tumor liver
tissues (Fig. 1A and B). Next, to
further validate the observed MED15 dysregulation in HCC, MED15
gene expression was analyzed in patients included in the TCGA-LIHC
and GSE14520 datasets. In the unpaired TCGA-LIHC cohort, MED15
expression was overexpressed in HCC tissues compared with that in
non-tumor liver tissues (P<0.001; Fig.
1C). In the paired TCGA-LIHC cohort, a similar result was
observed when MED15 expression was re-assessed in 50 paired HCC
samples and adjacent non-tumor liver samples (P<0.001; Fig. 1D). These findings were further
confirmed by assessing MED15 expression in the GSE14520 dataset,
which revealed that MED15 was upregulated in HCC tissues compared
with that in non-tumor liver tissues from both unpaired and paired
GSE14520 cohorts (P<0.05; Fig. 1E and
F). Furthermore, similar results were obtained by analyzing the
expression data from Oncomine gene expression array datasets (data
not shown). Collectively, these data indicated that MED15 mRNA
expression is elevated in HCC tissues.
MED15 protein is overexpressed in
human HCC specimens
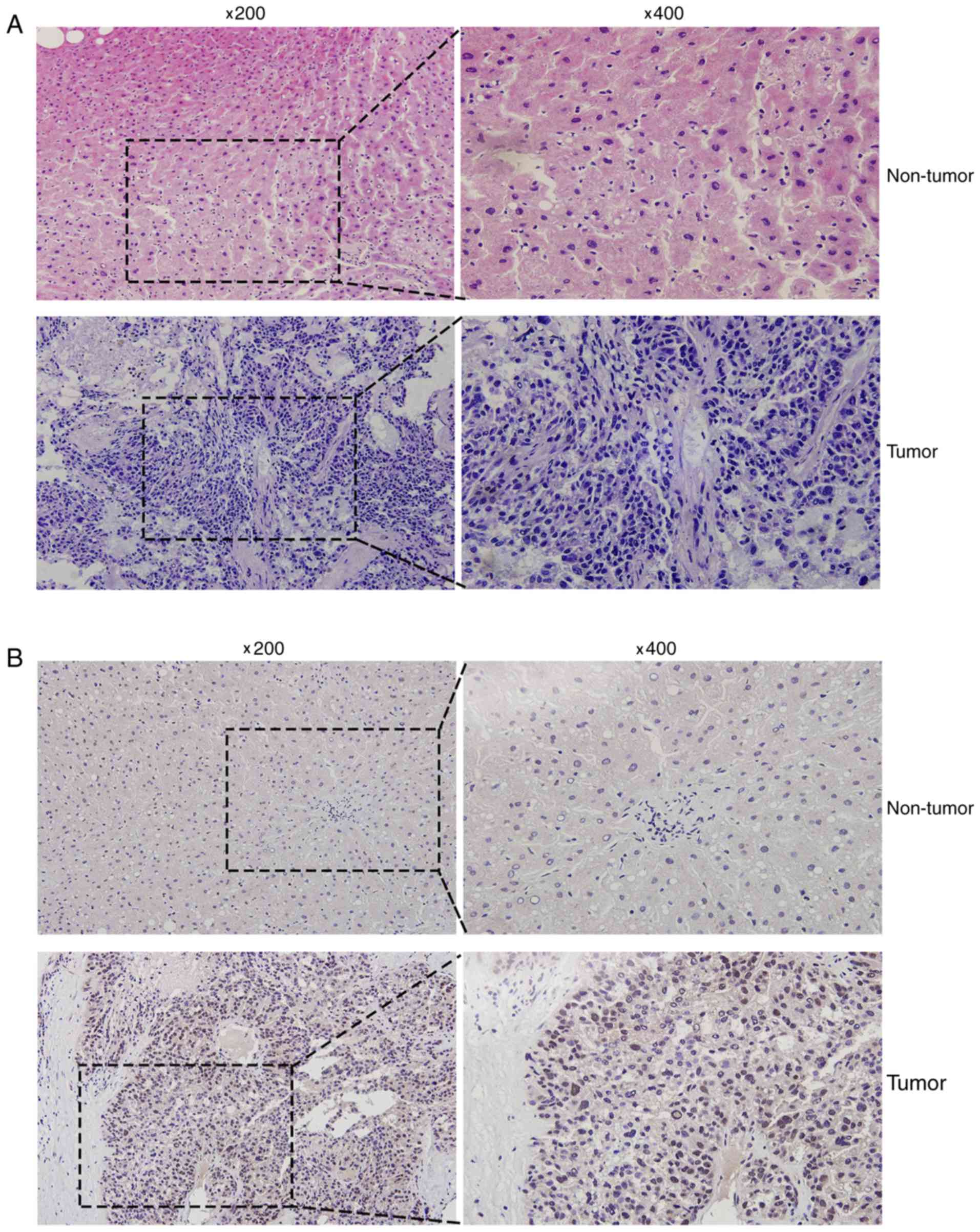
Hematoxylin and eosin staining was conducted to
distinguish HCC tissues from the corresponding adjacent non-tumor
liver tissues (Fig. 2A). In addition,
immunohistochemical analysis was used to detect the MED15 protein
expression levels in human liver cancer tissues, and varying
expression was observed. Positive immunostaining of MED15 appeared
as granular brown-colored staining, which was mainly located in the
nucleus of tumor cells (Fig. 2B). It
was also observed that MED15 protein was highly expressed in the
majority of HCC tissues as compared with that in the corresponding
non-tumor tissues, and 9 out of the 12 HCC samples investigated
were positive for MED15 immunostaining.
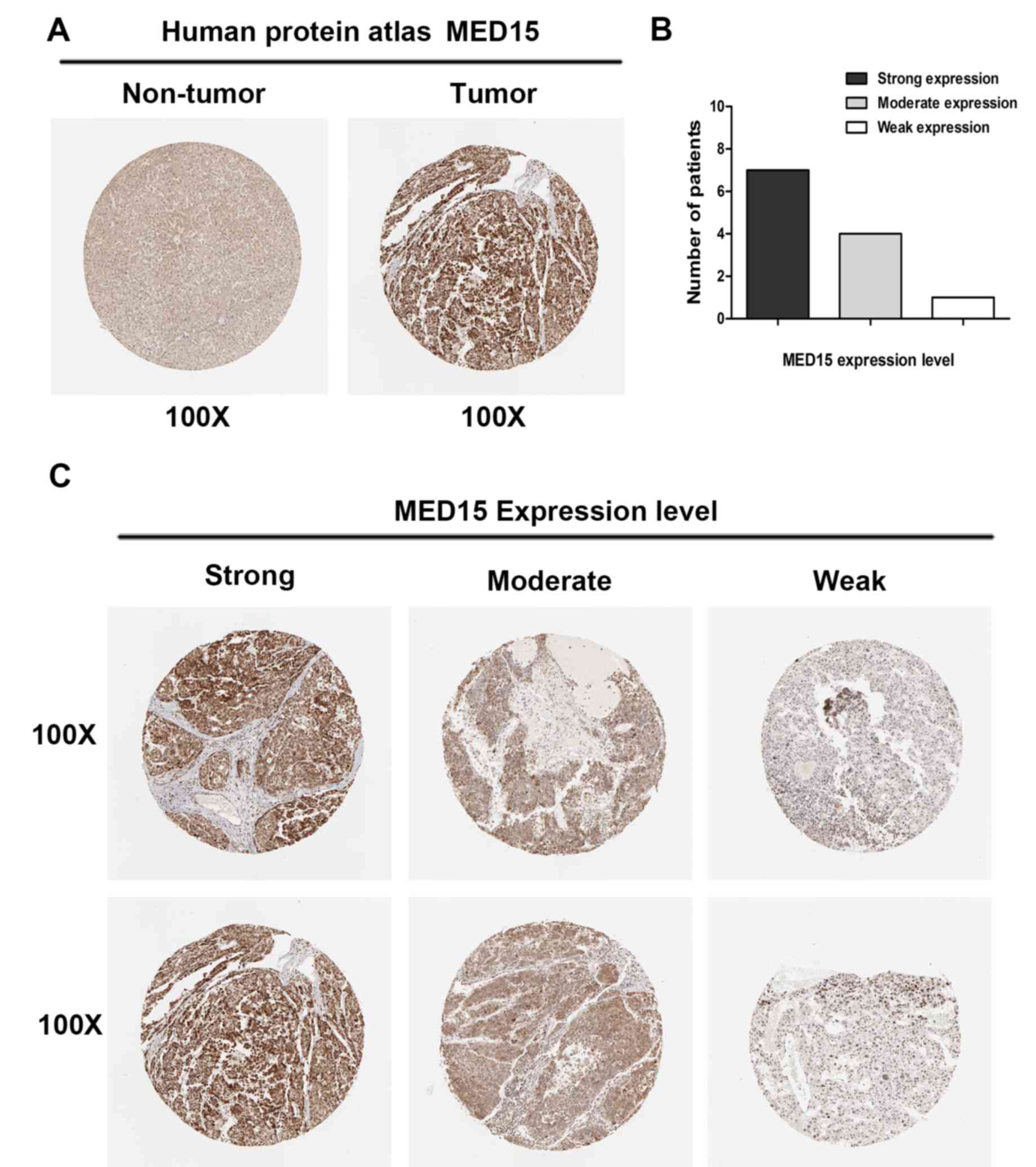
To further confirm the aforementioned results, MED15
protein expression was analyzed in clinical specimens from The
Human Protein Atlas. This analysis revealed that MED15 was
positively and strongly expressed in HCC samples, whereas it was
negatively or weakly expressed in normal hepatic tissues (Fig. 3A). Online immunohistochemical results
obtained from The Human Protein Atlas revealed that MED15 protein
was strongly expressed in 7 of the HCC tissues, moderately
expressed in 4 specimens and weakly expressed in 1 sample (Fig. 3B and C). These findings indicated that
MED15 protein is overexpressed in HCC.
Correlation between MED15 expression
and clinicopathological characteristics of HCC
Since MED15 was upregulated in HCC, the current
study attempted to explore the potential oncogenic role of MED15 in
HCC. Evidence of a correlation between MED15 expression and the
clinicopathological status of patients with HCC was searched in the
TCGA-LIHC and GSE14520 datasets. Statistical analysis of the
TCGA-LIHC cohort indicated that MED15 expression was positively
associated with the neoplasm grade (P=0.005) and α-fetoprotein
(AFP) levels (P=0.001). However, no statistically significant
correlation was observed between MED15 and other
clinicopathological features (Table
I). As summarized in Table II,
significant correlations were also detected between MED15
expression and certain clinicopathological features in the GSE14520
cohort, including the tumor size (P=0.033), Barcelona Clinic Liver
Cancer (BCLC) stage (P=0.031), AFP levels (P=0.002) and metastasis
risk (P=0.001). However, no statistically significant association
was identified between MED15 expression and other
clinicopathological characteristics, such as the patient sex, age,
tumor number, TNM staging, cirrhosis and relapse.
 | Table I.Correlation between MED15 expression
and hepatocellular carcinoma clinicopathological features in The
Cancer Genome Atlas-Liver Hepatocellular Carcinoma cohort. |
Table I.
Correlation between MED15 expression
and hepatocellular carcinoma clinicopathological features in The
Cancer Genome Atlas-Liver Hepatocellular Carcinoma cohort.
|
|
| MED15 expression,
n |
|
|---|
|
|
|
|
|
|---|
| Variable | Total, n | High | Low | P-value |
|---|
| Sex | 331 |
|
| 0.584 |
| Male | 219 | 140 | 79 |
|
|
Female | 112 | 75 | 37 |
|
| Age (years) | 331 |
|
| 0.139 |
| ≤50 | 66 | 48 | 18 |
|
|
>50 | 265 | 167 | 98 |
|
| Family cancer
history | 288 |
|
| 0.106 |
| Yes | 104 | 62 | 42 |
|
| No | 184 | 127 | 57 |
|
| Lymphocyte
infiltration | 212 |
|
| 0.455 |
|
Absent | 106 | 59 | 47 |
|
|
Mild | 91 | 60 | 31 |
|
|
Severe | 15 | 7 | 8 |
|
| Vascular tumor cell
invasion | 278 |
|
| 0.123 |
|
None | 183 | 109 | 74 |
|
|
Micro | 82 | 52 | 30 |
|
|
Macro | 13 | 12 | 1 |
|
| Neoplasm grade | 326 |
|
| 0.005a |
|
G1+G2 | 207 | 123 | 84 |
|
|
G3+G4 | 119 | 89 | 30 |
|
| Pathologic
stage | 310 |
|
| 0.544 |
|
I+II | 226 | 145 | 81 |
|
|
III+IV | 84 | 57 | 27 |
|
| AFP level
(ng/ml) | 245 |
|
|
<0.001a |
|
<400 | 189 | 110 | 79 |
|
|
≥400 | 56 | 47 | 9 |
|
| Child-Pugh
classification grade | 208 |
|
| 0.658 |
| A | 188 | 113 | 75 |
|
|
B+C | 20 | 11 | 9 |
|
| Liver fibrosis
Ishak score | 191 |
|
| 0.891 |
| ≤4 | 123 | 70 | 53 |
|
|
>4 | 68 | 38 | 30 |
|
| Relapse | 330 |
|
| 0.390 |
|
Yes | 133 | 83 | 50 |
|
| No | 197 | 132 | 65 |
|
 | Table II.Correlation between MED15 expression
and hepatocellular carcinoma clinicopathological features in the
GSE14520 cohort. |
Table II.
Correlation between MED15 expression
and hepatocellular carcinoma clinicopathological features in the
GSE14520 cohort.
|
|
| MED15 expression,
n |
|
|---|
|
|
|
|
|
|---|
| Variable | Total, n | High | Low | P-value |
|---|
| Sex | 241 |
|
| 0.756 |
|
Male | 210 | 148 | 62 |
|
|
Female | 31 | 21 | 10 |
|
| Age (years) | 241 |
|
| 0.768 |
|
≤50 | 124 | 88 | 36 |
|
|
>50 | 117 | 81 | 36 |
|
| No. of tumors | 241 |
|
| 0.455 |
|
Solitary | 189 | 129 | 60 |
|
|
Multiple | 52 | 40 | 12 |
|
| Tumor size
(cm) | 241 |
|
| 0.033a |
| ≤5 | 153 | 100 | 53 |
|
|
>5 | 88 | 69 | 19 |
|
| BCLC staging | 217 |
|
| 0.031a |
| A | 152 | 102 | 50 |
|
| B +
C | 65 | 53 | 12 |
|
| TNM staging | 225 |
|
| 0.127 |
| I | 96 | 60 | 36 |
|
| II +
III | 129 | 93 | 36 |
|
| AFP level
(ng/ml) | 237 |
|
| 0.002a |
|
≤300 | 128 | 78 | 50 |
|
|
>300 | 109 | 87 | 22 |
|
| Cirrhosis | 241 |
|
| 0.225 |
|
Yes | 222 | 158 | 64 |
|
| No | 19 | 11 | 8 |
|
| Metastasis
risk | 241 |
|
| 0.001a |
|
Low | 121 | 73 | 48 |
|
|
High | 120 | 96 | 24 |
|
| Relapse | 241 |
|
| 0.303 |
|
Yes | 136 | 99 | 37 |
|
| No | 105 | 70 | 35 |
|
MED15 expression levels predict the
clinical outcome of patients with HCC
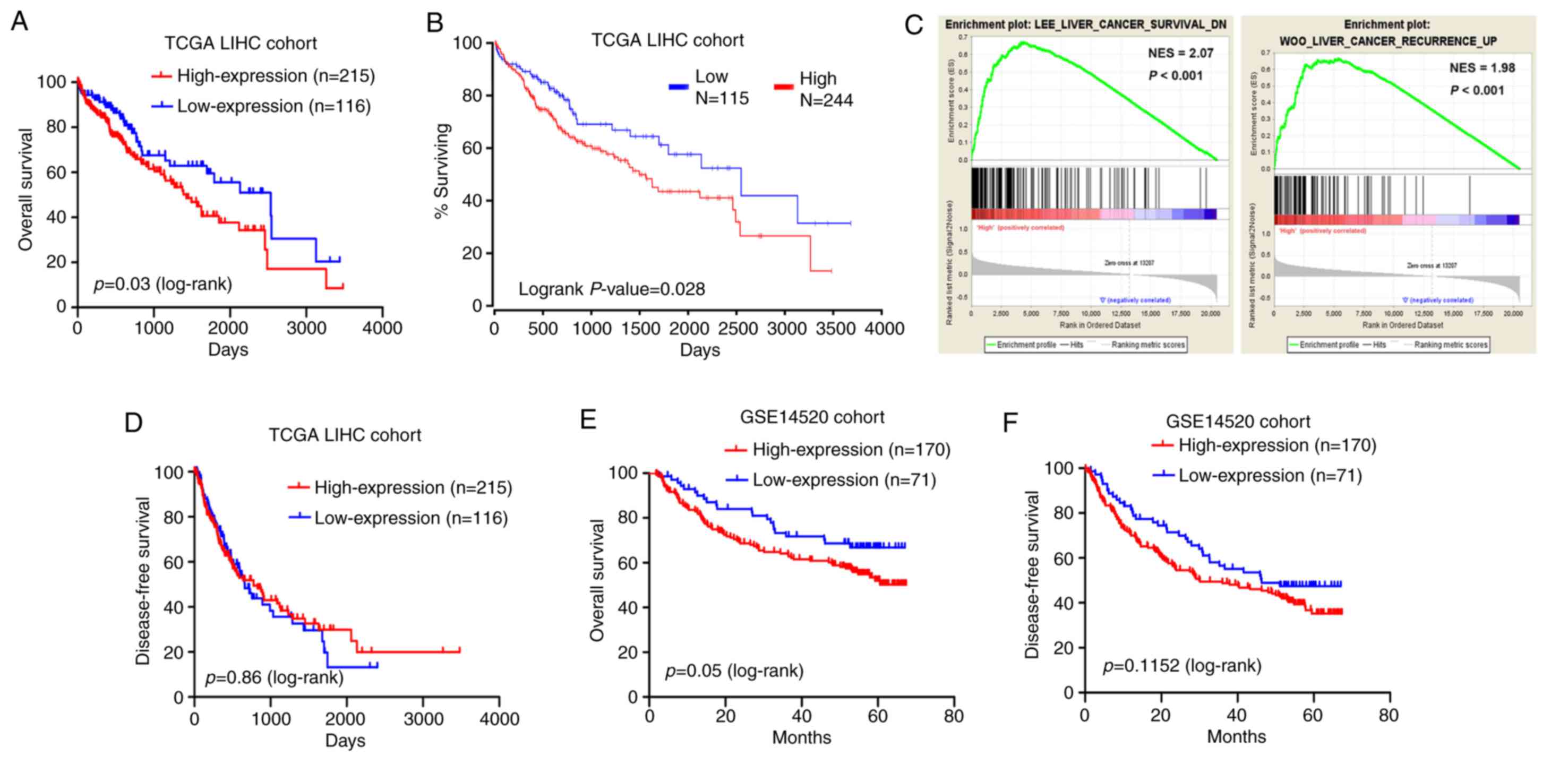
Kaplan-Meier and log-rank test analyses were
employed to test for an association between MED15 expression and
the OS or DFS of the patients. The results on the TCGA-LIHC cohort
data revealed that patients with highly expressed MED15 in tumor
tissues exhibited a significantly shorter survival time as compared
with those with low MED15 expression levels (P=0.03; Fig. 4A). This observation was supported by
the OncoLnc online system (http://www.oncolnc.org/), which was used to explore
links between TCGA-LIHC survival data and mRNA, miRNA or lncRNA
expression levels, and revealed that MED15 expression levels were
negatively associated with the OS (P=0.028; Fig. 4B). Furthermore, a comparative
microarray analysis of gene expression was performed in the
MED15-high and MED15-low expression groups using GSEA. This
analysis revealed that tumor samples with high MED15 expression
levels were enriched in the HCC survival reduction dataset and in
the HCC recurrence increase dataset (Fig.
4C). However, analysis of the TCGA-LIHC cohort revealed no
correlation between MED15 expression and DFS (Fig. 4D). Consistent with these findings,
Kaplan-Meier and log-rank test analyses in the GSE14520 cohort
demonstrated that high MED15 expression in tumor specimens was
associated with a poorer outcome of patients (P=0.05 for OS and
P=0.11 for DFS; Fig. 4E and F).
According to these findings, it can be deduced that a high MED15
expression predicts a worse prognosis for patients with HCC.
MED15 may be a valuable prognostic
marker for patients with HCC
Univariate and multivariate analyses were performed
to estimate the prognostic variables in patients included in the
TCGA-LIHC and GSE14520 cohorts. In the TCGA-LIHC cohort, univariate
analysis indicated that MED15 expression was significantly
associated with the OS rate of patients with HCC (P=0.032; Table III). Multivariate analysis was then
conducted using a Cox proportional hazards model and revealed that
MED15 expression may be an independent prognostic factor for
patients with HCC (hazard ratio (HR), 1.762; 95% confidence
interval (CI), 1.077–2.882; P=0.024). While in the GSE14520 cohort,
it was indicated that the number of tumors, tumor size, BCLC
staging, TNM staging, AFP level, Cirrhosis, metastasis risk and
relapse rather than MED15 expression were associated with OS rate
of patients with HCC via univariate analysis (Table IV). Multivariate analysis
demonstrated that the number of tumors (HR, 0.389; 95% CI,
0.203–0.745; P=0.004), BCLC staging (HR, 3.615; 95% CI,
1.982–6.593; P<0.001), metastasis risk (HR, 2.41; 95% CI,
1.523–3.814; P<0.001), cirrhosis (HR, 4.456; 95% CI,
1.081–18.367; P=0.039), and relapse (HR, 91.516; 95% CI,
12.664–661.334; P<0.001) were independent prognostic factors for
OS rate of patients with HCC. These preliminarily results indicate
that MED15 may have potential clinical value as a predictive
biomarker for OS in patients with HCC, but further studies are
needed to reach a clear conclusion.
 | Table III.Univariate and multivariate analyses
(Cox regression analysis) indicating the association between
overall survival and the characteristics of hepatocellular
carcinoma patients in The Cancer Genome Atlas-Liver Hepatocellular
Carcinoma cohort. |
Table III.
Univariate and multivariate analyses
(Cox regression analysis) indicating the association between
overall survival and the characteristics of hepatocellular
carcinoma patients in The Cancer Genome Atlas-Liver Hepatocellular
Carcinoma cohort.
| Variable | Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Sex | 0.754 | 0.510–1.114 | 0.156 |
|
|
|
| Age (years) | 0.943 | 0.574–1.552 | 0.819 |
|
|
|
| Family cancer
history | 1.298 | 0.859–1.961 | 0.216 |
|
|
|
| Lymphocyte
infiltration | 1.107 | 0.770–1.592 | 0.582 |
|
|
|
| Vascular tumor cell
invasion | 1.016 | 0.703–1.469 | 0.932 |
|
|
|
| Neoplasm grade | 1.310 | 0.883–1.945 | 0.180 |
|
|
|
| Pathologic
stage | 1.420 | 0.922–2.187 | 0.112 |
|
|
|
| AFP level
(ng/ml) | 1.566 | 0.976–2.512 | 0.063 |
|
|
|
| Child-Pugh
classification grade | 1.548 | 0.760–3.151 | 0.229 |
|
|
|
| Liver fibrosis
Ishak score | 1.059 | 0.591–1.898 | 0.846 |
|
|
|
| Relapse | 1.052 | 0.711–1.556 | 0.801 |
|
|
|
| MED15
expression | 1.581 | 1.039–2.404 | 0.032a | 1.762 | 1.077–2.882 | 0.024a |
 | Table IV.Univariate and multivariate analyses
(Cox regression analysis) indicating the association between
overall survival and the characteristics of hepatocellular
carcinoma patients in the GSE14520 cohort. |
Table IV.
Univariate and multivariate analyses
(Cox regression analysis) indicating the association between
overall survival and the characteristics of hepatocellular
carcinoma patients in the GSE14520 cohort.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Sex | 1.870 | 0.907–3.858 | 0.090 |
|
|
|
| Age (years) | 0.911 | 0.610–1.360 | 0.647 |
|
|
|
| No. of tumors | 1.642 | 1.057–2.551 | 0.027a | 0.389 | 0.203–0.745 | 0.004a |
| Tumor size
(cm) | 1.954 | 1.306–2.924 | 0.001a |
|
| 0.493 |
| BCLC staging | 3.357 | 2.211–5.096 |
<0.001a | 3.615 | 1.982–6.593 |
<0.001a |
| TNM staging | 3.084 | 1.883–5.053 |
<0.001a |
|
| 0.337 |
| AFP level
(ng/ml) | 1.710 | 1.141–2.562 | 0.009a |
|
| 0.589 |
| Cirrhosis | 5.126 | 1.263–20.804 | 0.022a | 4.456 | 1.081–18.367 | 0.039 |
| Metastasis
risk | 2.281 | 1.504–3.459 |
<0.001a | 2.41 | 1.523–3.814 | 0.001a |
| Relapse | 122.187 | 17.008–877.810 |
<0.001a | 91.516 | 12.664–661.334 |
<0.001a |
| MED15
expression | 1.517 | 0.950–2.425 | 0.081 |
|
|
|
MED15 expression is positively
associated with HIF-1α in HCC
The potential prognostic value of MED15
overexpression prompted the investigation of the mechanism that
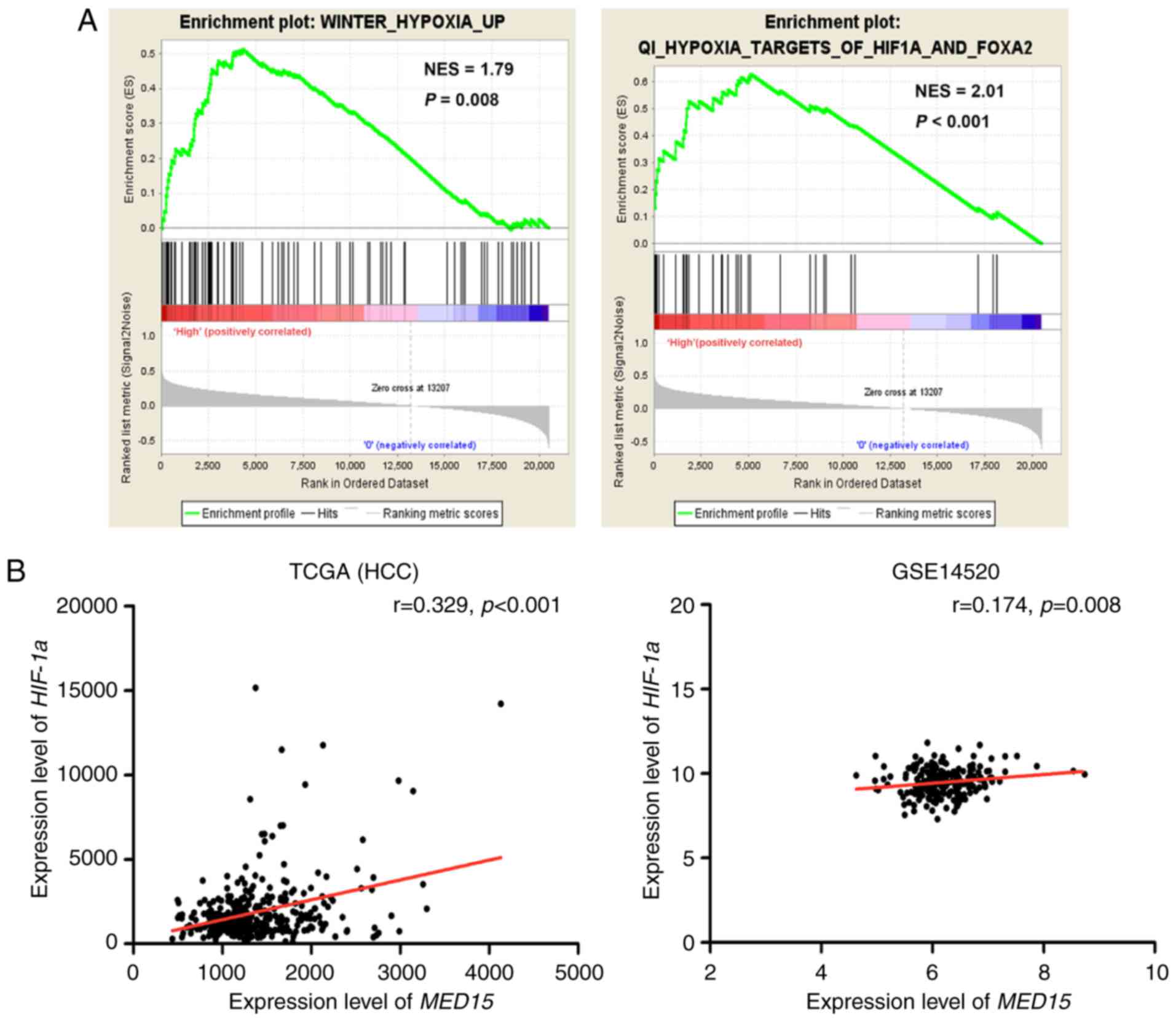
underlies its upregulation. GSEA was conducted on the MED15
overexpression group. The results revealed that Gene Ontology
terms, including ‘HYPOXIA’ and ‘HIF1A and FOXA2’, were
significantly enriched in MED15 overexpression group (P<0.01),
which indicated that MED15 was regulated by HIF-1α (Fig. 5A). Furthermore, a positive correlation
between HIF-1α expression and MED15 expression was determined via
analysis of the TCGA-LIHC and GSE14520 datasets (P<0.01;
Fig. 5B); therefore, it is
hypothesized that MED15 may be regulated by HIF-1α in HCC.
Discussion
The present study reported that MED15 is
overexpressed in HCC tissues and is closely associated with the
neoplasm grade, tumor size, BCLC stage and metastasis risk of
patients with HCC. In addition, patients with high MED15 expression
levels exhibited a poor disease prognosis. These observations
indicated that MED15 serves a role in liver tumorigenesis and may
be a potential prognostic factor for HCC.
In the present study, MED15 was observed to be
upregulated in human HCC specimens when compared with its
expression in the corresponding adjacent non-tumor specimens. It
has been reported that MED15 is dysregulated in several
malignancies, including breast cancer, prostate cancer and
testicular germ cell tumors (6,11–13). For instance, MED15 is overexpressed in
tissues obtained from primary tumors, metastasizing lymph nodes and
recurrent tumors, when compared with benign tissues in HNSCC
(10). Meanwhile, a previous study
demonstrated that the methylation levels of the MED15
promoter were significantly elevated in DNA samples from HNSCC
tissues compared with those in adjacent normal tissues. Notably,
hypermethylation of the MED15 promoter in HNSCC tissues
appeared to promote malignant transformation instead of functioning
as a typical tumor suppressor gene (16). These findings further confirm the
oncogenic potential of MED15 in HNSCC. To date, research into the
role of MED15 in cancer consistently reported its overexpression in
tumor tissues and its oncogenic functions in regulating the
initiation and progression of several malignancies. Furthermore,
consistent with the localization of MED15 in testicular germ cell
tumor tissues, prostate cancer tissues and HNSCC tissues (10,11,13), the
present study observed that MED15 was primarily located in the
nucleus of cells in HCC tissues.
The analysis conducted in the current study revealed
a significant association between MED15 overexpression and a poor
clinical outcome. Accumulating evidence has suggested that MED15
overexpression is a predictor of poor prognosis for several types
of human cancer, including prostate cancer, HNSCC and breast cancer
(6,10,11,16).
Shaikhibrahim et al (11) had
identified MED15 nuclear overexpression to be associated with poor
prognosis in castration-resistant prostate cancer. Furthermore, it
has been reported that MED15 overexpression correlates with a high
mortality rate in patients with HNSCC (10). In the current study, these findings
were extended to liver cancer, since MED15 overexpression was
observed to be indicative of a highly lethal phenotype in HCC. The
analysis of clinical data from patients with HCC patients in the
TCGA-LIHC cohort further suggested that MED15 expression may be an
independent prognostic survival indicator. However, similar results
were not observed in the GSE14520 cohort analysis in the present
study, possibly owing to inconsistent enrollment of patients at
different research centers. The number of cases may also be
important in the accuracy of these results, since the number of
patients included in the TCGA-LIHC cohort was larger in comparison
with that in the GSE14520 cohort. Nonetheless, the present study
proposes that MED15 may be regarded as a novel indicator for
identifying patients with a poor prognosis in HCC.
Previous studies demonstrated that certain
transcription factors regulate and control the expression of
specific genes (17–20). For instance, HMG box-containing
protein 1 has been reported to participate in Ras-induced premature
senescence by regulating p16INK4A expression (21). In addition, HIF-1α regulates the
expression of genes encoding proteins with key roles in cancer
biology (22), which can directly
activate vascular endothelial growth factor (VEGF) and VEGF
receptor 1 transcription via hypoxia response element binding
(23). In the current study, GSEA was
used to explore pathways that may be responsible for high MED15
expression levels, and HIF-1α was identified as one of the
potential regulators. HIF-1α is known to be an important
transcription factor in hypoxia response and to function in the
development and progression of tumors, including liver cancer
(24–26). The results of the present study
demonstrated that the expression levels of MED15 in the TCGA-LIHC
and GSE14520 datasets were positively associated with HIF-1α
expression, suggesting that MED15 may be a target of HIF-1α.
In conclusion, the present study reported that MED15
was upregulated in HCC tissues and that high MED15 expression was
correlated with poor prognosis in patients with HCC. To the best of
our knowledge, it was revealed for the first time that an
association exists between MED15 and human liver cancer, providing
valuable information for further studies on HCC. However, detailed
investigations are required to better understand the roles of MED15
in liver tumorigenesis and cancer progression.
Acknowledgements
The authors would like to thank Professor Xin Wei
Wang for sharing information regarding the GSE14520 dataset online.
The current study was supported by the National Nature Science
Foundation of China (grant nos. 81372283, 81472711, 81401180,
81672756 and 91540111), the Guangdong Province Universities and
Colleges Pearl River Scholar Funded Scheme (grant no. 2015), and
the Natural Science Foundation of Guangdong Province (grant no.
2014A030311013).
References
|
1
|
Sia D, Villanueva A, Friedman SL and
Llovet JM: Liver cancer cell of origin, molecular class, and
effects on patient prognosis. Gastroenterology. 152:745–761. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Siegel RL, Ward EM and Jemal A:
Global cancer incidence and mortality rates and trends-An update.
Cancer Epidemiol Biomarkers Prev. 25:16–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Llovet JM, Zucman-Rossi J, Pikarsky E,
Sangro B, Schwartz M, Sherman M and Gores G: Hepatocellular
carcinoma. Nat Rev Dis Primers. 2:160182016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kato Y, Habas R, Katsuyama Y, Näär AM and
He X: A component of the ARC/Mediator complex required for TGF
beta/Nodal signalling. Nature. 418:641–646. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang F, Vought BW, Satterlee JS, Walker
AK, Jim Sun ZY, Watts JL, DeBeaumont R, Saito RM, Hyberts SG, Yang
S, et al: An ARC/Mediator subunit required for SREBP control of
cholesterol and lipid homeostasis. Nature. 442:700–704. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao M, Yang X, Fu Y, Wang H, Ning Y, Yan
J, Chen YG and Wang G: Mediator MED15 modulates transforming growth
factor beta (TGFβ)/Smad signaling and breast cancer cell
metastasis. J Mol Cell Biol. 5:57–60. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang X and Yang F: Mediating lipid
biosynthesis: Implications for cardiovascular disease. Trends
Cardiovasc Med. 23:269–273. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nakatsubo T, Nishitani S, Kikuchi Y, Iida
S, Yamada K, Tanaka A and Ohkuma Y: Human mediator subunit MED15
promotes transcriptional activation. Drug Discov Ther. 8:212–217.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Berti L, Mittler G, Przemeck GK, Stelzer
G, Günzler B, Amati F, Conti E, Dallapiccola B, Hrabé de Angelis M,
Novelli G and Meisterernst M: Isolation and characterization of a
novel gene from the DiGeorge chromosomal region that encodes for a
mediator subunit. Genomics. 74:320–332. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shaikhibrahim Z, Offermann A, Halbach R,
Vogel W, Braun M, Kristiansen G, Bootz F, Wenzel J, Mikut R,
Lengerke C, et al: Clinical and molecular implications of MED15 in
head and neck squamous cell carcinoma. Am J Pathol. 185:1114–1122.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shaikhibrahim Z, Menon R, Braun M,
Offermann A, Queisser A, Boehm D, Vogel W, Rüenauver K, Ruiz C,
Zellweger T, et al: MED15, encoding a subunit of the mediator
complex, is overexpressed at high frequency in castration-resistant
prostate cancer. Int J Cancer. 135:19–26. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Offermann A, Vlasic I, Syring I, Vogel W,
Ruiz C, Zellweger T, Rentsch CA, Hagedorn S, Behrends J, Nowak M,
et al: MED15 overexpression in prostate cancer arises during
androgen deprivation therapy via PI3K/mTOR signaling. Oncotarget.
8:7964–7976. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Klümper N, Syring I, Offermann A,
Shaikhibrahim Z, Vogel W, Müller SC, Ellinger J, Strauß A, Radzun
HJ, Ströbel P, et al: Differential expression of Mediator complex
subunit MED15 in testicular germ cell tumors. Diagn Pathol.
10:1652015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schiano C, Casamassimi A, Rienzo M, de
Nigris F, Sommese L and Napoli C: Involvement of Mediator complex
in malignancy. Biochim Biophys Acta. 1845:66–83. 2014.PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ovchinnikov DA, Wan Y, Coman WB, Pandit P,
Cooper-White JJ, Herman JG and Punyadeera C: DNA methylation at the
novel CpG sites in the promoter of MED15/PCQAP gene as a biomarker
for head and neck cancers. Biomarker Insights. 9:53–60. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gañán-Gómez I, Wei Y, Yang H,
Boyano-Adánez MC and García-Manero G: Oncogenic functions of the
transcription factor Nrf2. Free Radic Biol Med. 65:750–764. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Luo D, Wang Z and Wu J, Jiang C and Wu J:
The role of hypoxia inducible factor-1 in hepatocellular carcinoma.
Biomed Res Int. 2014:4092722014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Monga SP: β-catenin signaling and roles in
liver homeostasis, injury, and tumorigenesis. Gastroenterology.
148:1294–1310. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rogacki K, Kasprzak A and Stępiński A:
Alterations of Wnt/β-catenin signaling pathway in hepatocellular
carcinomas associated with hepatitis C virus. Pol J Pathol. 1:9–21.
2015. View Article : Google Scholar
|
|
21
|
Li H, Wang W, Liu X, Paulson KE, Yee AS
and Zhang X: Transcriptional factor HBP1 targets P16(INK4A),
upregulating its expression and consequently is involved in
Ras-induced premature senescence. Oncogene. 29:5083–5094. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Semenza GL: Defining the role of
hypoxia-inducible factor 1 in cancer biology and therapeutics.
Oncogene. 29:625–634. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liao D and Johnson RS: Hypoxia: A key
regulator of angiogenesis in cancer. Cancer Metastasis Rev.
26:281–290. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu XZ, Xie GR and Chen D: Hypoxia and
hepatocellular carcinoma: The therapeutic target for hepatocellular
carcinoma. J Gastroenterol Hepatol. 22:1178–1182. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiang ZL, Zeng ZC, Fan J, Tang ZY, He J,
Zeng HY and Chang JY: The expression of HIF-1α in primary
hepatocellular carcinoma and its correlation with radiotherapy
response and clinical outcome. Mol Biol Rep. 39:2021–2029. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin D and Wu J: Hypoxia inducible factor
in hepatocellular carcinoma: A therapeutic target. World J
Gastroenterol. 21:12171–12178. 2015. View Article : Google Scholar : PubMed/NCBI
|