Introduction
The human body possesses a circadian clock, which
serves an important role in regulating normal physiological
functions (1). Clock genes are at the
core of this circadian clock. To date, 14 clock genes have been
identified: Clock circadian regulator (CLOCK), brain and
muscle ARNT-like 1 (BMAL1), period circadian clock
(PER)1, PER2, PER3, deleted in esophageal cancer
(DEC)1, DEC2, cryptochrome circadian clock
(CRY)1, CRY2, timeless circadian clock (TIM),
casein kinase 1 epsilon (CKIε), retinoic acid
receptor-related orphan receptor-α (RORα), neuronal PAS
domain protein 2 (NPAS2) and nuclear receptor subfamily 1
group D member 1 (REV-ERBα) (2–5), all of
which exist in the majority of human cells (6). In mammals, 2–10% of the genome is
regulated by these clock genes (7,8), including
numerous key cell cycle genes and cancer-associated genes (9–11); these
are termed clock-controlled genes (CCGs). Different clock genes
regulate different downstream CCGs and, thus, clock genes are
important in regulating cell biochemical and physiological
functions (12).
PER2 is an important clock gene, and previous
research has indicated that PER2 serves a notable function
in the occurrence and progression of cancer (13,14). The
expression of PER2 has been revealed to be downregulated in
various types of cancer, including breast cancer, gastric
carcinoma, head and neck cancer and oral cancer (15–18).
PER2 regulates a number of downstream cell cycle genes and
cancer-associated genes, including cyclin D1, cyclin A, tumor
protein p53, v-Myc avian myelocytomatosis viral oncogene homolog,
mouse double minute 2 homolog proto-oncogene and B-cell lymphoma 2
apoptosis regulator (18,19). However, it remains unclear whether the
decreased expression of PER2 in cancer cells is able to
affect the expression of other clock genes.
The present study used short hairpin RNA (shRNA)
interference to knockdown PER2 in human oral squamous cell
carcinoma (OSCC) SCC15 cells, and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) was
used to assess the altered mRNA expression of other clock genes.
The mRNA expression levels of many clock genes, proliferation and
apoptosis of tumor cells were altered. This research demonstrates
that the effect of PER2 on tumor occurrence and progression
is induced not only through the regulation of downstream CCGs, but
also through the regulation of other clock genes in the network.
This is of great significance to further illustrating the molecular
functions and tumor-suppressive mechanisms of PER2.
Materials and methods
Construction of lentiviral shRNA
plasmids
The mRNA sequence of hPER2 was acquired from
the GenBank Database (http://www.ncbi.nlm.nih.gov/genbank/; accession no.
NM_022817). BLAST screening (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to
ascertain three interference target sequences of PER2
(20): PER2-I, CAGAGTCCAGATACCTTTA;
PER2-II, ATCCATATTTCACTGTAAA; and PER2-III, CACACACAAAGAACTGATA.
Based on the design principles of Chernolovskaya and Zenkova
(21), three RNA interference target
sequences of PER2 were designed and synthesized: PER2-shRNA-I,
PER2-shRNA-II and PER2-shRNA-III (Table
I). The restriction enzymes AgeI/EcoRI (New
England BioLabs, Inc., Ipswich, MA, USA) were used to double digest
the lentiviral vector (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), and the double enzyme-restricted vector was
connected with PER2-shRNA-I–III, prior to the double-strand DNA
being annealed using T4 DNA ligase in order to construct
PER2-shRNA-I–III lentiviral plasmids. Scramble plasmids
(Invitrogen; Thermo Fisher Scientific, Inc.), which exerted no
interference effects on any genes, served as the controls
(control-shRNA). Subsequently, these lentiviral plasmids were
transformed into freshly prepared Escherichia coli DH5α
cells (Sangon Biotech Co., Ltd., Shanghai, China), selected in
lysogeny broth medium containing the antibiotic ampicillin and then
cultured overnight at 37°C. Plasmids were extracted using a Qiagen
Plasmid Midi kit (Qiagen GmbH, Hilden, Germany). qPCR was performed
using a SYBR Premix Ex TaqII kit (Takara Bio, Inc., Otsu, Japan) in
a reaction system comprising 5 µl SYBR Premix Ex TaqII, 0.5 µl each
of the forward and reverse primer (0.4 µmol/l; Sangon Biotech Co.,
Ltd.), (forward primer sequence: 5′-CCATGATTCCTTCATATTTGC-3′ and
reverse primer sequence: 5′-GTAATACGGTTATCCACGCG-3′), 2 µl DNA
template (50 ng/µl) and 2 µl double distilled H2O
(ddH2O). The ddH2O is the template for blank
control, and the negative control was a template for an empty
carrier that is not inserted in the target gene. The thermocycling
conditions were as follows: Pre-denaturation at 95°C for 3 min;
denaturation for 30 sec at 95°C; annealing extension for 30 sec at
55°C and 30 sec at 72°C, which for 22 cycles. Fluorescence signals
were recorded during the 72°C annealing extension phase. The
amplification products from PCR were assessed using a DNA
sequencing assay (3730 DNA Analyzer; Applied Biosystems; Thermo
Fisher Scientific, Inc.), and the results of DNA sequencing were
analyzed using Chromas v2.4 software (Technelysium Pty Ltd.,
Brisbane, Australia).
 | Table I.Seque nces of PER2-shRNAs. |
Table I.
Seque nces of PER2-shRNAs.
| Group | Sense strand | Antisense
strand |
|---|
| PER2-shRNA-I |
5′-CCGGGCCAGAGTCCAGATACCTTTACTCG |
5′-AATTCAAAAAGCCAGAGTCCAGATACCTT |
|
|
AGTAAAGGTATCTGGACTCTGGCTTTTTG-3′ |
TACTCGAGTAAAGGTATCTGGACTCTGGC-3′ |
| PER2-shRNA-II |
5′-CCGGGCATCCATATTTCACTGTAAACTCG |
5′-AATTCAAAAAGCATCCATATTTCACTGTAA |
|
|
AGTTTACAGTGAAATATGGATGCTTTTTG-3′ |
ACTCGAGTTTACAGTGAAATATGGATGC-3′ |
| PER2-shRNA-III |
5′-CCGGGACACACACAAAGAACTGATACTC |
5′-AATTCAAAAAGACACACACAAAGAACTG |
|
|
GAGTATCAGTTCTTTGTGTGTGTCTTTTTG-3′ |
ATACTCGAGTATCAGTTCTTTGTGTGTGTC-3′ |
Lentiviral PER2-shRNA plasmid
packing
Each PER2-shRNA-I–III and scramble plasmid (8 µg)
was mixed with 20 µl Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) and incubated for 15 min at room
temperature. The mixture was then applied drop-wise to 293 cells
(Institute of Life Sciences, Chongqing Medical University,
Chongqing, China), which had been cultured to 70–80% growth
density, and incubated in Dulbecco's modified Eagle's medium (DMEM;
Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum
(FBS; Natocor Industria Biologica, Cordoba, Argentina) for 48 h at
37°C and 5% CO2. Between 48 and 72 h after transfection,
the culture supernatant was collected and centrifuged for 10 min at
4°C and 12,000 × g to remove the cell debris, prior to being
centrifuged again for 1 h at 4°C and 25,000 × g in overspeed
centrifuge tubes. The sediment was then mixed with virus
preservation solution and centrifuged for 5 min at 4°Cand 10,000 ×
g. The supernatant was divided and four groups of lentiviral
plasmids were obtained: PER2-shRNA-I, PER2-shRNA-II, PER2-shRNA-III
and control-shRNA.
Cell transfection
SCC15 cells (Institute of Life Sciences, Chongqing
Medical University) were cultured in Dulbecco's modified Eagle's
medium/F-12 (Gibco; Thermo Fisher Scientific, Inc.) containing FBS
and penicillin 100 U/ml and streptomycin 0.1 mg/ml (Yocon
Biotechnology Co., Beijing, China) in cell culture flasks, at 37°C
in an atmosphere containing 5% CO2. Lentiviral plasmids
(5 µl) were applied drop-wise to SCC15 cells during the logarithmic
growth phase, and 100 µl Polybrene infection reagent
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added. Cells
were incubated in DMEM containing 10% FBS without antibiotics for
24 h (37°C, 5% CO2), following which the medium was
replaced every day with DMEM containing 10% FBS and puromycin (2
µg/ml). Stably transfected SCC15 cells, in which PER2 was
knocked down, were obtained following continuous cultivation for 7
days. The cells were divided into five experimental groups:
PER2-shRNA-I, PER2-shRNA-II and PER2-shRNA-III groups (transfected
with lentiviral plasmids PER2-shRNA-I to III, respectively); a
control group (transfected with control-shRNA, expressing scramble
shRNA); and an SCC15 group (untransfected SCC15 cells as a blank
control).
Western blot analysis
Cells in the logarithmic growth phase were lysed at
4°C in radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Haimen, China) for 30 min and collected by a cell
scraper, following which the cell suspension was centrifuged for 5
min at 4°C and 12,000 × g to collect the supernatant. Protein
concentration was determined using a Bicinchoninic Acid Protein
Assay kit (Beyotime Institute of Biotechnology), according to the
manufacturer's protocols. Subsequently, the sample solution,
containing 50 µg/lane protein, was subjected to SDS-PAGE (12% gel).
The proteins were transferred to polyvinylidene fluoride (PVDF)
membranes (EMD Millipore, Billerica, MA, USA), which were
subsequently blocked with 5% dried skimmed milk for 1 h at room
temperature. Following blocking, the membranes were probed with a
rabbit polyclonal anti-hPER2 antibody (1:300; ab180655; Abcam,
Cambridge, UK) and a mouse monoclonal anti-hGAPDH antibody
(1:1,000; 51332; Cell Signaling Technology, Inc., Danvers, MA, USA)
overnight at 4°C, followed by three washes in PBS. Subsequently,
the membranes were incubated with a horseradish
peroxidase-conjugated goat anti-rabbit IgG antibody (Cell Signaling
Technology, Inc.) for 2 h at room temperature, followed by three
further washes in PBS. Under dark conditions, the enhanced
chemiluminescent color-substrate solution (Beyotime Institute of
Biotechnology) was applied to the PVDF membranes and exposed using
chemiluminescence apparatus. A gel imaging system GelDoc 2000
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used for
visualization and protein detection. The ratio of the grey value of
each PER2 band to that of GAPDH was calculated by ImageJ (version
1.48u; National Institutes of Health, Bethesda, MA, USA). All
assays were performed in triplicate.
RT-qPCR
RT-qPCR was performed using the PrimeScript RT
Reagent kit (cat no. RR037A; Takara Biotechnology Co., Ltd.)
according to the manufacturers' protocols. Total RNA was extracted
from cells using RNAiso Plus (Takara Biotechnology Co., Ltd.),
prior to the RNA concentration and quality being determined using a
UV/Visible spectrophotometer (GE Healthcare Life Sciences, Little
Chalfont, UK) by measuring absorbance at 260 and 280 nm. RNA was
reverse transcribed to cDNA using a PrimeScript RT Reagent kit
(Takara Biotechnology Co., Ltd.), according to the manufacturer's
protocols. The reaction system comprised 5X Primer Script Buffer (4
µl), Primer Script RT Enzyme mix (1 µl), Oligo-dT Primers (1 µl),
Random Hexamers (1 µl) and RNase Free dH2O (13 µl). The
reverse transcription reaction conditions were 37°C for 15 min and
85°C for 5 sec. The qPCR primers for CLOCK, BMAL1, PER1, PER2,
PER3, DEC1, DEC2, CRY1, CRY2, TIM, CKIε, RORα, NPAS2 and
REV-ERBα were designed using Oligo 7.0 primer analysis
software (Molecular Biology Insights, Inc., Colorado Springs, CO,
USA) and are listed in Table II.
β-actin served as the normalization control. The reaction system
comprised 5 µl SYBR Premix Ex TaqII, 0.5 µl each forward and
reverse primer (0.4 µmol/l), 2 µl cDNA template (50 ng/µl) and 2 µl
double-distilled H2O. The reaction conditions were as
follows: Pre-denaturation at 95°C for 1.5 min, followed by 40
cycles of denaturation for 10 sec at 95°C and annealing extension
for 30 sec at 60°C. Fluorescence signals were recorded during the
60°C annealing extension phase. The data were analyzed using the
2−ΔΔCq method (22). All
assays were performed in triplicate.
 | Table II.Primer sequences for clock genes used
for reverse transcription-quantitative polymerase chain
reaction. |
Table II.
Primer sequences for clock genes used
for reverse transcription-quantitative polymerase chain
reaction.
| Gene
abbreviation | Gene name | Forward primer
sequence | Reverse primer
sequence |
|---|
| PER1 | Period circadian
clock 1 |
5′-CTGCTACAGGCACGTTCAAG-3′ |
5′-CTCAGGGACCAAGGCTAGTG-3′ |
| PER2 | Period circadian
clock 2 |
5′-TTGGACAGCGTCATCAGGTA-3′ |
5′-TCCGCTTATCACTGGACCTT-3′ |
| PER3 | Period circadian
clock 3 |
5′-GCAGGTCTATGCCAGTGTGA-3′ |
5′-ACCACCACCATTCGGTTCT-3′ |
| CLOCK | Clock circadian
regulator |
5′-CAGCCAGTGATGTCTCAAGC-3′ |
5′-ATGCGTGTCCGTTGTTCC-3′ |
| BMAL1 | Brain and muscle
ARNT-like 1 |
5′-TGCCACCAATCCATACACAG-3′ |
5′-TTCCCTCGGTCACATCCTAC-3′ |
| DEC1 | Deleted in
esophageal cancer 1 |
5′-CAGCTTTCGGATGATGAAGG-3′ |
5′-GCTGAAGGTGGGATCAGGTA-3′ |
| DEC2 | Deleted in
esophageal cancer 2 |
5′-GGGACCAACTGCTTCACACT-3′ |
5′-TAATCTGTGGGACGGTAGGC-3′ |
| CRY1 | Cryptochrome
circadian clock 1 |
5′-TGTGATTCGTGGACAACCAG-3′ |
5′-TAGCTGCGTCTCGTTCCTTT-3′ |
| CRY2 | Cryptochrome
circadian clock 2 |
5′-AGGAGAACCACGACGAGA-3′ |
5′-TCCGCTTCACCTTTTTATAC-3′ |
| CKIε | Casein kinase 1
epsilon |
5′-TGAGTATGAGGCTGCACAGG-3′ |
5′-CTTCCCGAGATGGTCAAATG-3′ |
| TIM | Timeless circadian
clock |
5′-GATAGAGGCCCATTCCTGCAT-3′ |
5′-GAAGGGCTGGGGAACTTAGAC-3′ |
| NPAS2 | Neuronal PAS domain
protein 2 |
5′-AACCTCGGCAGCACTTTAAC-3′ |
5′-GGTTCTGACATGGCTGTGTG-3′ |
| RORα | Retinoic acid
receptor-related orphan receptor-α |
5′-CTATCCCTCCAAGGCACAAG-3′ |
5′-AACACAAGACTGACGAGCACA-3′ |
|
REV-ERBα | Nuclear receptor
subfamily 1 group D member 1 |
5′-ACAGAATCGAACTCTGCACTTCT-3′ |
5′-GGGGAGGGAGGCAGGTATT-3′ |
| β-actin | β-actin |
5′-AGCGAGCATCCCCCAAAGTT-3′ |
5′-GGGCACGAAGGCTCATCATT-3′ |
Flow cytometry assay
To prepare a single-cell suspension, cells in the
logarithmic growth phase were harvested by 0.25% trypsinization and
centrifuged for 5 min at 800 × g and 4°C, following which the
supernatant was discarded, and the cell pellets were washed twice
with pre-cooled PBS and were resuspended in PBS at a density of
1×106 cells/ml. The suspension was then divided into
1-ml aliquots in EP tubes. For the detection of cell cycle
distribution and proliferation, 1-ml single-cell suspensions were
centrifuged for 5 min at 800 × g and 4°C) and the supernatants were
discarded. Subsequently, 70% pre-cooled ethyl alcohol (−20°C, 1 ml)
was added, mixed repeatedly and incubated overnight at 4°C. The
cells were resuspended in 1 ml PBS, and 1 ml RNase ribonuclease was
added to the cell suspensions, followed by incubation for 40 min at
37°C. Propidium iodide solution (1 ml) was added and the mixture
was kept in the dark for 10 min at 4°C. Proliferation was analyzed
using a FACS Vantage flow cytometer (BD Biosciences, Franklin
Lakes, NJ, USA), and the ModFit LT 2.0 program (Verity Software
House, Inc., Topsham, ME, USA) was used to analyze the proportion
of cells in each phase of the cell cycle. The tumor cell
proliferation index (PI) was calculated as follows: PI=[S + (G2 and
M)]/[(G0 and G1) + S + (G2 and M)] ×100.
For the detection of cell apoptosis, the single-cell
suspensions (1 ml) were fixed in PBS for 2 h at 4°C and were then
resuspended in Binding Buffer (0.5 ml). The suspensions were
incubated with Annexin V-FITC (20 µg/ml) reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) for 15 min at 4°C in the dark
according to the manufacturer's protocols, and were then stained
with 1 ml propidium iodide solution for 2 min at room temperature
in the dark. Apoptosis was analyzed using the FACS Vantage flow
cytometer (BD FACS Aria II software; BD Biosciences, Franklin
Lakes, NJ, USA). The tumor cell apoptotic index (AI) was calculated
as follows: AI=(number of apoptotic cells/total number of cells)
×100. Each experiment was performed in triplicate.
Statistical analysis
All statistical analyses were performed using SPSS
19.0 software (IBM Corp., Armonk, NY, USA). Comparisons between
multiple groups were made using one-way analysis of variance,
followed by the least significant difference test. Data are
expressed as the mean ± standard deviation. P<0.05was considered
to indicate a statistically significant difference.
Results
Construction and sequencing of shRNA
lentiviral plasmids
The sequencing results obtained from the lentiviral
plasmids, PER2-shRNA-I–III, are presented in Table III. These sequences exactly matched
the interference target sequences of the positive-sense strands of
PER2-shRNA-I–III, indicating that three interference target
sequences of PER2 were successfully constructed.
 | Table III.Sequences of PER2-shRNA
interference. |
Table III.
Sequences of PER2-shRNA
interference.
| Group | Sense strand |
|---|
| PER2-shRNA-I |
5′-CCGGGCCAGAGTCCAGATACCTTTACTCGAGTAAAGGTATCTGGACTCTGGCTTTTTG-3′ |
| PER2-shRNA-II |
5′-CCGGGCATCCATATTTCACTGTAAACTCGAGTTTACAGTGAAATATGGATGCTTTTTG-3′ |
| PER2-shRNA-III |
5′-CCGGGCACACACACAAAGAACTGATACTCGAGTATCAGTTCTTTGTGTGTGTCTTTTTG-3′ |
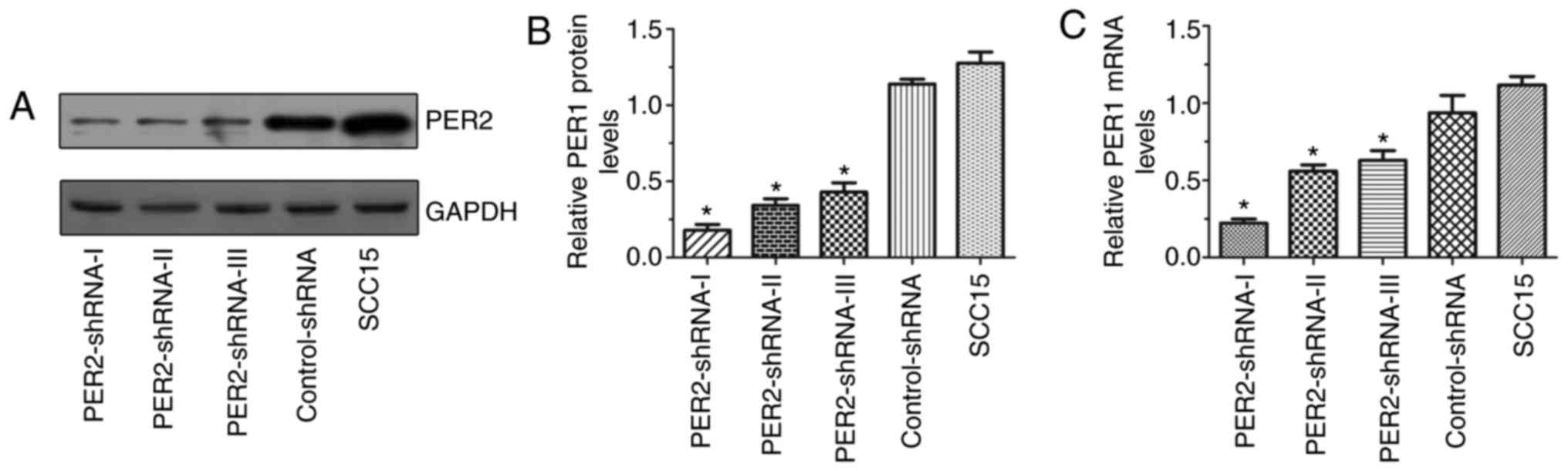
mRNA and protein expression of PER2 in
SCC15 cells following transfection
The mRNA and protein expression of PER2 in
the PER2-shRNA-I–III, control-shRNA and untransfected SCC15 groups
are presented in Fig. 1. The
PER2 mRNA and protein expression in the PER2-shRNA-I group
was significantly lower than in the other groups (P<0.05),
indicating that the PER2 knockdown was most effective in the
PER2-shRNA-I group. Therefore, this shRNA was selected for use in
the subsequent experiments.
mRNA expression of clock genes in
SCC15 cells following PER2 knockdown
RT-qPCR analysis demonstrated that the mRNA
expression levels of PER1 and REV-ERBα were
significantly upregulated in the PER2-shRNA-I group compared with
in the control-shRNA and SCC15 groups (P<0.05), while the mRNA
expression levels of PER3, BMAL1, DEC1, DEC2, CRY2, TIM,
RORα and NPAS2 were significantly downregulated
(P<0.05). There was no significant difference between mRNA
expression levels in the control-shRNA and the SCC15 groups
(P>0.05). In addition, there was no significant difference in
the mRNA expression levels of CLOCK, CRY1 or CKIε
among the three groups (P>0.05; Fig.
2).
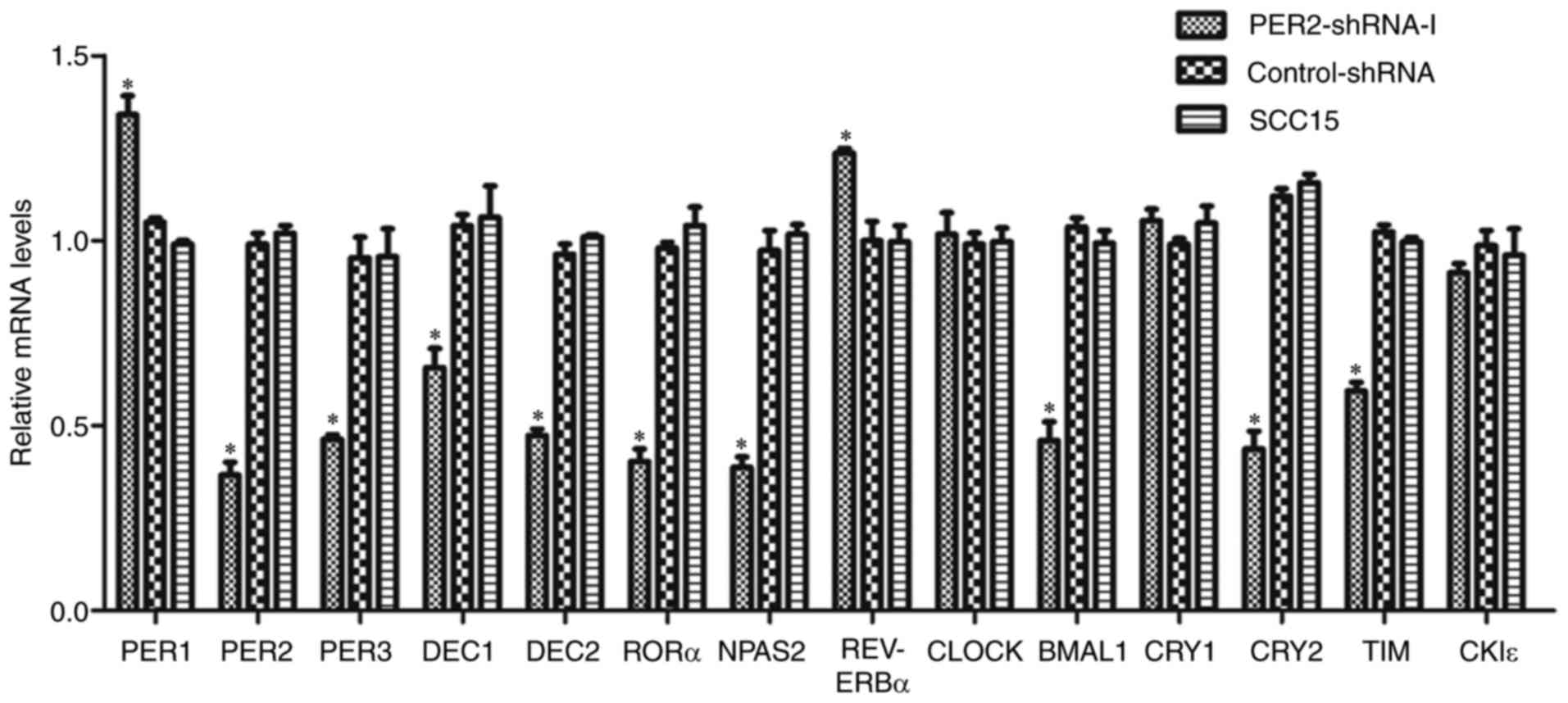 | Figure 2.Alteration of mRNA expression of
clock genes in SCC15 cells following PER2 knockdown. mRNA
expression levels of PER1 and REV-ERBα were
significantly upregulated in the PER2-shRNA-I group when compared
with in the control-shRNA and SCC15 groups, while the mRNA
expression of PER3, BMAL1, DEC1, DEC2, CRY2, TIM, RORα and
NPAS2 was significantly downregulated. There was no notable
difference between the control-shRNA and SCC15 groups. The mRNA
expression levels of CLOCK, CRY1 and CKIε were not
significantly different among the three groups. Data are presented
as the mean ± standard deviation. *P<0.05. PER, period
circadian clock; REV-ERBα, nuclear receptor subfamily 1
group D member 1; shRNA, short hairpin RNA; BMAL1, brain and
muscle ARNT-like 1; DEC, deleted in esophageal cancer;
CRY, cryptochrome circadian clock; TIM, timeless
circadian clock; RORα, retinoic acid receptor-related orphan
receptor-α; NPAS2, neuronal PAS domain protein 2;
CLOCK, clock circadian regulator; CKIε, casein kinase
1 epsilon. |
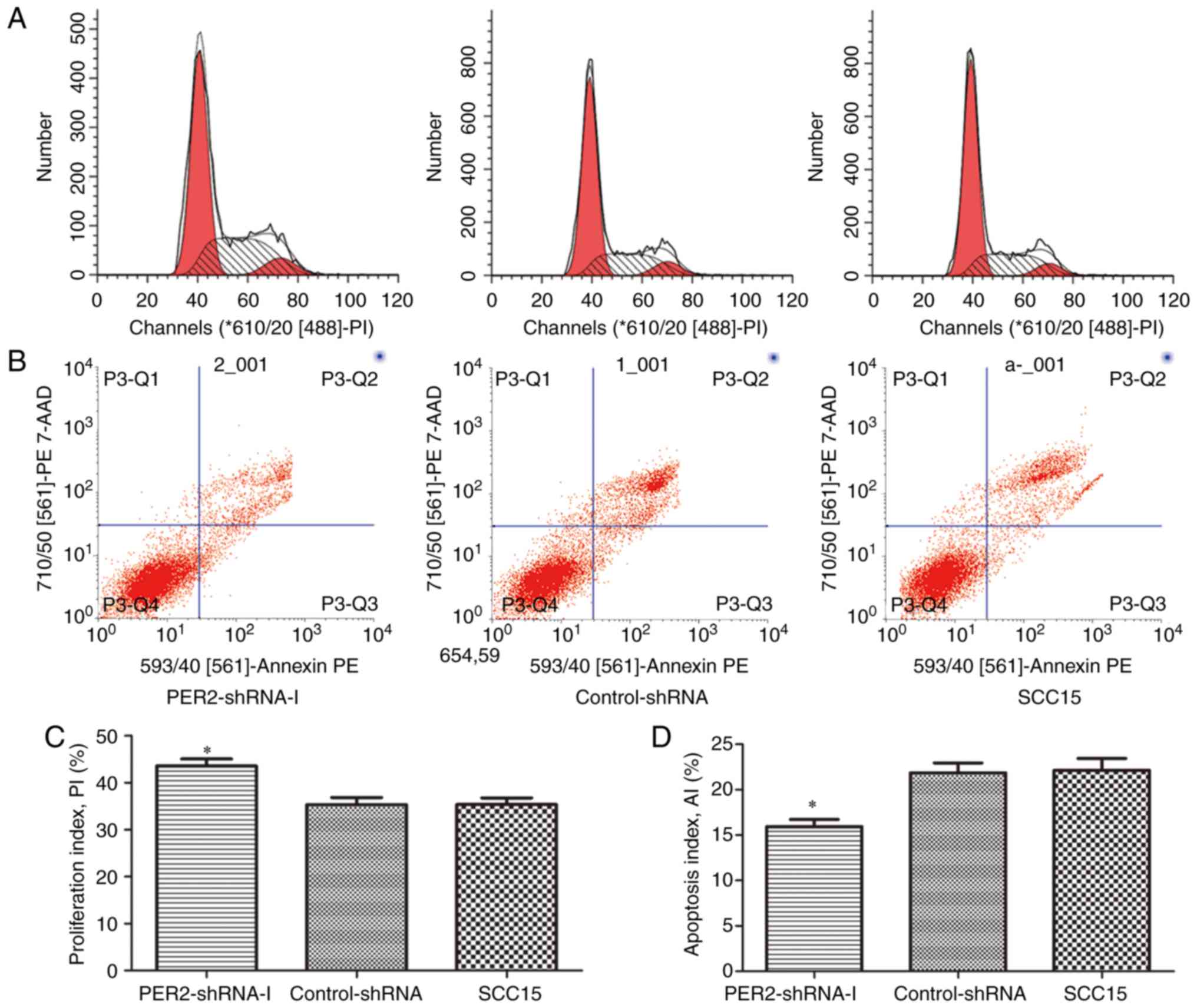
Proliferation and apoptosis of SCC15
cells following PER2 knockdown
Flow cytometric analysis indicated that the cell PIs
of the PER2-shRNA-I group, control-shRNA group and untreated SCC15
group were 43.55±2.64, 35.32±2.64 and 35.36±2.46%, respectively.
The cell AIs were 15.94±1.36, 21.85±1.90 and 22.13±2.29%,
respectively. The cell PI of the PER2-shRNA-I group was markedly
higher than that of the other two groups, while the cell AI was
significantly lower than that of the other two groups (P<0.05).
There was no significant difference between the control-shRNA group
and the SCC15 group with regards to PI or AI (P>0.05; Fig. 3).
Discussion
Previous studies have indicated that different clock
genes regulate various downstream CCGs, and therefore clock genes
are considered to be important for regulating cell biochemical and
physiological functions (12). The
clock gene, PER2, is downregulated in various types of
cancer, and is associated with cancer occurrence and progression
via the regulation of numerous downstream cell cycle genes
(15–18). Although PER2 is known to be an
important clock gene (23), whether
its reduced expression in cancer may lead to the alteration of
other clock genes remains unclear at present.
Research has demonstrated that there is a network
structure, including three feedback loops, between clock genes at
the translation-transcription levels in normal cells (4,24–26). To begin with, BMAL1 combines with
CLOCK or NPAS2 to form heterodimers, and acts as a transcription
factor to activate the expression of PER genes (PER1,
PER2 and PER3), CRY genes (CRY1 and
CRY2), DEC genes (DEC1 and DEC2),
TIM, RORα and REV-ERBα. PER and CRY proteins then
combine to form heterodimers and translocate into the cell nucleus
to repress the activity of CLOCK/BMAL1 or NPAS2/BMAL1, thereby
generating the first negative feedback loop, which is the primary
feedback loop in the network (24,25). In
the same way, DEC1 and DEC2 proteins form dimers or heterodimers
and translocate to the nucleus to inhibit the activity of
CLOCK/BMAL1, thereby comprising the second negative feedback loop
(26). REV-ERBα and RORα proteins
translocate to the nucleus to suppress and promote the expression
of BMAL1, respectively, thereby generating the third
feedback loop (4). In normal feedback
loops involving the aforementioned clock genes, PER2 is a
component of the first, main negative feedback loop, serving as a
negative regulatory factor. When the mRNA expression of PER2
is downregulated, its inhibitory effect on the positive-regulatory
factors, CLOCK/BMAL1 and NPAS2/BMAL1, is reduced; therefore,
PER2 downregulation may lead to higher mRNA expression
levels of negative regulatory genes, including PER1, PER3, CRY1,
CRY2, DEC1, DEC2, TIM, REV-ERBα and RORα. However, in
the present study, following PER2 knockdown, the mRNA
expression levels of NPAS2 and BMAL1 were
significantly downregulated, suggesting that the
transcription-activating effect of the positive regulatory factor,
CLOCK/BMAL1, is suppressed. Among the negative regulatory genes,
only the mRNA expression levels of PER1 and REV-ERBα
were significantly upregulated, which is consistent with the
features of circadian feedback regulation in normal cells. By
contrast, the mRNA expression of remaining negative regulatory
genes, including PER3, DEC1, DEC2, CRY2, TIM and
RORα, was downregulated, and the mRNA expression of
CRY1 was not significantly altered, which was not consistent
with the features of circadian feedback regulation in normal cells.
We hypothesized that the reason for this difference may be that the
three known feedback loops have been researched in normal cells
(4,24–26), while
the present study was conducted using cancer cells. Therefore, the
regulatory effects of circadian positive- and negative-feedback
networks in cancer cells may have been altered, or there may exist
other compensatory regulatory approaches. However, this requires
further investigation.
In the present study, the mRNA expression levels of
the CLOCK gene were not notably altered following
PER2 knockdown in human OSCC cells. A previous study by Lee
et al (27), conducted in
normal mouse liver cells, demonstrated that CLOCK was not
changed by the alteration of PER2 mRNA expression.
Additionally, Shearman et al (3) reported that, in normal mouse liver
cells, PER2 knockdown did not alter the expression of CLOCK
protein, but instead promoted the nuclear translocation of the
CLOCK/BMAL1 heterodimer to act as a transcription factor.
Therefore, we hypothesized that the regulatory effect of silencing
PER2 on the positive regulatory factor CLOCK may
result from changes in its intracellular distribution. In addition,
PER2 may exert no regulation on CLOCK at the
transcriptional level.
In summary, the clock gene, PER2, has
previously been reported to be downregulated in various types of
cancer, and to be involved in the occurrence and progression of
cancer via the regulation of downstream cell cycle genes (18,19). The
present study has demonstrated that PER2 is also important
in the regulation of the clock gene network, which contributes to
the previously established knowledge regarding PER2
function. To the best of our knowledge, the present study is the
first to report that the reduced expression of PER2
significantly decreases the mRNA expression levels of PER3,
BMAL1, DEC1, DEC2, CRY2, TIM, RORα and NPAS2 in cancer
cells, while the mRNA expressions of PER1 and
REV-ERBα were significantly upregulated. These observations
also indicated that that PER2 knockdown enhances the
proliferation and reduces the apoptosis of OSCC cells. In
conclusion, PER2 serves an important role in regulating
other clock genes in the clock gene network in cancer cells. This
is of great importance in further illustrating the molecular
functions and tumor-suppressor mechanisms of PER2. However,
the results of the present study identified the effects on
expression at the transcriptional level, and further studies at the
translational level are required in the future.
Acknowledgements
The authors would like to thank Master of Medicine
(MM) Xiao-Juan Fu and Han-Xue Li of The First Affiliated Hospital
of Chongqing Medical University for providing technical and
statistical assistance. The authors would also like to thank Dr
Qing-Qing Wang of Stomatological Hospital of Chongqing Medical
University, for aiding in the conduction of certain
experiments.
Glossary
Abbreviations
Abbreviations:
|
CCGs
|
clock-controlled genes
|
|
shRNA
|
short hairpin RNA
|
|
OSCC
|
oral squamous cell carcinoma
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
PVDF
|
polyvinylidene fluoride
|
|
ECL
|
enhanced chemiluminescence
|
References
|
1
|
Fu L and Lee CC: The circadian clock:
Pacemaker and tumour suppressor. Nature Rev Cancer. 3:350–361.
2003. View
Article : Google Scholar
|
|
2
|
Ye H, Yang K, Tan XM, Fu XJ and Li HX:
Daily rhythm variations of the clock gene PER1 and cancer-related
genes during various stages of carcinogenesis in a golden hamster
model of buccal mucosa carcinoma. Onco Targets Ther. 8:1419–1426.
2015.PubMed/NCBI
|
|
3
|
Shearman LP, Sriram S, Weaver DR, Maywood
ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings
MH and Reppert SM: Interacting molecular loops in the mammalian
circadian clock. Science. 288:1013–1019. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Preitner N, Damiola F, Lopez-Molina L,
Zakany J, Duboule D, Albrecht U and Schibler U: The orphan nuclear
receptor REV-ERBalpha controls circadian transcription within the
positive limb of the mammalian circadian oscillator. Cell.
110:251–260. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao N, Yang K, Yang G, Chen D, Tang H,
Zhao D and Zhao C: Aberrant expression of clock gene period1 and
its correlations with the growth, proliferation and metastasis of
buccal squamous cell carcinoma. PLoS One. 8:e558942013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lowrey PL and Takahashi JS: Genetics of
the mammalian circadian system: Photic entrainment, circadian
pacemaker mechanisms, and posttranslational regulation. Annu Rev
Genet. 34:533–562. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rana S and Mahmood S: Circadian rhythm and
its role in malignancy. J Circadian Rhythms. 8:32010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bozek K, Relogio A, Kielbasa SM, Heine M,
Dame C, Kramer A and Herzel H: Regulation of clock-controlled genes
in mammals. PLoS One. 4:e48822009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chu G, Yoshida K, Narahara S, Uchikawa M,
Kawamura M, Yamauchi N, Xi Y, Shigeyoshi Y, Hashimoto S and Hattori
MA: Alterations of circadian clockworks during differentiation and
apoptosis of rat ovarian cells. Chronobiol Int. 28:477–487. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Filipski E, King VM, Li X, Granda TG,
Mormont MC, Liu X, Claustrat B, Hastings MH and Lévi F: Host
circadian clock as a control point in tumor progression. J Natl
Cancer Inst. 94:690–697. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hayashida S, Kuramoto Y, Koyanagi S, Oishi
K, Fujiki J, Matsunaga N, Ikeda E, Ohdo S, Shimeno H and Soeda S:
Proxisome proliferator-activated receptor-α mediates high-fat,
diet-enhanced daily oscillation of plasminogen activator
inhibitor-1 activity in mice. Chronobiol Int. 27:1735–1753. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Panda S, Antoch MP, Miller BH, Su AI,
Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS and
Hogenesch JB: Coordinated transcription of key pathways in the
mouse by the circadian clock. Cell. 109:307–320. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Miyazaki K, Wakabayashi M, Hara Y and
Ishida N: Tumor growth suppression in vivo by overexpression of the
circadian component, PER2. Genes Cells. 15:351–358. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang X, Wood PA, Ansell C and Hrushesky
WJ: Circadian time-dependent tumor suppressor function of period
genes. Integr Cancer Ther. 8:309–316. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ
and Chang JG: Deregulated expression of the PER1, PER2 and PER3
genes in breast cancers. Carcinogenesis. 26:1241–1246. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao H, Zeng ZL, Yang J, Jin Y, Qiu MZ, Hu
XY, Han J, Liu KY, Liao JW, Xu RH and Zou QF: Prognostic relevance
of Period1 (Per1) and Period2 (Per2) expression in human gastric
cancer. Int J Clin Exp Pathol. 7:619–630. 2014.PubMed/NCBI
|
|
17
|
Hsu CM, Lin SF, Lu CT, Lin PM and Yang MY:
Altered expression of circadian clock genes in head and neck
squamous cell carcinoma. Tumour Biol. 33:149–155. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Q, Ao Y, Yang K, Tang H and Chen D:
Circadian clock gene Per2 plays an important role in cell
proliferation, apoptosis and cell cycle progression in human oral
squamous cell carcinoma. Oncol Rep. 35:3387–3394. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tan XM, Ye H, Yang K, Chen D, Wang QQ,
Tang H and Zhao NB: Circadian variations of clock gene Per2 and
cell cycle genes in different stages of carcinogenesis in golden
hamster buccal mucosa. Sci Rep. 5:99972015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
O'Driscoll A, Belogrudov V, Carroll J,
Kropp K, Walsh P, Ghazal P and Sleator RD: HBLAST: Parallelised
sequence similarity-A Hadoop MapReducable basic local alignment
search tool. J Biomed Inform. 54:58–64. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chernolovskaya EL and Zenkova MA: Design
of nuclease-resistant fork-like small interfering RNA (fsiRNA).
Methods Mol Biol. 942:153–168. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Militi S, Maywood ES, Sandate CR, Chesham
JE, Barnard AR, Parsons MJ, Vibert JL, Joynson GM, Partch CL,
Hastings MH and Nolan PM: Early doors (Edo) mutant mouse reveals
the importance of period 2 (PER2) PAS domain structure for
circadian pacemaking. Proc Natl Acad Sci USA. 113:pp. 2756–2761.
2016; View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gekakis N, Staknis D, Nguyen HB, Davis FC,
Wilsbacher LD, King DP, Takahashi JS and Weitz CJ: Role of the
CLOCK protein in the mammalian circadian mechanism. Science.
280:1564–1569. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Borgs L, Beukelaers P, Vandenbosch R,
Belachew S, Nguyen L and Malgrange B: Cell ‘circadian’ cycle: New
role for mammalian core clock genes. Cell Cycle. 8:832–837. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nakashima A, Kawamoto T, Honda KK, Ueshima
T, Noshiro M, Iwata T, Fujimoto K, Kubo H, Honma S, Yorioka N, et
al: DEC1 modulates the circadian phase of clock gene expression.
Mol Cell Biol. 28:4080–4092. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee C, Etchegaray JP, Cagampang FR, Loudon
AS and Reppert SM: Posttranslational mechanisms regulate the
mammalian circadian clock. Cell. 107:855–867. 2001. View Article : Google Scholar : PubMed/NCBI
|