Introduction
Chemokines are a superfamily of small (~8-14 kDa)
molecules that mediate numerous cellular functions by activating G
protein-coupled receptors (1).
Chemokines and their respective receptors are also associated with
metastasis in different types of cancer, including osteosarcoma
(2) and neuroblastoma (3), as well as prostate (4) and breast (5) cancer. The chemokine CXC ligand 12
(CXCL12; also known as stromal cell-derived factor-1) is implicated
in numerous cellular processes that are important in aspects of
tumour progression. It interacts with its cognate receptors CXC
chemokine receptor (R) type 4 (1) and
CXCR7 (6) to regulate cell
trafficking and adhesion, tumour vascularisation, cell
proliferation and survival (7,8). CXCL12
enhances the invasiveness and migratory properties of breast cancer
cells, particularly when these cells also express CXCR4 (9). Indeed, CXCR4 expression is upregulated
in primary breast tumours compared with normal mammary epithelial
cells (5) indicating that it serves
an important function in the progression and metastasis of breast
cancer (10,11).
CXCL12 responses are regulated by other factors
beyond its receptors CXCR4 and CXCR7. Among these factors is
cluster of differentiation (CD)24, a glycosylated cell surface
protein that acts as a signal transducer in modulating responses to
B cell activation (12). Schabath
et al (13) demonstrated that
MDA-MB-231 breast cancer cells with low CD24 expression exhibit
augmented CXCL12/CXCR4-mediated cell migration and enhanced tumour
growth compared with MDA-MB-231 cells that express high exogenous
levels of CD24, suggesting that higher CD24 expression decreases
CXCL12 responses in breast cancer cells. Hypoxia-inducible
factor-2α (HIF2α) also regulates CXCR4 expression (14) and may therefore influence CXCL12
responsiveness.
Certain cells respond to CXCL12 activation by
releasing Ca2+ from the endoplasmic reticulum internal
Ca2+ store via G-protein coupled receptor, triggering
phospholipase C activation and the generation of inositol
trisphosphate and diacylglycerol (7).
Ca2+ signalling is associated with processes that occur
during metastasis, including cell migration and invasion (15,16), as
well as the induction of an increasingly invasive phenotype by
stimulating the epithelial-mesenchymal transition (EMT) (17). EMT is a process whereby epithelial
cells undergo conversion to an increasingly mesenchymal (invasive)
phenotype (18). However, the nexus
between CXCL12, Ca2+ signalling, CXCL12 modulators and
receptors and EMT has not yet been fully evaluated.
The nature of Ca2+ store release as a
result of CXCL12/CXCR4 interaction may be tissue-dependent and vary
between cell types (7). Changes in
Ca2+ signalling and/or the expression of specific
modulators of Ca2+ signalling are a feature of some
subtypes of breast cancer and these changes often differ between
different breast cancer subtypes. For example, the ratio of the
calcium release-activated calcium channel protein (Orai)1 calcium
influx pathway activators stromal interaction molecule 1/2 is
higher in the basal molecular breast cancer subtype than in other
subtypes (19). It has been
demonstrated that Orai3 regulates store-operated Ca2+
entry in oestrogen receptor-positive breast cancer cell lines such
as MCF-7 but does not in oestrogen receptor-negative breast cancer
cell lines such as MDA-MB-231 (20).
Elevated transient receptor potential cation channel V6 levels are
more common in types of breast cancer that are oestrogen
receptor-negative (21). Oestrogen
receptor-negative breast cancer, particularly those of the
triple-negative subtype, exhibit a significant overlap with
molecularly defined basal breast cancer (22).
Basal breast cancer cell lines possess gene
signatures that allow them to be divided into basal A and basal B
subtypes (23). In the present study,
Ca2+ signalling induced by CXCL12 was compared between
two triple-negative basal breast cancer cell lines, MDA-MB-468
(basal A) and MDA-MB-231 (basal B). mRNA levels of CXCL12 receptors
and their response modulators in EMT and in breast cancer cell
lines of different molecular subtypes were also characterised. The
present study therefore aimed to assess the potential heterogeneity
of responses to CXCL12 in the context of induced Ca2+
increases in basal breast cancer.
Materials and methods
Cell culture
The human basal-like triple-negative breast cancer
cell lines MDA-MB-231 and MDA-MB-468 were obtained from the
American Type Culture Collection (Manassas, VA, USA) and The
Brisbane Breast Bank, University of Queensland Centre for Clinical
Research, (Brisbane, Australia) respectively, and maintained in
Dulbecco's modified Eagle's medium (DMEM; D6546; Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) supplemented with 10% foetal bovine
serum (FBS; Sigma-Aldrich; Merck KGaA), L-glutamine (4 mM;
Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
penicillin 100 U/ml and streptomycin 100 µg/ml (Invitrogen; Thermo
Fisher Scientific, Inc.) in a humidified incubator at 37°C in an
atmosphere containing 5% CO2. Cells were routinely
screened for mycoplasma contamination using the MycoAlert
Mycoplasma Detection kit (LT07-218; Lonza Group, Ltd., Basel,
Switzerland) and validated by short tandem repeat profiling using
the StemElite ID Profiling kit (Promega Corporation, Madison, WI,
USA).
Intracellular Ca2+
measurement
For Ca2+ measurements, MDA-MB-231
(7.5×103 cells/well) or MDA-MB-468 (1.5 ×104
cells/well) cells were seeded in a 96-well CellBIND plate (Corning
Life Sciences, Corning, NY, USA) in antibiotic-free DMEM containing
L-glutamine (4 mM) and 10% FBS (MDA-MB-468 cells were seeded at a
higher density due to their slower proliferation rate). At 24 h
post-plating, the FBS concentration was decreased to 8%. At 72 h
post-plating, Ca2+ assays were performed using a
fluorescence imaging plate reader, FLIPRTETRA (Molecular
Devices, LLC, Sunnyvale, CA, USA) and 4 µM Fluo-4 AM dye (Molecular
Probes; Thermo Fisher Scientific, Inc.) in physiological salt
solution, as previously described (24). Cells were excited at 470–495 nm and
emission was assessed at 515–575 nm over an 800 sec period.
Relative cytoplasmic (CYT) [Ca2+]CYT was
determined in the presence of 300 and 100 ng/ml recombinant human
CXCL12 (R&D Systems, Inc., Minneapolis, MN, USA) or 100 µM
adenosine 5′-triphosphate (ATP; Sigma-Aldrich; Merck KGaA). Data
were acquired using ScreenWorks™ software (v2.0.0.27, Molecular
Devices, LLC) and are presented as the response over baseline,
which is a measure of relative [Ca2+]CYT.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated using the RNeasy®
Plus Mini kit (Qiagen GmbH, Hilden, Germany). RT reactions were
performed using an Omniscript Reverse Transcriptase kit (Qiagen
GmbH) with random primers and RNase inhibitor (Promega
Corporation), according to the manufacturer's protocol. qPCR was
conducted using Applied Biosystems TaqMan gene expression assays
and TaqMan Universal PCR Master mix (Applied Biosystems; Thermo
Fisher Scientific, Inc.). Assays included CD24 (Assay ID:
Hs02379687_s1), CXCR4 (Assay ID: Hs00237052_m1), CXCR7 (Assay ID:
Hs00664172_s1), HIF2α (Assay ID: Hs01026149_m1) and the endogenous
control 18S ribosomal RNA (4319413E). All amplifications were
performed using universal cycling conditions [20 sec at 95°C
(holding stage)], followed by 40 cycles of denaturation for 1 sec
at 95°C and combined annealing and extension steps for 20 sec at
60°C) in a StepOnePlus™ Real-Time PCR System Thermal Cycling Block
(Applied Biosystems; Thermo Fisher Scientific, Inc.). Data were
normalised to 18S ribosomal RNA and analysed using the comparative
Cq method as previously described (25).
Epidermal growth factor (EGF)-induced
EMT
For assessment of EGF-induced EMT, MDA-MB-468 cells
were plated at a density of 2×104 into a 96-well plate
and serum-starved (0.5% FBS) for 24 h prior to treatment with 50
ng/ml EGF (Sigma-Aldrich; Merck KGaA), as previously described
(26). Total RNA was isolated at 24 h
post-EGF treatment and subjected to RT-qPCR following the
aforementioned protocol to assess the changes in CXCR4, CXCR7, CD24
and HIF2α expression.
Hypoxia-induced EMT
For hypoxia-induced EMT, MDA-MB-468 cells were
seeded at a density of 2×104 in a 96-well plate and
serum-deprived (0.5% FBS) for 24 h. Cells were then exposed to
hypoxic conditions (1% O2) in a Sanyo MCO-18M multi-gas
incubator (Sanyo Electric Co., Ltd., Tokyo, Japan). Normoxic
control MDA-MB-468 cells were incubated in a humidified incubator
(37°C, 5% CO2) with normal atmosphere (21%
O2). Total RNA was isolated at 24 h following normoxic
or hypoxic conditions to assess changes in CXCR4, CXCR7, CD24 and
HIF2α expression, following the aforementioned protocol.
Analysis of CXCR4 and CXCR7 expression
in breast tumours
Breast tumour gene expression data were sourced from
the METABRIC breast cancer data (www.cbioportal.org, last accessed April 3, 2017)
(27–29). This dataset comprises gene expression
profiles from 1,860 tumours from six molecular subtypes including
198 basal-like (Basal), 182 Claudin-low (C-low), 218 human
epidermal growth factor receptor 2-enriched (HER2), 673 luminal A
(LumA), 454 luminal B (LumB) and 135 normal-like (N-Like).
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 7 (GraphPad Software, Inc., La Jolla, CA, USA). Results are
expressed as the mean ± standard deviation from the specified
number of independent experiments. The statistical tests used are
stated in each figure legend, and included one-way analysis of
variance followed by Tukey's multiple comparison and an unpaired
t-test, where appropriate.
Results
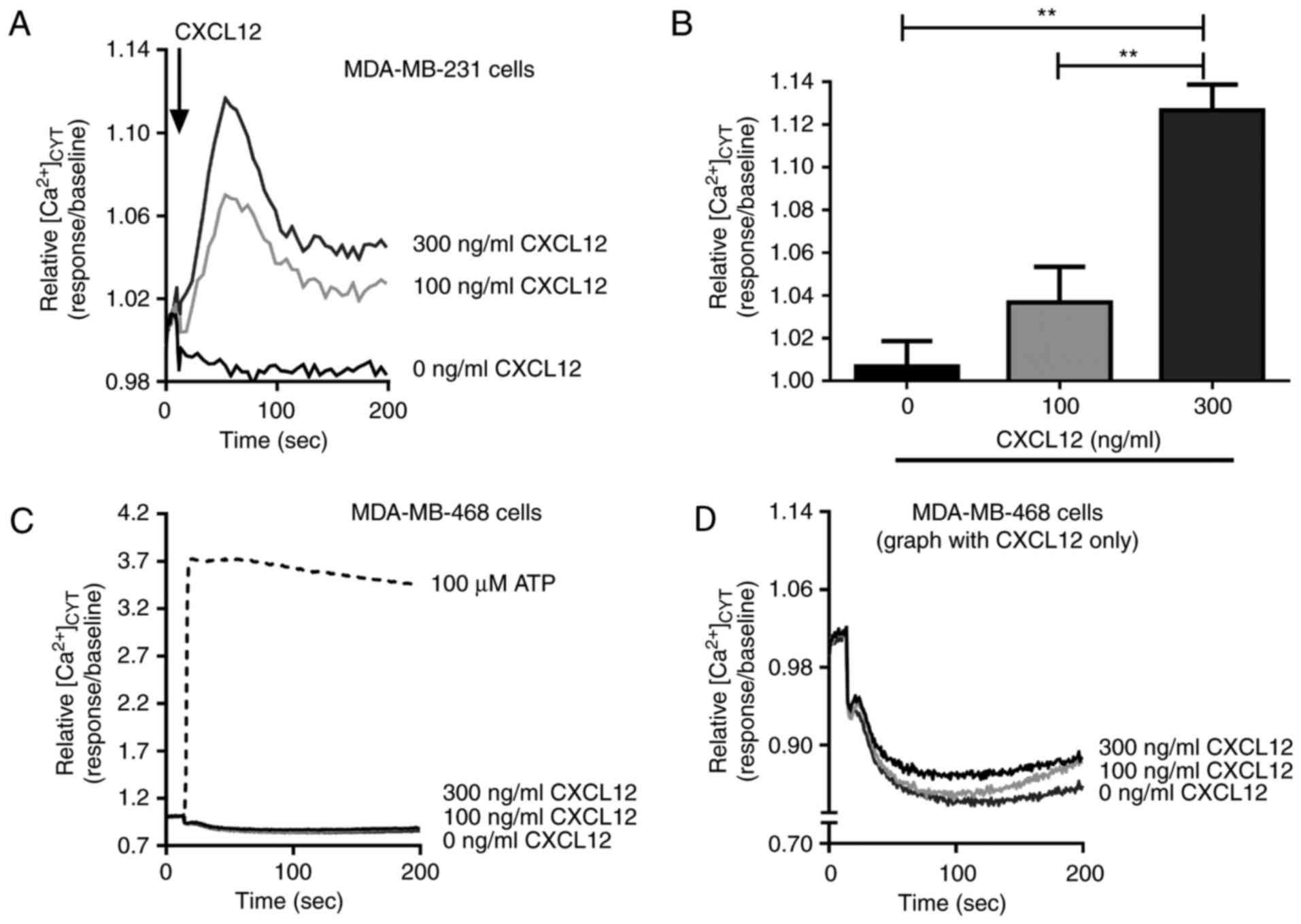
CXCL12-induced intracellular free
Ca2+ increases in breast cancer cells
The effects of CXCL12 on intracellular
Ca2+ concentrations were assessed in two basal-like
breast cancer cell lines, MDA-MB-231 (basal B) and MDA-MB-468
(basal A). A significant concentration-dependent increase in
[Ca2+]CYT was observed following CXCL12
treatment in MDA-MB-231 breast cancer cells (P<0.01; Fig. 1A and B) compared with the untreated
control. However, no significant increase in
[Ca2+]CYT was observed in MDA-MB-468 cells
treated with CXCL12 compared with the untreated control, despite a
pronounced elevation in intracellular Ca2+ levels
observed during stimulation with the purinergic receptor activator
ATP (Fig. 1C and D).
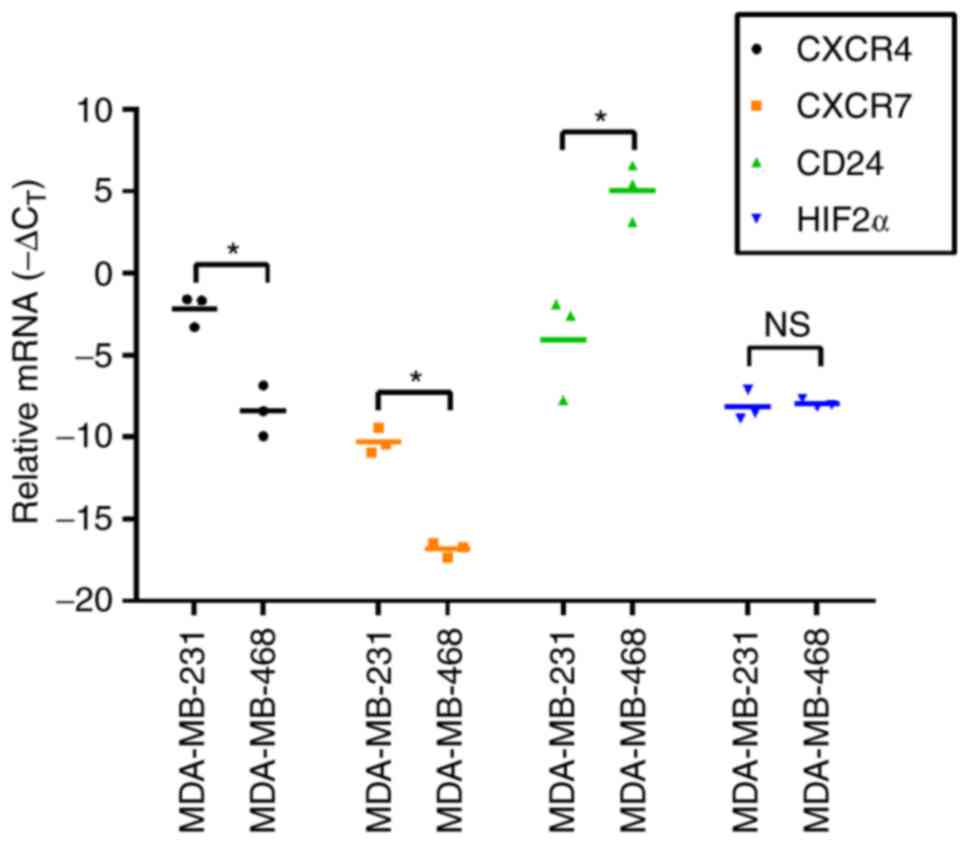
Levels of CXCR4, CXCR7, CD24 and HIF2α
mRNA in MDA-MB-231 and MDA-MB-468 breast cancer cells
To explore the potential reasons for the lack of
CXCL12-induced [Ca2+]CYT in MDA-MB-468 basal
A breast cancer cells, compared with the significant increases
observed in more metastatic MDA-MB-231 basal B breast cancer cells,
mRNA expression of the potential regulators of CXCL12 responses
were measured. Levels of mRNA for the CXCL12 receptor CXCR4 were
significantly increased in MDA-MB-231 cells compared with
MDA-MB-468 cells (P<0.05; Fig. 2).
Similarly, levels of CXCR7, another receptor for CXCL12 (6), were significantly increased in
MDA-MB-231 cells compared with MDA-MB-468 cells (P<0.05;
Fig. 2). CD24 is a negative regulator
of CXCL12 responses in MDA-MB-231 cells (13) and in the present study, it was
revealed that there were significantly higher levels of CD24 mRNA
in MDA-MB-468 cells (P<0.05; Fig.
2), which were not responsive to CXCL12 as assessed by
increases in [Ca2+]CYT (Fig. 1C), compared with MDA-MB-231 cells.
Given that an association between HIF2α and CXCR4 expression has
been identified (14), HIF2α levels
were also assessed in the two cell lines in the present study;
however, no significant difference was observed (Fig. 2).
Assessment of CXCR4, CXCR7, CD24 and
HIF2α during hypoxia- and EGF-induced EMT in MDA-MB-468 breast
cancer cells
MDA-MB-231 is a basal B cell line and exhibits
mesenchymal features, including vimentin expression and a lack of
E-cadherin expression (30). Given
that CXCL12-mediated calcium signalling may be influenced by CXCR4,
CXCR7 and CD24 (7,13), and that their expression differed
between the more epithelial MDA-MB-468 (basal A) and the more
mesenchymal MDA-MB-231 (basal B) breast cancer cell lines, the
effect of EMT induction on CXCR4, CXCR7, CD24 and HIF2α expression
in MDA-MB-468 cells was assessed. In the present study EMT
induction with two distinct inducers was assessed to define the
changes associated with EMT rather than the stimuli. Our previous
studies of EMT in EGF and hypoxia models in this cell line produced
increases in levels of the mesenchymal markers N-cadherin, zinc
finger protein SNAI1, zinc finger E-box binding homeobox 1, CD44,
twist-related protein and vimentin, downregulation of the
epithelial markers E-cadherin and Claudin-4, as well as CD24, and
changes to a more spindle morphology (31–33). In
the present study, EGF and hypoxia produced opposing effects on
CXCR4 levels and did not affect CXCR7 levels (Fig. 3A and B). Only the mRNA level of the
known EMT marker CD24 was significantly decreased following
induction of the EMT by the two inducers (P<0.01; Fig. 3C). HIF2α mRNA levels significantly
increased following hypoxia (P<0.05), however they were
unaffected by EGF (Fig. 3D).
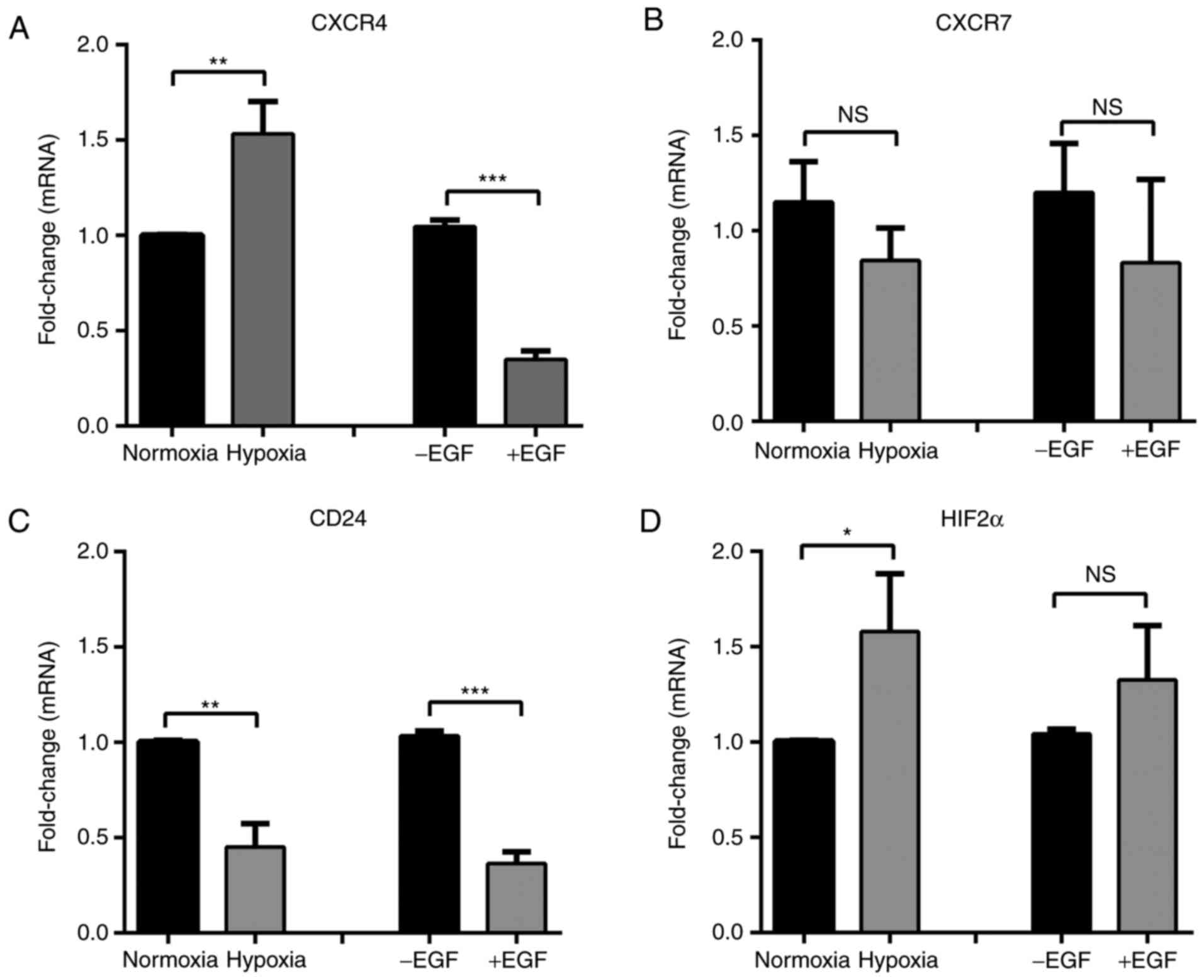 | Figure 3.mRNA levels of CXCR4, CXCR7, CD24 and
HIF2α mRNA following hypoxia- and EGF-induced EMT in MDA-MB-468
breast cancer cells expressed as fold change. (A) CXCR4, (B) CXCR7,
(C) CD24, and (D) HIF2α mRNA levels in MDA-MB-468 cells incubated
under normoxic or hypoxic (24 h) conditions or stimulated with EGF
(24 h) to induce EMT. Results are presented as the mean ± standard
deviation (n=3). Statistical analysis was performed using an
unpaired t-test; *P<0.05, **P<0.01, ***P<0.001. NS, not
significant; CXCR, CXC chemokine receptor; CD, cluster of
differentiation; HIF, hypoxia-inducible factor; EMT,
epithelial-mesenchymal transition; EGF, epidermal growth
factor. |
CXCR4 and CXCR7 expression is enriched
in breast tumours with mesenchymal features
Assessment of gene expression profiles in breast
tumours classified based on the differential expression of 50 genes
(PAM50) (29), demonstrated that
CXCR4 and CXCR7 levels were higher in the C-Low subtype compared
with the other subtypes (Fig. 4). The
C-Low subtype is markedly associated with the EMT and stem
cell-like features (34).
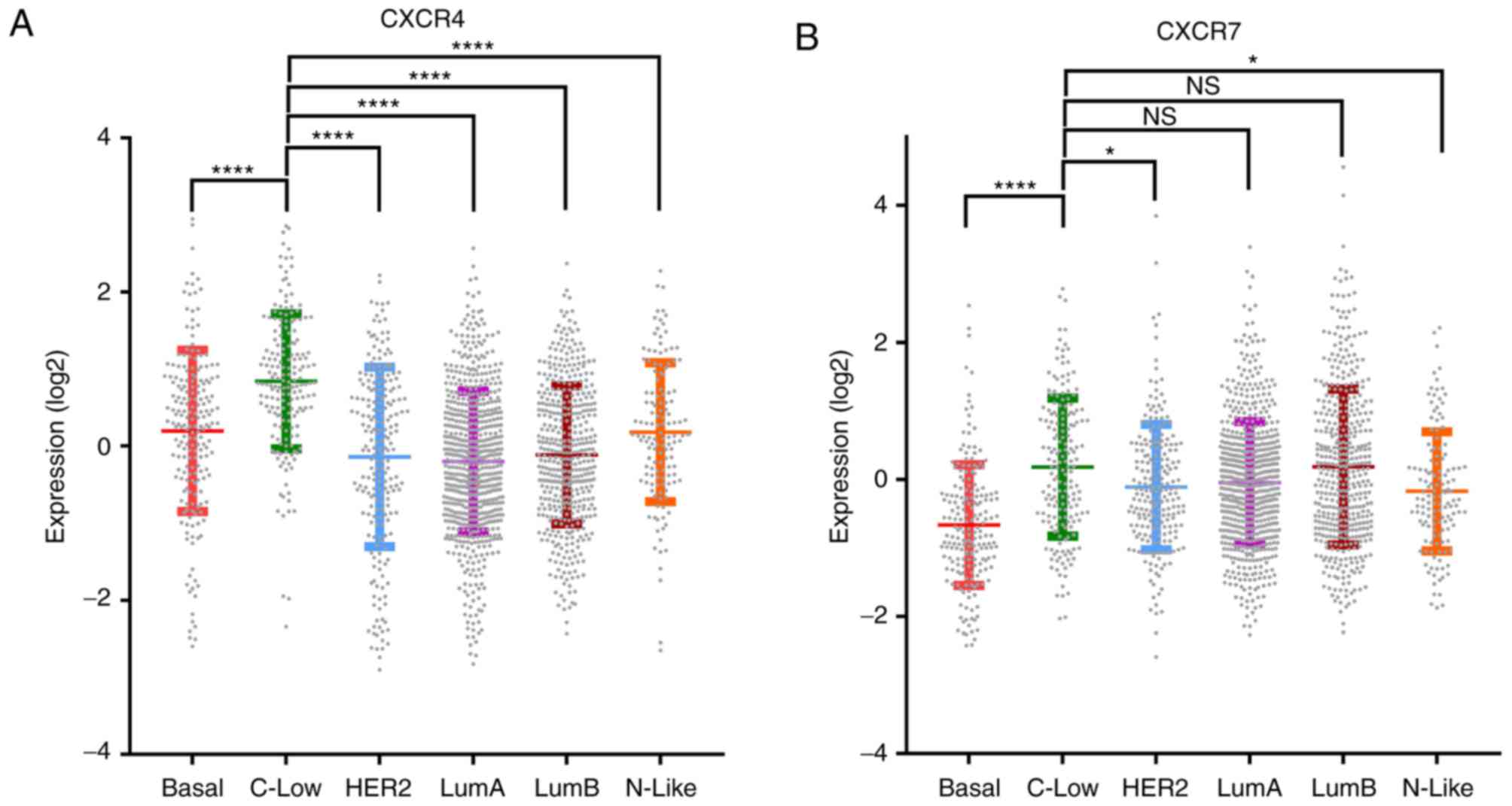 | Figure 4.The expression of CXCR4 and CXCR7 is
enriched in the C-Low molecular subtype of breast tumours compared
with the basal molecular subtype. Log2 expression values of (A)
CXCR4 and (B) CXCR7 in 6 subtypes of breast tumours, including
Basal, C-Low, HER2, LumA, LumB and N-Like. Data were sourced from
the METABRIC breast cancer data (29)
and analysed using www.cbioportal.org (27,28).
Statistical analysis was performed using one-way analysis of
variance followed by Tukey's multiple comparisons post-hoc test.
*P<0.05, ****P<0.0001. NS, not significant; Basal,
Basal-Like; C-Low, Claudin-Low; HER2, human epidermal growth factor
receptor 2-enriched; LumA, Luminal A; LumB, Luminal B; N-Like,
Normal-like; CXCR, CXC chemokine receptor. |
Discussion
The present study identified distinct CXCL12-induced
Ca2+ responses between two basal-like breast cancer cell
lines, MDA-MB-231 (basal B) and MDA-MB-468 (basal A).
CXCL12-mediated Ca2+ responses were observed in
MDA-MB-231 cells but not in MDA-MB-468 cells. It has been
demonstrated that CXCL12/CXCR4-mediated Ca2+ signalling
is enhanced in invasive breast cancer cell lines, such as
MDA-MB-231 and BT-549, compared with non-metastatic cell lines, due
to differences at the level of G protein subunit coupling that may
prevent the activation of CXCR4 in less metastatic cell lines
(35). The results of the present
study expand upon these findings, as it was demonstrated that
CXCL12-induced increases in [Ca2+]CYT were
more pronounced in the more metastatic MDA-MB-231 cell line
compared with less metastatic MDA-MB-468 cells.
The potential cause of differential CXCL12-induced
Ca2+ signalling between MDA-MB-231 and MDA-MB-468 cells
was explored by assessing the expression of the CXCL12 receptors
CXCR4 and CXCR7, as well as potential regulators of CXCL12
responses, specifically CD24 and HIF2α (13,14). The
results of the present study demonstrate that the expression of
CXCR4 and CXCR7 is increased in MDA-MB-231 cells compared with
MDA-MB-468 cells. The presence of CXCR4 and CXCR7 in the two breast
cancer cell lines supports the results of previous studies which
state that CXCR4 and CXCR7 are expressed in a variety of human
malignancies, including in breast and lung cancer (36,37). The
expression of CXCR4 and CXCR7 suggests that these receptors may
contribute to the CXCL12-induced Ca2+ responses in
MDA-MB-231 cells, which were observed in the present study. The
non-responsiveness of MDA-MB-468 cells to CXCL12 stimulation (as
measured by increases in Ca2+ levels) may be due in part
to the decreased expression of CXCR4 and CXCR7 in this cell line
compared with MDA-MB-231 cells. Analysis of CD24 mRNA expression
indicated that CD24 was abundantly expressed in non-responsive
MDA-MB-468 cells compared with MDA-MB-231 cells, a known feature of
basal A cell lines compared with basal B (38). The differential CD24 expression
detected in the present study is also consistent with the results
of a previous study by Schindelmann et al (39); although this study did not assess
MDA-MB-468 cells, it was reported that CD24 mRNA levels are
generally increased in non-invasive breast cancer cell lines
compared with invasive cell lines. The significant upregulation of
CD24 observed in MDA-MB-468 cells may have also contributed to the
attenuation of CXCL12-mediated Ca2+ signalling in this
cell line, given that CD24 interferes with CXCL12/CXCR4-mediated
cell migration and tumour growth in pre-B lymphocytes and breast
cancer cells (13). By contrast,
levels of HIF2α, which has been demonstrated to modulate CXCR4
expression (14), did not differ
significantly between the two cell lines, therefore it is unlikely
to have contributed to any differences in CXCL12-mediated
Ca2+ signalling.
Having revealed that CXCR4, CXCR7 and CD24 were
differentially expressed in more mesenchymal MDA-MB-231 cells
(40) compared with MDA-MB-468 cells
in their more epithelial state (17),
the expression of these targets were investigated in MDA-MB-468
cells following EGF- and hypoxia-induced EMT. Induction of EMT with
EGF in MDA-MB-468 cells decreased CXCR4 mRNA levels while the
hypoxia-induced EMT was associated with a significant increase in
CXCR4 levels. This suggests that the induction of the EMT via EGF
and hypoxia differentially affects CXCR4 expression in MDA-MB-468
cells. Bertran et al (41)
previously demonstrated an increase in CXCR4 expression in rat
hepatoma cells treated with transforming proliferation factor-β to
induce the EMT. Hence, transcriptional regulation of CXCR4 may
differ depending on the EMT stimuli and may not be a fundamental
characteristic of a more mesenchymal state. In the present study,
CXCR7 levels were unaltered during hypoxia- and EGF-induced EMT in
MDA-MB-468 cells. Hypoxia- and EGF-induced EMT in MDA-MB-468 cells
produced a significant decrease in levels of CD24, consistent with
its known association as a marker of the more epithelial state
(31). CXCR4 and CXCR7 levels were
enriched in the C-Low molecular subtype of breast tumours compared
with the basal molecular subtype. The C-Low subtype is highly
associated with metaplastic, EMT and stem cell-like features
(34,42). Hence, increased levels of CXCR4 and
CXCR7 in C-Low sub-types of breast cancer is consistent with the
increased levels of these two receptors in the basal B MDA-MB-231
breast cancer cell line, which usually exhibits increased levels of
mesenchymal markers (30) compared
with the less mesenchymal basal A MDA-MB-468 cell line (30). Hence, significant differences in
CXCL12-induced Ca2+ signalling may also be a feature of
different types of breast cancer and influence their
invasive/metastatic properties. This should be the primary focus of
future in vivo studies as methods of assessing
Ca2+ signalling in xenografts continue to progress. In
conclusion, these studies have defined distinct differences in
CXCL12-mediated Ca2+ signalling between MDA-MB-468 and
MDA-MB-231 breast cancer cells. The present study also provides
evidence for the occasional differential remodelling of potential
Ca2+ signalling regulators as a consequence of EGF and
hypoxia in MDA-MB-468 breast cancer cells. It also provides
evidence that CXCR4 and CXCR7 expression is enriched in breast
tumours with mesenchymal features.
Acknowledgements
The present study was supported by the Ministry of
Higher Education Malaysia Scholarship, a National Collaborative
Research Program of the National Breast Cancer Foundation,
Australia (grant no. GC-10-04), the National Health and Medical
Research Council (grant no. 1022263), Queensland Cancer Council
(grant no. 104281), Mater Research and the Australian
Government.
Glossary
Abbreviations
Abbreviations:
|
C-Low
|
Claudin-Low
|
|
EGF
|
epidermal growth factor
|
|
EMT
|
epithelial-mesenchymal transition
|
|
HER2
|
human epidermal growth factor receptor
2
|
|
HIF2α
|
hypoxia-inducible factor-2α
|
|
FBS
|
foetal bovine serum
|
|
FLIPR
|
fluorescence imaging plate reader
|
|
LumA
|
luminal A
|
|
LumB
|
luminal B
|
|
N-Like
|
normal-like
|
References
|
1
|
Rossi D and Zlotnik A: The biology of
chemokines and their receptors. Annu Rev Immunol. 18:217–242. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Perissinotto E, Cavalloni G, Leone F,
Fonsato V, Mitola S, Grignani G, Surrenti N, Sangiolo D, Bussolino
F, Piacibello W and Aglietta M: Involvement of chemokine receptor
4/stromal cell-derived factor 1 system during osteosarcoma tumor
progression. Clin Cancer Res. 11:490–497. 2005.PubMed/NCBI
|
|
3
|
Geminder H, Sagi-Assif O, Goldberg L,
Meshel T, Rechavi G, Witz IP and Ben-Baruch A: A possible role for
CXCR4 and its ligand, the CXC chemokine stromal cell-derived
factor-1, in the development of bone marrow metastases in
neuroblastoma. J Immunol. 167:4747–4757. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mochizuki H, Matsubara A, Teishima J,
Mutaguchi K, Yasumoto H, Dahiya R, Usui T and Kamiya K: Interaction
of ligand-receptor system between stromal-cell-derived factor-1 and
CXC chemokine receptor 4 in human prostate cancer: A possible
predictor of metastasis. Biochem Biophys Res Commun. 320:656–663.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Muller A, Homey B, Soto H, Ge N, Catron D,
Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al:
Involvement of chemokine receptors in breast cancer metastasis.
Nature. 410:50–56. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Balabanian K, Lagane B, Infantino S, Chow
KY, Harriague J, Moepps B, Arenzana-Seisdedos F, Thelen M and
Bachelerie F: The chemokine SDF-1/CXCL12 binds to and signals
through the orphan receptor RDC1 in T lymphocytes. J Biol Chem.
280:35760–35766. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Teicher BA and Fricker SP: CXCL12
(SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 16:2927–2931.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sun X, Cheng G, Hao M, Zheng J, Zhou X,
Zhang J, Taichman RS, Pienta KJ and Wang J: CXCL12/CXCR4/CXCR7
chemokine axis and cancer progression. Cancer Metastasis Rev.
29:709–722. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kang H, Watkins G, Parr C, Douglas-Jones
A, Mansel RE and Jiang WG: Stromal cell derived factor-1: Its
influence on invasiveness and migration of breast cancer cells in
vitro, and its association with prognosis and survival in human
breast cancer. Breast Cancer Res. 7:R402–R410. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mukherjee D and Zhao J: The role of
chemokine receptor CXCR4 in breast cancer metastasis. Am J Cancer
Res. 3:46–57. 2013.PubMed/NCBI
|
|
11
|
Xu C, Zhao H, Chen H and Yao Q: CXCR4 in
breast cancer: Oncogenic role and therapeutic targeting. Drug Des
Dev Ther. 9:4953–4964. 2015.
|
|
12
|
Kay R, Rosten PM and Humphries RK: CD24, a
signal transducer modulating B cell activation responses, is a very
short peptide with a glycosyl phosphatidylinositol membrane anchor.
J Immunol. 147:1412–1416. 1991.PubMed/NCBI
|
|
13
|
Schabath H, Runz S, Joumaa S and Altevogt
P: CD24 affects CXCR4 function in pre-B lymphocytes and breast
carcinoma cells. J Cell Sci. 119:314–325. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Imtiyaz HZ, Williams EP, Hickey MM, Patel
SA, Durham AC, Yuan LJ, Hammond R, Gimotty PA, Keith B and Simon
MC: Hypoxia-inducible factor 2alpha regulates macrophage function
in mouse models of acute and tumor inflammation. J Clin Invest.
120:2699–2714. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Prevarskaya N, Skryma R and Shuba Y:
Calcium in tumour metastasis: New roles for known actors. Nat Rev
Cancer. 11:609–618. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Azimi I, Roberts-Thomson SJ and Monteith
GR: Calcium influx pathways in breast cancer: Opportunities for
pharmacological intervention. Br J Pharmacol. 171:945–960. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Davis FM, Kenny PA, Soo ET, van Denderen
BJ, Thompson EW, Cabot PJ, Parat MO, Roberts-Thomson SJ and
Monteith GR: Remodeling of purinergic receptor-mediated Ca2+
signaling as a consequence of EGF-induced epithelial-mesenchymal
transition in breast cancer cells. PLoS One. 6:e234642011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Azimi I and Monteith GR: Plasma membrane
ion channels and epithelial to mesenchymal transition in cancer
cells. Endocr Relat Cancer. 23:R517–R525. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
McAndrew D, Grice DM, Peters AA, Davis FM,
Stewart T, Rice M, Smart CE, Brown MA, Kenny PA, Roberts-Thomson SJ
and Monteith GR: ORAI1-mediated calcium influx in lactation and in
breast cancer. Mol Cancer Ther. 10:448–460. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Motiani RK, Abdullaev IF and Trebak M: A
novel native store-operated calcium channel encoded by Orai3:
Selective requirement of Orai3 versus Orai1 in estrogen
receptor-positive versus estrogen receptor-negative breast cancer
cells. J Biol Chem. 285:19173–19183. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Peters AA, Simpson PT, Bassett JJ, Lee JM,
Da Silva L, Reid LE, Song S, Parat MO, Lakhani SR, Kenny PA, et al:
Calcium channel TRPV6 as a potential therapeutic target in estrogen
receptor-negative breast cancer. Mol Cancer Ther. 11:2158–2168.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Badve S, Dabbs DJ, Schnitt SJ, Baehner FL,
Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR,
et al: Basal-like and triple-negative breast cancers: A critical
review with an emphasis on the implications for pathologists and
oncologists. Mod Pathol. 24:157–167. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Neve RM, Chin K, Fridlyand J, Yeh J,
Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al: A
collection of breast cancer cell lines for the study of
functionally distinct cancer subtypes. Cancer Cell. 10:515–527.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Aung CS, Ye W, Plowman G, Peters AA,
Monteith GR and Roberts-Thomson SJ: Plasma membrane calcium ATPase
4 and the remodeling of calcium homeostasis in human colon cancer
cells. Carcinogenesis. 30:1962–1969. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stewart TA, Azimi I, Brooks AJ, Thompson
EW, Roberts-Thomson SJ and Monteith GR: Janus kinases and Src
family kinases in the regulation of EGF-induced vimentin expression
in MDA-MB-468 breast cancer cells. Int J Biochem Cell Biol.
76:64–74. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pereira B, Chin SF, Rueda OM, Vollan HK,
Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ,
et al: Erratum: The somatic mutation profiles of 2,433 breast
cancers refine their genomic and transcriptomic. Nat Commun.
7:119082016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Charafe-Jauffret E, Ginestier C, Monville
F, Finetti P, Adélaïde J, Cervera N, Fekairi S, Xerri L, Jacquemier
J, Birnbaum D and Bertucci F: Gene expression profiling of breast
cell lines identifies potential new basal markers. Oncogene.
25:2273–2284. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Davis FM, Azimi I, Faville RA, Peters AA,
Jalink K, Putney JW Jr, Goodhill GJ, Thompson EW, Roberts-Thomson
SJm and Monteith GR: Induction of epithelial-mesenchymal transition
(EMT) in breast cancer cells is calcium signal dependent. Oncogene.
33:2307–2316. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Azimi I, Beilby H, Davis FM, Marcial DL,
Kenny PA, Thompson EW, Roberts-Thomson SJ and Monteith GR: Altered
purinergic receptor-Ca2 signaling associated with
hypoxia-induced epithelial-mesenchymal transition in breast cancer
cells. Mol Oncol. 10:166–178. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Azimi I, Milevskiy MJG, Kaemmerer E,
Turner D, Yapa KTDS, Brown MA, Thompson EW, Roberts-Thomson SJ and
Monteith GR: TRPC1 is a differential regulator of hypoxia-mediated
events and Akt signalling in PTEN-deficient breast cancer cells. J
Cell Sci. 130:2292–2305. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hennessy BT, Gonzalez-Angulo AM,
Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, Fridlyand J,
Sahin A, Agarwal R, Joy C, et al: Characterization of a naturally
occurring breast cancer subset enriched in
epithelial-to-mesenchymal transition and stem cell characteristics.
Cancer Res. 69:4116–4124. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Holland JD, Kochetkova M, Akekawatchai C,
Dottore M, Lopez A and McColl SR: Differential functional
activation of chemokine receptor CXCR4 is mediated by G proteins in
breast cancer cells. Cancer Res. 66:4117–4124. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Miao Z, Luker KE, Summers BC, Berahovich
R, Bhojani MS, Rehemtulla A, Kleer CG, Essner JJ, Nasevicius A,
Luker GD, et al: CXCR7 (RDC1) promotes breast and lung tumor growth
in vivo and is expressed on tumor-associated vasculature. Proc Natl
Acad Sci USA. 104:pp. 15735–15740. 2007; View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Salvucci O, Bouchard A, Baccarelli A,
Deschênes J, Sauter G, Simon R, Bianchi R and Basik M: The role of
CXCR4 receptor expression in breast cancer: A large tissue
microarray study. Breast Cancer Res Treat. 97:275–283. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Blick T, Hugo H, Widodo E, Pinto C, Mani
SA, Weinberg RA, Neve RM, Lenburg ME and Thompson EW: Epithelial
mesenchymal transition traits in human breast cancer cell lines
parallel the CD44(hi/)CD24(lo/-) stem cell phenotype in human
breast cancer. J Mammary Gland Biol Neoplasia. 15:235–252. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Schindelmann S, Windisch J, Grundmann R,
Kreienberg R, Zeillinger R and Deissler H: Expression profiling of
mammary carcinoma cell lines: Correlation of in vitro invasiveness
with expression of CD24. Tumor Biol. 23:139–145. 2002. View Article : Google Scholar
|
|
40
|
Blick T, Widodo E, Hugo H, Waltham M,
Lenburg ME, Neve RM and Thompson EW: Epithelial mesenchymal
transition traits in human breast cancer cell lines. Clin Exp
Metastasis. 25:629–642. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bertran E, Caja L, Navarro E, Sancho P,
Mainez J, Murillo MM, Vinyals A, Fabra A and Fabregat I: Role of
CXCR4/SDF-1 alpha in the migratory phenotype of hepatoma cells that
have undergone epithelial-mesenchymal transition in response to the
transforming growth factor-beta. Cell Signal. 21:1595–1606. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Prat A, Parker JS, Karginova O, Fan C,
Livasy C, Herschkowitz JI, He X and Perou CM: Phenotypic and
molecular characterization of the claudin-low intrinsic subtype of
breast cancer. Breast Cancer Res. 12:R682010. View Article : Google Scholar : PubMed/NCBI
|