Introduction
The global incidence and mortality rates of
colorectal cancer (CRC) are high. In 2012, the estimated global
incidence of novel CRC cases diagnosed was 1.4 million, with
693,900 mortalities, and the number of novel cases and mortalities
is continuously increasing due to economic developments and
lifestyle changes in developing countries (1). Rectal cancer accounts for a large
proportion of CRC cases and is accountable for 28% of CRC cases in
America (2). The standard treatment
for locally advanced rectal cancer is neoadjuvant chemoradiotherapy
with radical surgery, and this treatment pattern may help achieve a
complete pathological response (pCR) (3). Thus, the rates of surgical resection and
anal retention are increasing. However, treatments for rectal
cancer remain far from satisfactory due to the occurrence of
post-treatment cancer recurrence and metastasis.
The phenomenon of radiotherapy or chemotherapy
altering invasion and metastasis in tumor cells has begun to
attract increased attention. Certain studies have demonstrated that
treatment with chemotherapy drugs and radiotherapy may result in
the opposite to the desired effect in patients with tumors;
ignoring the potential additional side effects (4–6). However,
the details of this phenomenon in CRC remain uncertain. Certain
studies have verified that radiotherapy may result in CRC cell
adhesion to the vascular endothelium, and radiotherapy may cause
increased extracellular matrix deposition and the expression of
matrix metalloproteinases (MMPs) in CRC cells (7,8). As a
result, the invasive potential of residual tumor cells may be
increased following ionizing radiation treatment. Other studies
have reported opposing results (9).
Furthermore, the effects of radiotherapy on the invasion and
metastasis of tumor cells vary with radiation therapy style and the
tumor cell type. In addition, the effects of radiation on the
expression of CRC-dependent proteins have been reported to be time-
and dose-dependent (10,11).
The prognosis of patients with locally advanced
rectal cancer who underwent neoadjuvant radiotherapy and
chemotherapy and achieved a pCR was revealed to be significantly
improved compared with patients who did not achieve a pCR (12). However, the rate of pCR following
neoadjuvant chemoradiotherapy is only ~10% (13). Therefore, the majority of these
patients do not achieve a pCR, meaning that residual tumor cells
are present. These patients must then wait 6–8 weeks for surgery.
During this period, the invasive and metastatic potentials of
residual rectal tumor cells may have increased, thus affecting the
overall therapeutic effect in patients with rectal cancer.
The key mechanisms underlying this change remain
uncertain. Long non-coding RNAs (lncRNAs) are non-coding RNA
molecules (>200 nt) that affect corresponding metabolic
processes or directly affect mRNA expression (14). LncRNAs have previously been
demonstrated to be associated with invasion, metastasis and
prognosis in CRC (15). Therefore,
certain lncRNAs may account for the biological changes in the
characteristics of residual CRC cells following chemoradiotherapy.
Further studies are required to explore these changes and
associated mechanisms, and this was the goal of the present
study.
Materials and methods
Cell cultures
The human CRC HCT116 and HT29 cell lines were
purchased from the Kunming Institute of Zoology, Chinese Academy of
Sciences (Kunming, China). Cells were cultured in RPMI-1640 medium
(Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) with 10%
fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and 1% penicillin and streptomycin. Cells were
cultured in an incubator at 37°C in an atmosphere containing 5%
CO2.
Concurrent chemoradiation of CRC cells
in vitro
The chemoradiation model was established by
culturing cells in RPMI-1640 medium (at 37°C) treated with 10
µmol/l 5-fluorouracil (5-FU) (Shanghai Aladdin Bio-Chem Technology
Co., Ltd., Shanghai, China) for 24 h and simultaneously exposing
them to 4 Gy of 6 MV X-ray irradiation (PrecisePlan A-C, iView-GT;
Elekta Instrument AB, Stockholm, Sweden) when the cells had grown
in cell culture flask to between 80 and 100% confluence, as
previously described (16). The
chemical and irradiation treatments were repeated once the cells
had reached >80% confluence. A residual CRC cell model was
established following 4 treatment rounds in the HCT116 and HT29
cells.
Migration and invasion assays using
original and residual cells
According to the methods described in a previous
study (17), a Transwell assay was
used to study cell migration and invasion. For the migration study,
200 µl single cells suspension (2×104 cells/chamber)
with serum-free RPMI-1640 medium was added to the upper chamber,
and 600 µl RPMI-1640 medium with 10% FBS was added to the lower
chamber in each well of a 24-well plate. Then, the Transwell
chambers (24-well, 8-µm pore size; EMD Millipore, Billerica, MA,
USA) and the 24-well plates were incubated at 37°C in a 5%
CO2 atmosphere. After 48 h, the plate was removed from
the incubator and a wet cotton swab was used to remove cells from
the top of the upper chamber. Next, the remaining cells were fixed
for analysis using pure methanol for 30 min at room temperature,
and then stained with 0.1% crystal violet for 15 min at room
temperature. Then, cells in at least 5 fields of view were counted
under an inverted microscope, and images were captured using the
Leica 3000 Suite Application (version 3.8.0; Leica Microsystems
GmbH, Wetzlar, Germany).
For the invasion study, the Transwell inserts were
first covered with 80 µl Matrigel (200–300 µg/ml, Corning
Incorporated, Corning, NY, USA). Then, 10×104 cells were
seeded in the upper chamber. The subsequent steps were the same as
those for the migration assay.
lncRNA expression profile
detection
The original and residual cancer cells derived from
the HCT116 cell line (HCT116N and HCT116CR, respectively), were
routinely digested with 0.25% trypsin-EDTA (Invitrogen; Thermo
Fisher Scientific, Inc.) and centrifuged at 100 × g for 5 min at
room temperature. The cells were subsequently collected, treated
with TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) according
to the manufacturer's protocol, and sent to the biological company
KangChen Bio-tech, Inc. (Shanghai, China) for lncRNA expression
profile detection with the Human LncRNA Array v3.0 (8×60 K;
Arraystar, Inc., Rockville, MD, USA). Differentially expressed
lncRNAs were screened with a threshold of fold-change >2 and
P<0.05. In addition, Gene Ontology (18,19) and
Kyoto Encyclopedia of Genes and Genomes (20–22)
analyses were used to classify the differentially expressed lncRNAs
and analyze the associated signaling pathways with false discovery
rate <1%.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RT-qPCR was performed to detect the expression of
MIR22 host gene (MIR22HG), long-chain non-protein-coding RNA 152
(LINC00152) and Pvt1 oncogene (PVT1) in cells. RT-qPCR was also
used to identify the effects of silencing the expression of these
three lncRNAs in HCT116CR cells. The primer sequences are listed in
Table I. β-Actin was used as the
internal control. All the following operations were performed
according to the manufacturer's protocol. TRIzol reagent was used
to extract total RNA from cells. Total RNA was converted to cDNA at
37°C for 60 min by using Quantscript RT Kit (Tiangen Biotech Co.,
Ltd., Beijing, China) according to the manufacturer's protocol.
Each sample had three replicates, was amplified in a 10-µl reaction
mixture (containing 1 µl cDNA and 0.6 µl primer) by applying FS
Universal SYBR Green Master (Roche Diagnostics, Basel,
Switzerland). The thermocycling conditions were: initial
denaturation at 95°C for 10 min, followed by 45 cycles of; 95°C for
15 sec and 60°C for 60 sec. Samples were analyzed using an ABI
QuantStudio 6 Flex system (Applied Biosystems; Thermo Fisher
Scientific). The relative gene expression levels in cells were
calculated using the 2−∆∆Cq method (23).
 | Table I.Primer sequences for PVT1, LINC00152
and MIR22HG. |
Table I.
Primer sequences for PVT1, LINC00152
and MIR22HG.
| Name | Primer sequences
(5′-3′) |
|---|
| β-αctin-F |
AGCACAGAGCCTCGCCTTTG |
| β-αctin-R |
CTTCTGACCCATGCCCACCA |
| PVT1-F |
GAGAGAATCCTGTTACACCTGGG |
| PVT1-R |
CGACCTGGTTTCTCGTGAGC |
| LINC00152-F |
ACAAGCGGTGCCTGAGCC |
| LINC00152-R |
CCGACTCTCCTACACATCCACAG |
| MIR22HG-F |
TGGGAAGGTCCGAACAGCA |
| MIR22HG-R |
GGGAGAATTTCCTGTCTGCACA |
siRNA transfection of HCT116CR
cells
Specific small interfering (si)RNAs against the
selected genes were provided by Shanghai GenePharma Co., Ltd.
(Shanghai, China). The siRNA sequences are listed in Table II. HCT116CR cells in the logarithmic
growth phase were seeded in 24-well plates at a density of
5×104 cells per well. The culture medium consisted of
500 µl antibiotic-free Opti-minimum essential medium (MEM;
Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% FBS.
Cells were cultured in a CO2 incubator for 24 h until
reaching ~70% confluence. Then, 1 µl Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) was diluted with 50 µl
Opti-MEM, and 20 pmol siRNA was diluted with 50 µl Opti-MEM. The
diluted Lipofectamine reagent and siRNA were then mixed, and then
the siRNA and lipofectamine mixture (100 µl) was added to each well
of the prepared cells. Next, the cells were incubated (37°C, 5%
CO2) for 6 h. The culture medium was then replaced with
normal RPMI-1640 medium with 10% FBS, and the cells were cultured
for an additional 48 h in the same incubator. The cells were then
collected for use in subsequent steps.
 | Table II.SiRNA sequences against PVT1,
LINC00152 and MIR22HG. |
Table II.
SiRNA sequences against PVT1,
LINC00152 and MIR22HG.
| siRNA | Sequence
(5′-3′) |
|---|
| PVT1-444-s |
GCUUCAAGCUCACGAGAAATT |
| PVT1-444-as |
UUUCUCGUGAGCUUGAAGCTT |
| PVT1-368-s |
GGACUUGAGAACUGUCCUUTT |
| PVT1-368-as |
AAGGACAGUUCUCAAGUCCTT |
| PVT1-302-s |
CCUGUUACACCUGGGAUUUTT |
| PVT1-302-as |
AAAUCCCAGGUGUAACAGGTT |
|
LINC00152-481-s |
GCAGAAGACAAAGCCGAAATT |
|
LINC00152-481-as |
UUUCGGCUUUGUCUUCUGCTT |
|
LINC00152-738-s |
GCAUGAUUGGAUGAUGUUUTT |
|
LINC00152-738-as |
AAACAUCAUCCAAUCAUGCTT |
|
LINC00152-619-s |
GGGAGACAGUUCACAGAUATT |
|
LINC00152-619-as |
UAUCUGUGAACUGUCUCCCTT |
| MIR22HG-111-s |
CCCUGGGAACAAGUCAGUUTT |
| MIR22HG-111-as |
AACUGACUUGUUCCCAGGGTT |
| MIR22HG-457-s |
GAAGGCUCAAACAACCCAATT |
| MIR22HG-457-as |
UUGGGUUGUUUGAGCCUUCTT |
| MIR22HG-470-s |
ACCCAAGGUGGUAUGUGAUTT |
| MIR22HG-470-as |
AUCACAUACCACCUUGGGUTT |
| Negative
control-s |
UUCUCCGAACGUGUCACGUTT |
| Negative
control-as |
ACGUGACACGUUCGGAGAATT |
Migration and invasion assays of
HCT116CR and siRNA-transfected HCR116CR cells
Transwell assays were used to study the migration of
HCT116CR and siRNA-transfected HCT116CR cells. Cells were collected
and resuspended in serum-free RPMI-1640 medium. In the lower
chamber, 600 µl medium containing 10% FBS was added. Each prepared
cell suspension (150 µl; 3×104 cells/chamber) was added
to the upper chambers, and the cells were incubated at 37°C and 5%
CO2 in an incubator for 24 h. The chambers were then
removed, and the fluid was aspirated. Then, the cells were fixed
with pure methanol (800 µl/chamber) for 30 min and stained with
diluted Giemsa Stain solution (10×Stock solution; Beijing Solarbio
Science & Technology Co., Ltd., Beijing, China) for 20 min. A
wet cotton swab was used to wipe the cells off the bottom of the
upper chamber. Tweezers were used to peel off the membrane and turn
it upside down to dry. The dried membrane was then transferred to a
glass slide and mounted with a piece of neutral gum. Finally, the
cells were counted in at least 5 fields of view and images were
captured under a light microscope.
For the invasion assays, Matrigel was diluted to a
final concentration of 1 mg/ml with 4°C precooled serum-free
RPMI-1640 medium. A volume of 100 µl diluted Matrigel was added
vertically to the bottom of the upper chamber and incubated at 37°C
for 4–5 h to dry. The next steps were the same as those
aforementioned for the migration assay.
Statistical analysis
Where the data are normally distributed, they are
reported as the mean ± standard deviation. Non-normally distributed
data is presented as the median. Statistical analyses were
performed using SPSS 20.0 software (IBM Corp., Armonk, NY, USA).
Unpaired Student's t tests were used for comparison between two
groups, and one-way analysis of variance followed by Dunnett's
tests was used for comparisons between three or more groups. All
experiments were repeated at least three times. P<0.05 was
considered to indicate a statistically significant difference.
Results
Residual HCT116 and HT29 CRC cell
models
HCT116 and HT29 CRC cells were simultaneously
treated with 10 µmol/l 5-FU and irradiation (4 Gy 6 MV X-ray). The
cells consistently died from the fourth day to the tenth day
following treatment. Then, the remaining cells slowly proliferated
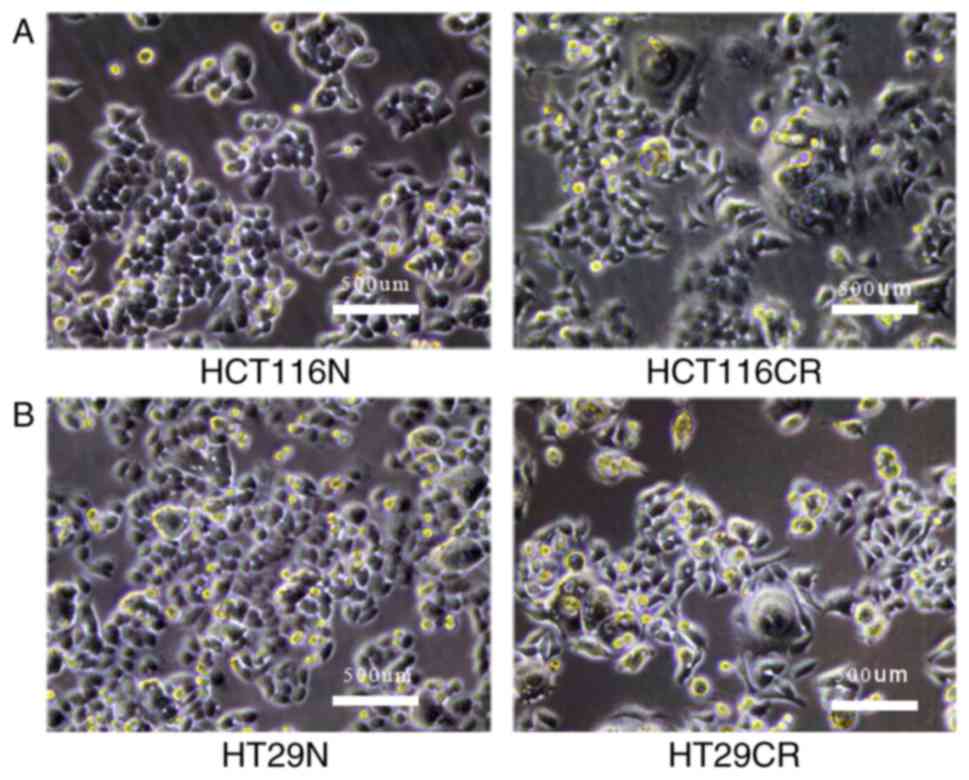
until reaching confluence. Morphological changes in the residual
CRC cells were determined. For example, a number of large cells
were observed, with volumes three or four times greater than the
normal cells, and the new cells growing around the large cells
exhibited diverse pseudopodia. In addition, larger intercellular
gaps were observed by light microscopy (Fig. 1). These residual CRC cells were
designated HCT116CR and HT29CR cells.
Residual CRC cells demonstrated
increased migration and invasion in vitro
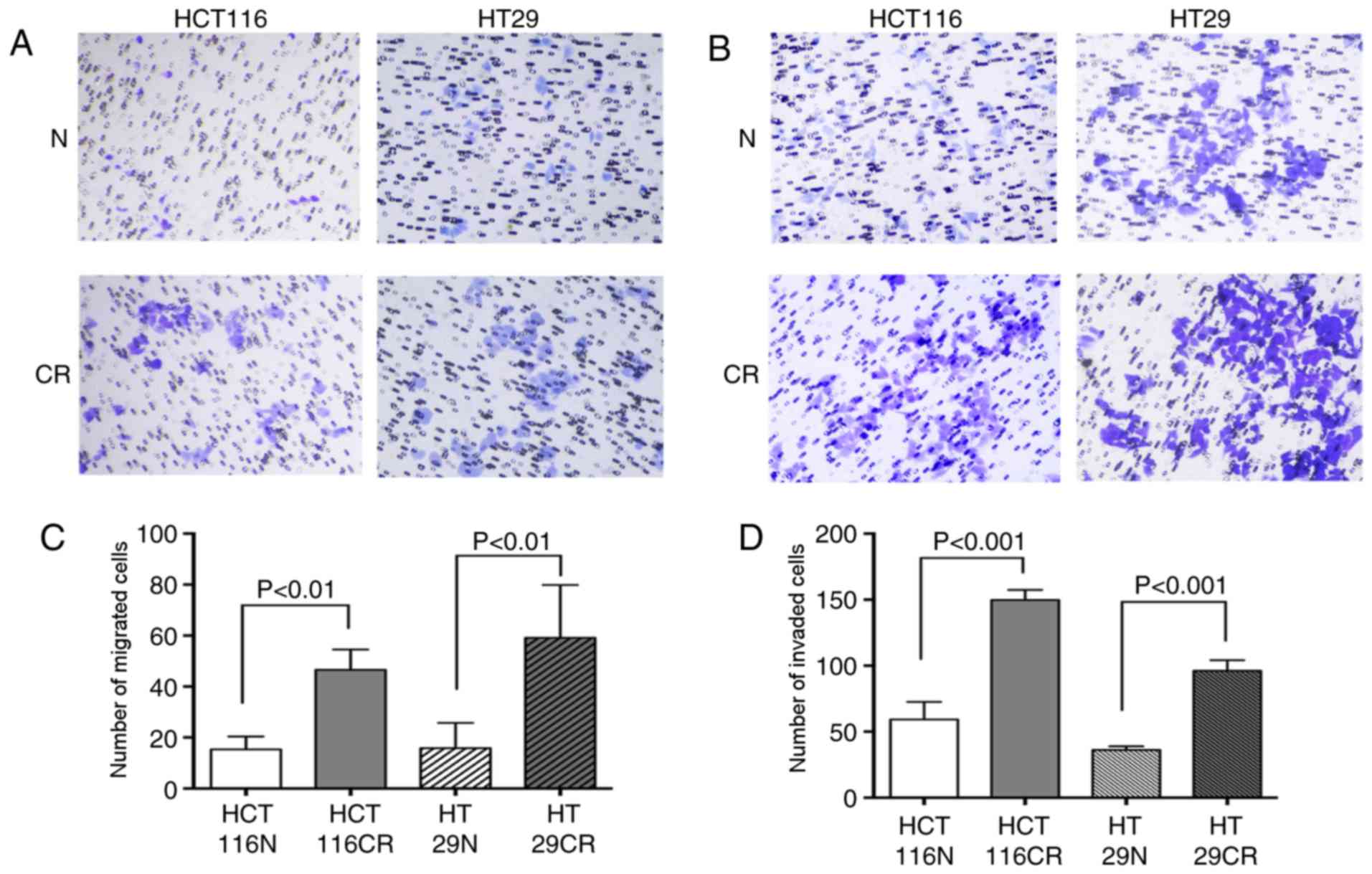
Once the residual CRC cell model was established,
the migration and invasion of the original and residual cells were
compared (Fig. 2). A significantly
increased number of residual cells migrated compared with original
cells; 15.33±5.07 HCT116N cells migrated, while 46.56±7.97 HCT116CR
cells migrated (Ρ=0.005; Fig. 2A and
C). A similar trend was observed for the HT29 cells, as
15.83±9.88 original cells and 59.16±20.73 residual cells migrated
(Ρ=0.003; Fig. 2A and C). The
invasion assay also revealed that more residual cells passed
through the membrane, as 59.17±13.34 HCT116N cells invaded and
149.63±7.65 HCT116CR cells invaded (P<0.001; Fig. 2B and D). In addition, 35.97±2.89 and
96.00±8.13 HT29N and HT29CR cells invaded, respectively
(P<0.001; Fig. 2B and D).
HCT116CR and HCT116N cell lncRNA
expression profiling
Having identified differences in the migratory and
invasive potentials of HCT116N and HCT116CR cells, lncRNA
expression levels in HCT116N and HCT116CR cells were then detected
and analyzed. A total of 18,928 lncRNAs were detected by biochip,
2,662 of which were differentially expressed. Among these, 1,245
lncRNAs were upregulated and 1,417 lncRNAs were downregulated.
Three lncRNAs, PVT1, LINC00152 and MIR22HG, which were upregulated
in HCT116CR cells compared with HCT116N cells were selected
according to associated reviews and analyses (24–26). The
expression levels of these three selected lncRNAs in the cell types
were confirmed by RT-qPCR, with the PCR results being consistent
with the biochip results.
Results of siRNA silencing as detected
by RT-qPCR
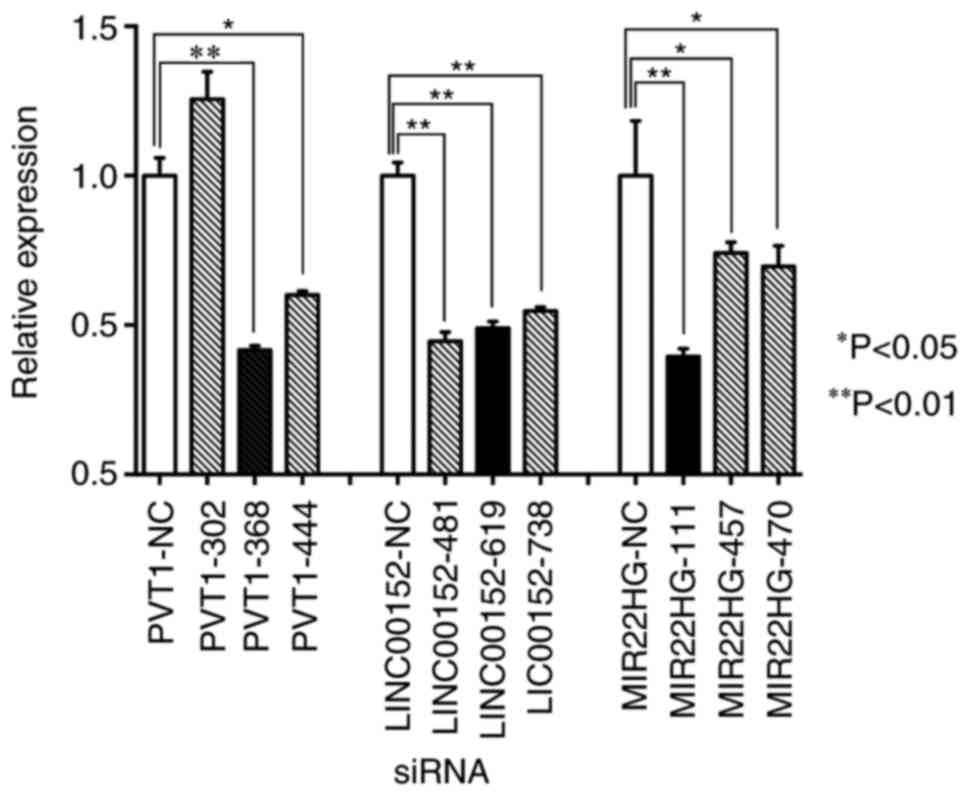
To investigate the role of the three lncRNAs (PVT1,
LINC00152 and MIR22HG) in residual CRC cells, HCT116CR cells were
transfected with siRNAs against the three selected lncRNAs (PVT1,
LINC00152 and MIR22HG). The results of siRNA transfection were
assessed by RT-qPCR. The most effective specific siRNAs for each
molecular target were PVT1-368, LINC00152-481 and MIR22HG-111. The
expression levels of PVT1, LINC00152 and MIR22HG in HCT116CR cells
were significantly decreased compared with the control group, with
the relative expression level of each target gene decreased by 58,
55 and 61%, respectively (P<0.01; Fig.
3).
Residual CRC cell biological
characteristics are modulated via LINC00152 silencing
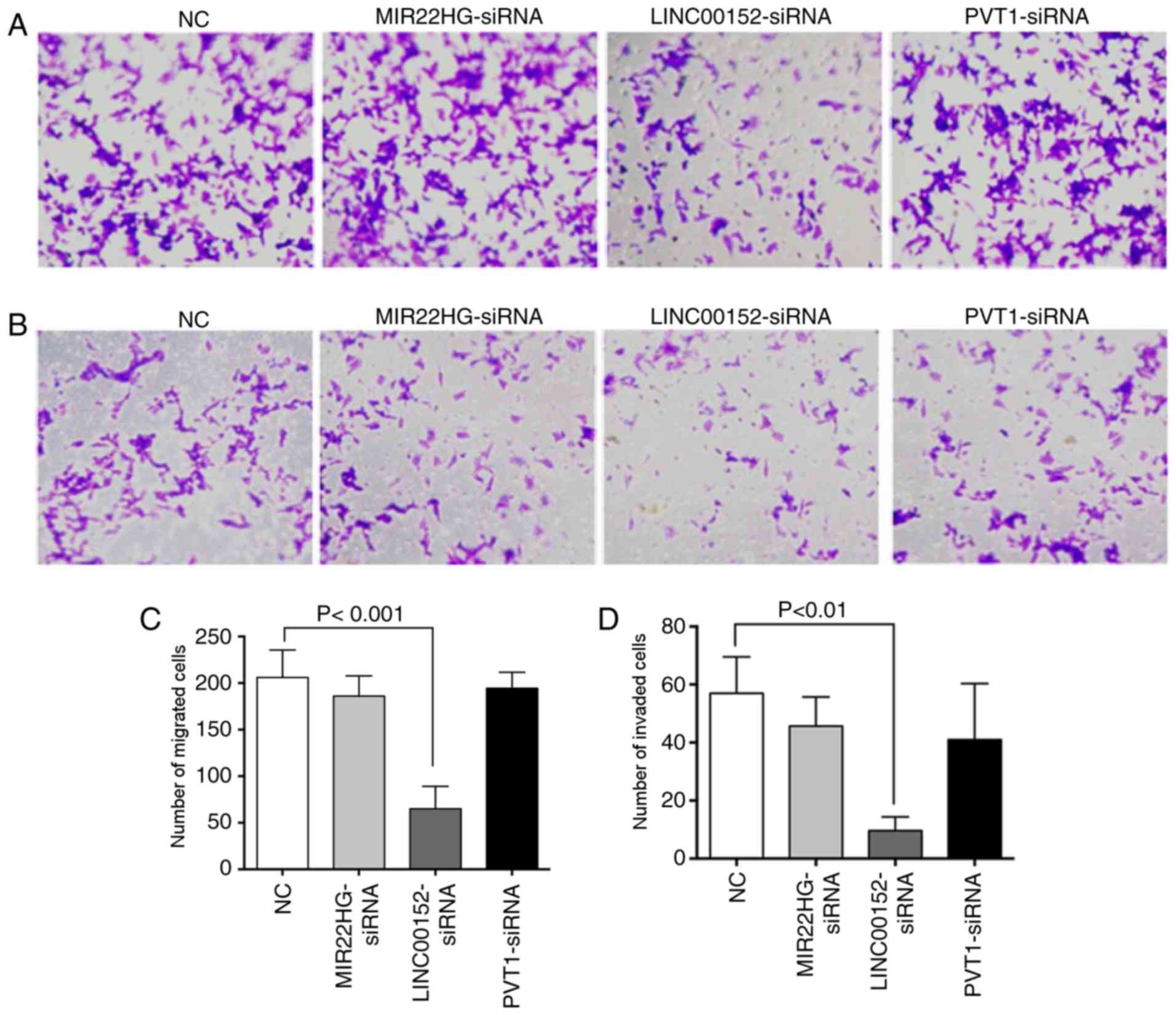
To compare the effects of siRNA transfection on the
migration and invasion of HCT116 residual cancer cells, the
Transwell experiments were performed again (Fig. 4). In the migration experiment, a total
of 206.00±29.46 cells passed through the chamber membrane in the
negative control (NC) group, 186.00±21.63 cells in the
MIR22HG-siRNA group (P=0.618 compared with the NC group),
65.00±24.06 cells in the LINC00152-siRNA group (P<0.001 compared
with the NC group) and 194.33±17.21 in the PVT1-siRNA group
(P=0.875 compared with the NC group; Fig.
4A and C). Only the number of cells in the LINC00152-siRNA
group was significantly reduced, suggesting a significant decline
in the mobility of HCT116CR cells transfected with LINC00152
siRNA.
Similar to the migration experiment, a significant
difference in invasion was only observed in the LINC00152-siRNA
group compared with the NC group. The number of invading cells in
the control group was 57.00±12.53, while the numbers in the
LINC00152-siRNA, MIR22HG-siRNA and PVT1-siRNA groups were 9.67±4.73
(P=0.005 compared with the NC group), 45.67±10.02 (P=0.590 compared
with the NC group) and 41.00±19.31 (P=0.347 compared with the NC
group), respectively (Fig. 4B and D).
There was no significant difference between number of invading
cells in the MIR22HG-siRNA or PVT1-siRNA groups and the control
group.
Discussion
In the cell experiments of the present study,
residual CRC cell models were successfully established via repeated
concurrent chemoradiotherapy, which was intended to mimic the
clinical treatment model as closely as possible. The morphological
changes observed in residual CRC cells following chemoradiation
therapy indicated that the biological characteristics may also be
altered. In addition, Transwell experiments demonstrated that the
migration and invasion of the residual cell lines were
significantly increased compared with the original cells. These
results agree with the results of several previous studies
(7,27–29). A
similar phenomenon has also been observed in multiple types of
cancer cell. Yamauchi et al (6) demonstrated that the invasion and
metastasis of human HT1080 fibrosarcoma cells was increased in
cancer cell-bearing host mice pretreated with cyclophosphamide.
Therefore, in addition to from side effects, chemotherapy drugs may
exert effects opposite to those which are desired. Jadhav et
al (4) irradiated human SK-N-AS
neuroblastoma cells with different irradiation doses, and observed
that irradiated cells had increased expression levels of
urokinase-type plasminogen activator, MMP-9 and vascular
endothelial growth factor compared with non-irradiated cells, as
well as increased capillary-like structure of microvascular
endothelial cells.
X-rays were used to treat the CRC cells in the
present study, and migration and invasion were increased in
residual cells compared with original cells in the two CRC cell
lines that were assessed, HCT116 and HT29. However, the use of
X-ray irradiation to treat HCT116 cells has also been demonstrated
to decrease invasion and metastasis of residual cancer cells via
upregulation of KiSS-1 metastasis suppressor expression (30), which disagrees with the results of the
present study. However, this previous study only observed the
effect of radiotherapy and did not fully simulate concurrent
clinical radiotherapy and chemotherapy. Therefore, further studies
are required. The results of other previous studies suggest that
the effects of radiotherapy on tumor cell invasion and metastasis
may vary depending on the radiation mode and tumor cell type, and
that the effect of radiotherapy on the migration of CRC cells is
time- and dose-dependent (10,11,31).
Therefore, studies investigating alterations in the invasion and
metastasis of residual cells following chemotherapy may need to
assess various irradiation patterns, doses, observation times and
types of CRC cell.
In addition, the mechanisms underlying these
phenomena remain uncertain. The epithelial-to-mesenchymal
transition (EMT) has been observed in CRC cells and rectal cancer
tissues following neoadjuvant radiotherapy and chemotherapy,
indicating that the invasion of residual rectal cancer cells was
increased (27,29). The morphological changes of the
residual CRC cancer cells observed in the present study suggest
that EMT occurred. In addition, a previous study demonstrated that
invasion and metastasis were increased in residual CRC cells due to
an increase in EPH receptor A4 expression levels and the extent of
the EMT in these cells (28). There
is also another view regarding the mechanism underlying the
biological changes observed in residual CRC cells following
radiotherapy. The degradation and destruction of the extracellular
matrix and the basement membrane are important processes in tumor
metastasis. An in vitro study revealed that radiotherapy
treatment resulted in upregulated MMP expression in CRC cells
(8). The increased expression of MMPs
in CRC cells following radiotherapy may indicate an increase in
cell invasion and metastasis. However, this change was transient,
not continuous (8).
To explore the key mechanisms underlying the
biological changes in residual CRC cells, the differential
expression of lncRNAs was investigated. Among the three selected
lncRNAs that were differentially expressed in residual CRC cells
compared with original CRC cells, that may have been potential
therapeutic molecular targets, only LINC00152 appeared to alter the
biological characteristics of residual CRC cells. The invasive and
metastatic potentials of residual CRC cells were decreased by
silencing the expression of LINC00152.
LINC00152, which is known to regulate the
cytoskeleton, may also be involved in cell cycle regulation and DNA
damage repair. LINC00152 is a potential oncogene which may be
involved in various types of cancer (32). In gastric cancer (GC), downregulation
of LINC00152 expression decreased migration and invasion and
suppressed proliferation and EMT progression in gastric cancer
cells (33). In addition, LINC00152
has potential as a prognostic biomarker and therapeutic target in
tongue squamous cell carcinoma, renal cell carcinoma, lung cancer,
and gallbladder cancer (GBC) (34–37). In
the present study, residual CRC cell migration and invasion were
demonstrated to be significantly modulated by LINC00152
interference. In CRC, increased expression of LINC00152 is
associated with clinical stage and lymph node metastasis, and may
serve as a molecular marker for colon cancer metastasis (38). Furthermore, LINC00152 has been
revealed to be associated with oxaliplatin (L-OHP) resistance in
vitro and in vivo, and LIN00152 contributes to L-OHP
resistance by acting as a competing endogenous RNA to regulate
microRNA (miR)-193a-3p and erb-b2 receptor tyrosine kinase 4
expression (39). In other types of
cancer, including hepatocellular carcinoma, LINC00152 activates the
mechanistic target of rapamycin signaling pathway to increase
tumorigenesis (40). In GC and lung
adenocarcinoma (LAC), LINC00152 interacts with enhancer of zeste
homologue 2, silencing the expression of p15 and p21
in GC and inhibiting interleukin 24 transcription in LAC, resulting
in acceleration of the cell cycle and cell proliferation (24,41).
LINC00152 has also been revealed to promote proliferation in GC
through an epidermal growth factor receptor-dependent pathway.
Other studies have demonstrated that LINC00152 may contribute to
renal cell carcinoma progression by epigenetically repressing P16
expression and interacting with miR-205 (35). In addition, the
LINC00152/miR-138/hypoxia-inducible factor-1α pathway potentiates
the progression of GBC (37). The
results of another previous study suggested that SP1 transcription
factor/LINC00152/phosphoinositide 3-kinase/protein kinase B may be
a potential therapeutic target in GBC (42). Overall, LINC00152 overexpression seems
to serve a critical function in cancer development, and the
associated mechanisms vary among different types of cancer.
However, the functional regulatory mechanisms of LINC00152 remain
uncertain, and the exact underlying mechanism requires further
exploration. While LINC00152 has been a major area of research in
multiple types of cancer, to the best of our knowledge the
association between LINC00152 and the biological characteristics of
residual CRC cells had not been previously reported. The results of
the present study provide additional scientific support for the
clinical value of LINC00152 in treating CRC.
However, patients with CRC patients with high
LINC00152 expression in tumor tissues have been reported to have an
improved prognosis than those with low LINC00152 expression
(43). Zhang et al (44) demonstrated that LINC00152
overexpression reduces CRC cell viability and increases apoptosis,
and LINC00152 may be downregulated by miR-367c-3p in CRC tissues
and cells, but this previous study did not examine changes in the
invasion and migration of CRC cells. Thus, further CRC cell
experiments in clinical contexts concerning LINC00152 are
merited.
The other two potential molecular targets that were
selected, MIR22HG and PVT1, were also differentially expressed
between the residual and original CRC cells. However, the migratory
and invasive abilities of residual HCT116 cells were not
significantly different from those of residual cells when MIR22HG
or PVT1 expression was silenced. MIR22HG is an indicator for
chemical stress response, and functions as an oncogene in ovarian
cancer (25,45). MIR22HG is also one of the most
upregulated tumor suppressor genes in CRC cells under microgravity
(46). In CRC cell lines,
transforming growth factor-β expression and the apoptotic signaling
pathway are activated when PVT1 expression is downregulated, the
proliferation and invasion of CRC cells are reduced, and high PVT1
expression in patients with CRC is associated with a poor prognosis
(26). In the present study, CRC
cells treated with radiotherapy and chemotherapy had relatively
high PVT1 and MIR22HG expression compared with untreated cells.
However, there were no significant differences in the invasion and
metastasis of residual CRC cells following PVT1 and MIR22HG
silencing. These results indicated that neither PVT1 nor MIR22HG is
a key diagnostic or therapeutic target in the process of biological
characteristic alterations caused by radiotherapy and chemotherapy
in CRC cells.
To the best of our knowledge, the present study is
the first to examine the effects of radiotherapy and chemotherapy
on the invasion and metastasis of CRC cells and reveal a relevant
potential biomarker. These results may help improve the therapeutic
effects of CRC treatments. However, there were two main limitations
with the present study. First, only two cell lines and limited
doses of one radiation therapy type were used. Second, this
research was conducted entirely in vitro. Therefore, the aim
of our future studies is to perform ethical animal experiments and
clinical research regarding LINC00152 and alterations of the
biological characteristics of residual CRC cells.
In conclusion, the invasion and metastasis of
residual CRC cells increased following radiotherapy and
chemotherapy, indicating that these biological characteristics of
residual cancer cells were altered by this treatment. In addition,
the lncRNA LINC00152 was revealed to be a potential biomarker that
modulates the alterations caused by these treatments in CRC. The
present study provides a robust scientific basis for further
research to improve the therapeutic effects of CRC treatments.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81301825), the
Training Program for Medical Reserve Talents of the Health and
Family Planning Commission of Yunnan Province (grant nos. H-201641,
H-201624), and the National Key Clinical Specialty (Oncology)
Fund.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Benson AB III, Venook AP, Bekaii-Saab T,
Chan E, Chen YJ, Cooper HS, Engstrom PF, Enzinger PC, Fenton MJ,
Fuchs CS, et al: Rectal cancer, version 2.2015. J Natl Compr Canc
Netw. 13:719–728. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jadhav U and Mohanam S: Response of
neuroblastoma cells to ionizing radiation: Modulation of in vitro
invasiveness and angiogenesis of human microvascular endothelial
cells. Int J Oncol. 29:1525–1531. 2006.PubMed/NCBI
|
|
5
|
Xiong W, Ren ZG, Qiu SJ, Sun HC, Wang L,
Liu BB, Li QS, Zhang W, Zhu XD, Liu L, et al: Residual
hepatocellular carcinoma after oxaliplatin treatment has increased
metastatic potential in a nude mouse model and is attenuated by
Songyou Yin. BMC Cancer. 10:2192010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yamauchi K, Yang M, Hayashi K, Jiang P,
Yamamoto N, Tsuchiya H, Tomita K, Moossa AR, Bouvet M and Hoffman
RM: Induction of cancer metastasis by cyclophosphamide pretreatment
of host mice: An opposite effect of chemotherapy. Cancer Res.
68:516–520. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Meineke V, Gilbertz KP, Schilperoort K,
Cordes N, Sendler A, Moede T and van Beuningen D: Ionizing
radiation modulates cell surface integrin expression and adhesion
of COLO-320 cells to collagen and fibronectin in vitro.
Strahlenther Onkol. 178:709–714. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Speake WJ, Dean RA, Kumar A, Morris TM,
Scholefield JH and Watson SA: Radiation induced MMP expression from
rectal cancer is short lived but contributes to in vitro invasion.
Eur J Surg Oncol. 31:869–874. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hasegawa T, Sato M, Kurimoto M, Takahashi
H, Kawashima T, Matsuo Y, Yamamoto M, Sawai H, Funahashi H, Okada
Y, et al: Biological effect of irradiation on adhesion molecules in
human colon cancer cells in vitro. Anticancer Res. 25:875–879.
2005.PubMed/NCBI
|
|
10
|
Goetze K, Scholz M, Taucher-Scholz G and
Mueller-Klieser W: The impact of conventional and heavy ion
irradiation on tumor cell migration in vitro. Int J Radiat Biol.
83:889–896. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Suetens A, Moreels M, Quintens R, Soors E,
Buset J, Chiriotti S, Tabury K, Gregoire V and Baatout S: Dose- and
time-dependent gene expression alterations in prostate and colon
cancer cells after in vitro exposure to carbon ion and
X-irradiation. J Radiat Res. 56:11–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jalilian M, Davis S, Mohebbi M, Sugamaran
B, Porter IW, Bell S, Warrier SK and Wale R: Pathologic response to
neoadjuvant treatment in locally advanced rectal cancer and impact
on outcome. J Gastrointest Oncol. 7:603–608. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wasmuth HH, Rekstad LC and Tranø G: The
outcome and the frequency of pathological complete response after
neoadjuvant radiotherapy in curative resections for advanced rectal
cancer: A population-based study. Colorectal Dis. 18:67–72. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Han D, Wang M, Ma N, Xu Y, Jiang Y and Gao
X: Long noncoding RNAs: Novel players in colorectal cancer. Cancer
Lett. 361:13–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ojima E, Inoue Y, Watanabe H, Hiro J,
Toiyama Y, Miki C and Kusunoki M: The optimal schedule for
5-fluorouracil radiosensitization in colon cancer cell lines. Oncol
Rep. 16:1085–1091. 2006.PubMed/NCBI
|
|
17
|
Li YJ, Dong BK, Fan M and Jiang WX: BTG2
inhibits the proliferation and metastasis of osteosarcoma cells by
suppressing the PI3K/AKT pathway. Int J Clin Exp Pathol.
8:12410–12418. 2015.PubMed/NCBI
|
|
18
|
The Gene Ontology Consortium: Expansion of
the gene ontology knowledgebase and resources. Nucleic Acids Res.
45(Database Issue): D331–D338. 2017.PubMed/NCBI
|
|
19
|
The Gene Ontology Consortium, ; Ashburner
M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP,
Dolinski K, Dwight SS, et al: Gene ontology: Tool for the
unification of biology. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kanehisa M, Sato Y, Kawashima M, Furumichi
M and Tanabe M: KEGG as a reference resource for gene and protein
annotation. Nucleic Acids Res. 44:D457–D462. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen WM, Huang MD, Sun DP, Kong R, Xu TP,
Xia R, Zhang EB and Shu YQ: Long intergenic non-coding RNA 00152
promotes tumor cell cycle progression by binding to EZH2 and
repressing p15 and p21 in gastric cancer. Oncotarget. 7:9773–9787.
2016.PubMed/NCBI
|
|
25
|
Li J, Yu H, Xi M and Lu X: Long noncoding
RNA C17orf91 is a potential prognostic marker and functions as an
oncogene in ovarian cancer. J Ovarian Res. 9:492016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takahashi Y, Sawada G, Kurashige J, Uchi
R, Matsumura T, Ueo H, Takano Y, Eguchi H, Sudo T, Sugimachi K, et
al: Amplification of PVT-1 is involved in poor prognosis via
apoptosis inhibition in colorectal cancers. Br J Cancer.
110:164–171. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kawamoto A, Yokoe T, Tanaka K, Saigusa S,
Toiyama Y, Yasuda H, Inoue Y, Miki C and Kusunoki M: Radiation
induces epithelial-mesenchymal transition in colorectal cancer
cells. Oncol Rep. 27:51–57. 2012.PubMed/NCBI
|
|
28
|
Marcondes PGD, Bastos LG, Rocha MR and
Morgado-Díaz JA: EphA4-mediated signaling regulates the aggressive
phenotype of irradiation survivor colorectal cancer cells. Tumour
Biol. 37:12411–12422. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tato-Costa J, Casimiro S, Pacheco T, Pires
R, Fernandes A, Alho I, Pereira P, Costa P, Castelo HB, Ferreira J
and Costa L: Therapy-induced cellular senescence induces
epithelial-to-mesenchymal transition and increases invasiveness in
rectal cancer. Clin Colorectal Cancer. 15:170–178. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ke CL, Chen ZH, Shi JF, et al: X-ray
irradiation on the expression of Kiss-1 gene and the potential of
cell invasion and migration in colorectal cancer cell line HCT116.
J Front Med. 1–15. 2014.
|
|
31
|
Moncharmont C, Levy A, Guy JB, Falk AT,
Guilbert M, Trone JC, Alphonse G, Gilormini M, Ardail D, Toillon
RA, et al: Radiation-enhanced cell migration/invasion process: A
review. Crit Rev Oncol Hematol. 92:133–142. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu Y, Yang J, Li Q, Xu B, Lian Y and Miao
L: LINC00152: A pivotal oncogenic long non-coding RNA in human
cancers. Cell Prolif. 50:2017. View Article : Google Scholar
|
|
33
|
Zhao J, Liu Y, Zhang W, Zhou Z, Wu J, Cui
P, Zhang Y and Huang G: Long non-coding RNA Linc00152 is involved
in cell cycle arrest, apoptosis, epithelial to mesenchymal
transition, cell migration and invasion in gastric cancer. Cell
Cycle. 14:3112–3123. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yu J, Liu Y, Guo C, Zhang S, Gong Z, Tang
Y, Yang L, He Y, Lian Y, Li X, et al: Upregulated long non-coding
RNA LINC00152 expression is associated with progression and poor
prognosis of tongue squamous cell carcinoma. J Cancer. 8:523–530.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang Y, Liu J, Bai H, Dang Y, Lv P and Wu
S: Long intergenic non-coding RNA 00152 promotes renal cell
carcinoma progression by epigenetically suppressing P16 and
negatively regulates miR-205. Am J Cancer Res. 7:312–322.
2017.PubMed/NCBI
|
|
36
|
Feng S, Zhang J, Su W, Bai S, Xiao L, Chen
X, Lin J, Reddy RM, Chang AC, Beer DG and Chen G: Overexpression of
LINC00152 correlates with poor patient survival and knockdown
impairs cell proliferation in lung cancer. Sci Rep. 7:29822017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cai Q, Wang Z, Wang S, Weng M, Zhou D, Li
C, Wang J, Chen E and Quan Z: Long non-coding RNA LINC00152
promotes gallbladder cancer metastasis and epithelial-mesenchymal
transition by regulating HIF-1α via miR-138. Open Biol. 7:pii:
1602472017. View Article : Google Scholar
|
|
38
|
Zhang XL, Zhu Y, Li SN, Fan SQ, Yuan P,
Gao XD, Wang CJ and Zhang FY: Expression of long non-coding RNA
LINC00152 in human colon cancer and its clinical significance. Chin
J Clin Laborat Sci. 354–358. 2015.(In Chinese).
|
|
39
|
Yue B, Cai D, Liu C, Fang C and Yan D:
Linc00152 functions as a competing endogenous RNA to confer
oxaliplatin resistance and holds prognostic values in colon cancer.
Mol ther. 24:2064–2077. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ji J, Tang J, Deng L, Xie Y, Jiang R, Li G
and Sun B: LINC00152 promotes proliferation in hepatocellular
carcinoma by targeting EpCAM via the mTOR signaling pathway.
Oncotarget. 6:42813–42824. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen QN, Chen X, Chen ZY, Nie FQ, Wei CC,
Ma HW, Wan L, Yan S, Ren SN and Wang ZX: Long intergenic non-coding
RNA 00152 promotes lung adenocarcinoma proliferation via
interacting with EZH2 and repressing IL24 expression. Mol Cancer.
16:172017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cai Q, Wang ZQ, Wang SH, Li C, Zhu ZG,
Quan ZW and Zhang WJ: Upregulation of long non-coding RNA LINC00152
by SP1 contributes to gallbladder cancer cell growth and tumor
metastasis via PI3K/AKT pathway. Am J Transl Res. 8:4068–4081.
2016.PubMed/NCBI
|
|
43
|
Qiu JJ and Yan JB: Long non-coding RNA
LINC01296 is a potential prognostic biomarker in patients with
colorectal cancer. Tumour Biol. 36:7175–7183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang YH, Fu J, Zhang ZJ, Ge CC and Yi Y:
LncRNA-LINC00152 down-regulated by miR-376c-3p restricts viability
and promotes apoptosis of colorectal cancer cells. Am J Transl Res.
8:5286–5297. 2016.PubMed/NCBI
|
|
45
|
Tani H and Torimura M: Identification of
short-lived long non-coding RNAs as surrogate indicators for
chemical stress response. Biochem Biophys Res Commun. 439:547–551.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Vidyasekar P, Shyamsunder P, Arun R,
Santhakumar R, Kapadia NK, Kumar R and Verma RS: Genome wide
expression profiling of cancer cell lines cultured in microgravity
reveals significant dysregulation of cell cycle and MicroRNA gene
networks. PLoS One. 10:e01359582015. View Article : Google Scholar : PubMed/NCBI
|