Introduction
Chronic myeloid leukemia (CML) is a clonal disorder
characterized by the reciprocal translocation between the long arms
of chromosomes 9 and 22 [t(9;22)(q34;q11)]. This translocation,
which is present in 95% of patients with CML, creates a BCR, RhoGEF
and GTPase activating protein (BCR)-ABL proto-oncogene 1
non-receptor tyrosine kinase (ABL) fusion gene that produces an
abnormal tyrosine kinase. The imatinib mesylate is commonly used as
the first-line oral treatment in patients with CML (1). It blocks the BCR-ABL tyrosine kinase
activity and subsequently induces apoptosis, followed by a
reduction in the proliferation of BCR-ABL-expressing cells
(2). Therefore, the treatment of
patients with CML with imatinib significantly increased survival
and improved quality of life (3).
A small proportion of patients with CML (5–8%)
present a more complex rearrangement of the Philadelphia (Ph)
chromosome (4,5). These complex variant translocations and
other mutations may be facilitated by genomic instability triggered
by the t(9;22)(q34;q11) translocation, resulting in accelerated
disease progression to blast crisis (6–8). How these
events occur in detail remains unknown. The present report
describes a patient with CML in B-lymphoid blast crisis who
presented with a rare three-way Ph variant translocation
t(3;9;22)(p21;q34;q11) in addition to isodicentric Ph
chromosomes.
Materials and methods
Patient
A 42-year-old Chinese male was admitted to The
Second Affiliated Hospital and Yuying Children's Hospital of
Wenzhou Medical University (Zhejiang, China) in June 2011 because
of neutrophilic granulocytosis and splenomegaly lasting the
previous 6 months. Written informed consent from the patient was
obtained for publication of this study. Hematological tests
revealed a white blood cell count of 69×109/l (normal
range, 4–10×109/l), consisting of 67% neutrophils
(normal range, 40–70%), 5% lymphocytes (normal range, 20–50%), 12%
metamyelocytes (normal range, 0%), 10% myelocytes (normal range,
0%), 1% promyelocyte (normal range, 4–10×109/l), 1%
eosinophils (normal range, 4–8%), 0.5% basophils (normal range,
0–1%) and 3.5% blasts (normal range, 0%); a platelet count of
499×109/l (normal range, 100–300×109/l); and
a hemoglobin concentration of 92 g/l (normal range, 120–160 g/l).
Bone marrow examination revealed predominant granulopoiesis, a
markedly elevated ratio of granulocytes to erythrocytes, and blasts
cells accounting for 23% of all nucleated cells. Flow cytometry
revealed high proportions of cells expressing CD10 (23%), CD19
(97%), CD13 (96.7%), CD33 (97.4%), HLA-DR (98.3%), and CD34
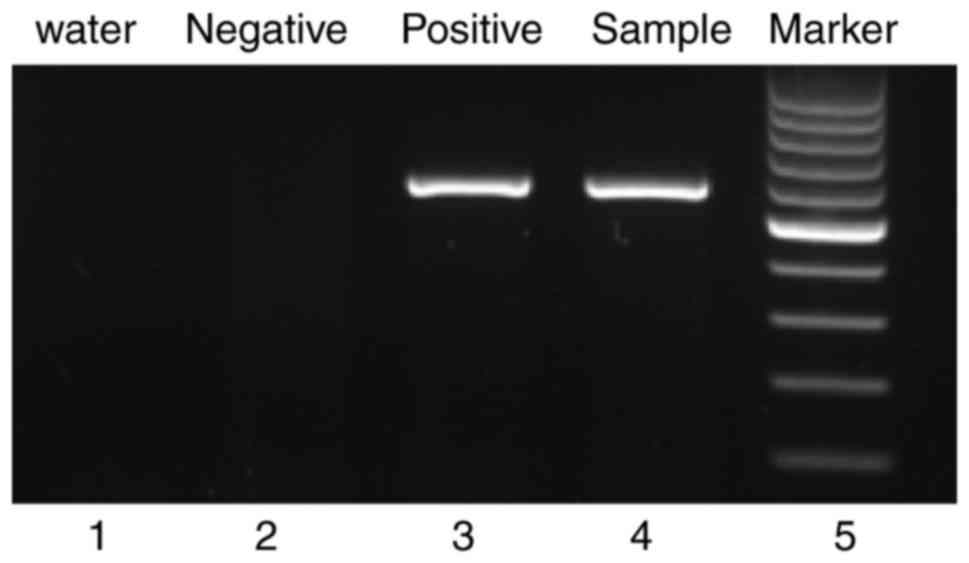
(85.8%). Reverse transcription-polymerase chain reaction (RT-PCR)
revealed the presence of the p210-type (major) BCR-ABL fusion
transcript (Fig. 1).
The patient was diagnosed with CML in B lymphoid
blast crisis. He was initially treated with orally administered
imatinib (600 mg daily), which was subsequently increased to 800
mg. This treatment was deemed ineffective after 65 days, when 13%
of nucleated cells in bone marrow were found to be blast cells.
Compared with the GeneBank sequence accession number NM_005157.5,
no T315I or F317L mutations were observed in the ABL1 region. The
patient was treated with dasatinib (140 mg daily) for 3 months,
following which the patient displayed complete hematologic
response. Later, the patient received hematopoietic stem cells from
an HLA-matched sibling donor, and he underwent myeloablative
conditioning. On day 126 following stem cell transplantation,
immunosuppressive therapy was withdrew and dasatinib therapy (140
mg daily) was again resumed.
At the last follow-up in September 2016, the patient
was alive and displayed clinical, hematological and cytogenetic
remission, with complete molecular response and complete donor
chimerism (Table I). Dasatinib
therapy was halted in May 2014.
 | Table I.Evaluation of treatment in the chronic
myeloid leukemia patient. |
Table I.
Evaluation of treatment in the chronic
myeloid leukemia patient.
|
| Hematological
response | Cytogenetic
response | Molecular
response |
|---|
| Imatinib | NR | NR | NR |
| Dasatinib | CHR | PCyR | <MMR |
| Allo-HSCT | CHR | CCyR | CMR |
Cytogenetic analysis
Unstimulated bone marrow was cultured for 24–48 h
and chromosomes were prepared from these cultures using standard
procedures. Chromosomes were analyzed in 20 metaphase cells using
G-banding and R-banding, and karyotypes were described according to
the International System for Human Cytogenetic Nomenclature
(9).
Fluorescence in situ hybridization
(FISH)
Chromosomes in 500 interphase nuclei cells were
analyzed using commercially available FISH probes according to the
manufacturers' instructions. The BCR-ABL fusion gene was detected
using the probes ES-BCR-ABL and DF-BCR-ABL (GP Gene Company,
Beijing, China), the region from the telomere of 22 to 22q11.1
downstream of the BCR breakpoint was detected using the breakpoint
probe EWSR1 (22q12) (Anbiping Gene Company, Guangzhou, China), and
the region from the centromere of chromosome 22 to 22q11.1 upstream
of the BCR breakpoint was detected using the probe CSP22 (22q11)
(GP Gene Company). The argininosuccinate synthase (ASS) probe
(9q34.1; GP Gene Company) was used to detect the deletion range of
ABL gene. The breakpoint probe BCL6 (3q27) (GP Gene Company) was
used to detect the presence of 3q end.
RT-PCR
Total RNA was extracted immediately from fresh bone
marrow cells of the patient. A negative control was used to monitor
RNA isolation. Primers and RT-PCR analyses for transcripts of P210
type BCR/ABL, p190-type BCR/ABL and β-actin were performed as
described previously (10).
Results
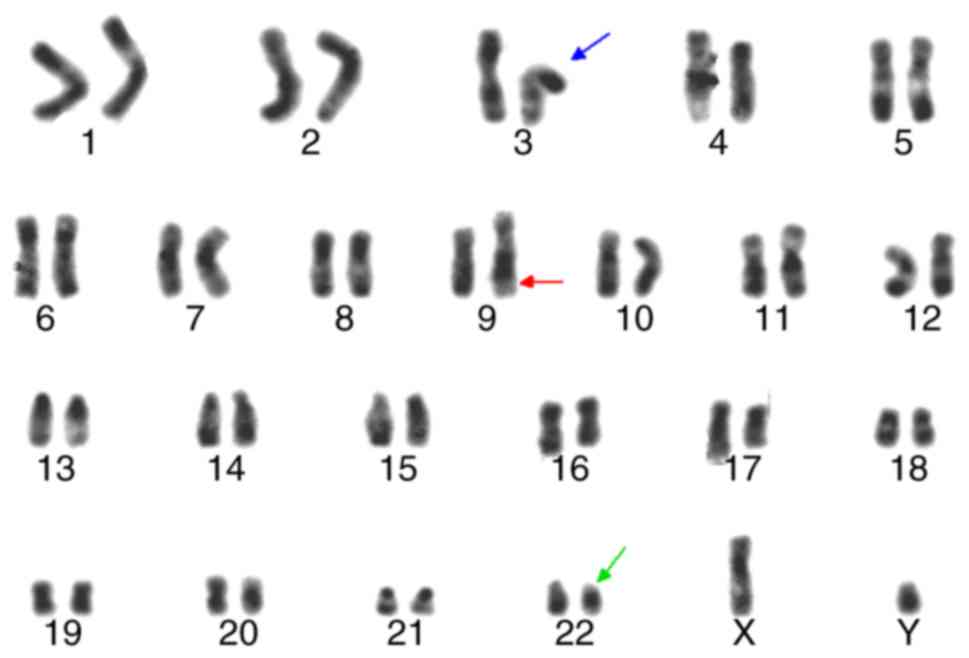
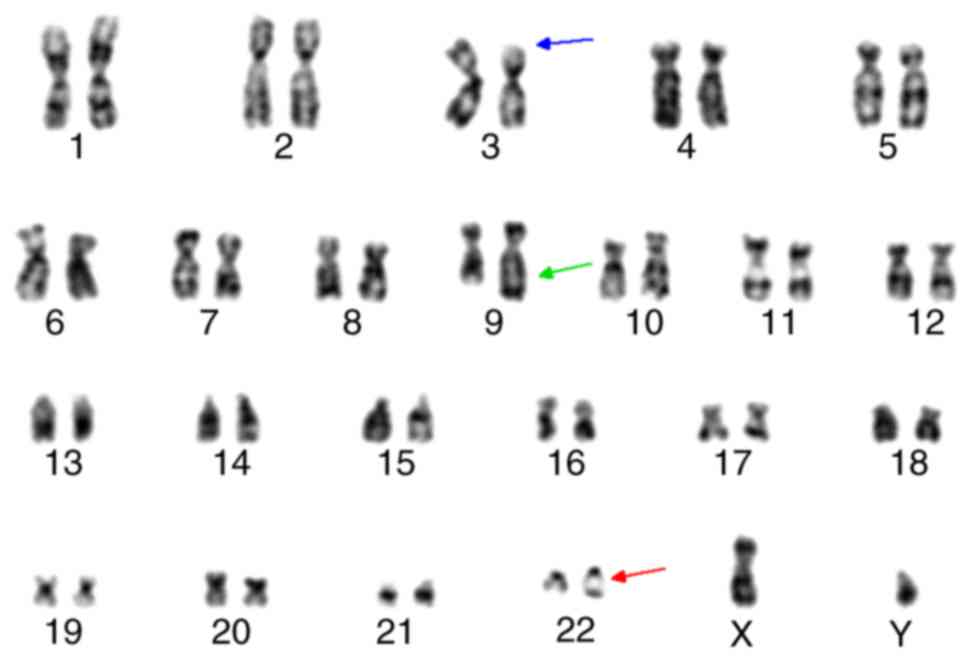
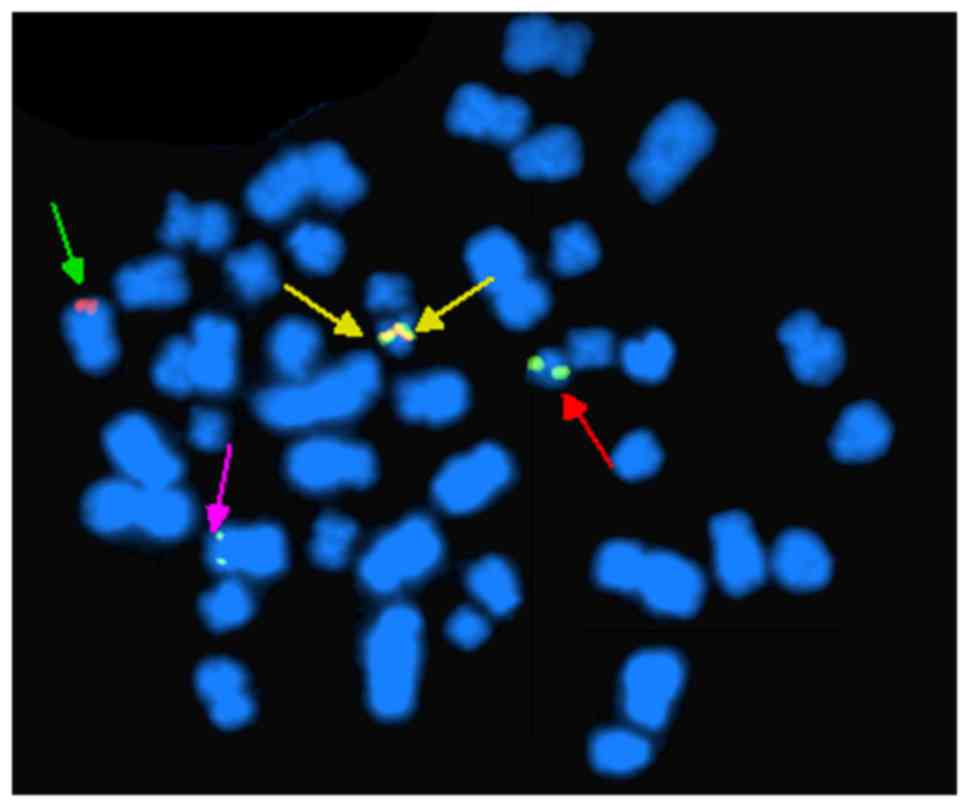
Chromosomal analysis at the time of diagnosis
revealed 46, XY, idicder(22) t(3;9;22)(p21;q34;q11)[18]/46, XY,
t(9;22)(q34;q11) (Figs. 2 and
3). These idic(Ph) chromosomes
appeared identical to normal chromosome 22 by R-banding, in
contrast to idic(Ph) chromosomes fused at satellite regions on the
p arms, which take on an equal length of two arms around
centromeres (Fig. 2). G-banding
analysis of the patient confirmed that the idic(Ph) chromosome was
spindle-shaped, implying two Ph chromosomes joined at the q
terminals (Fig. 3).
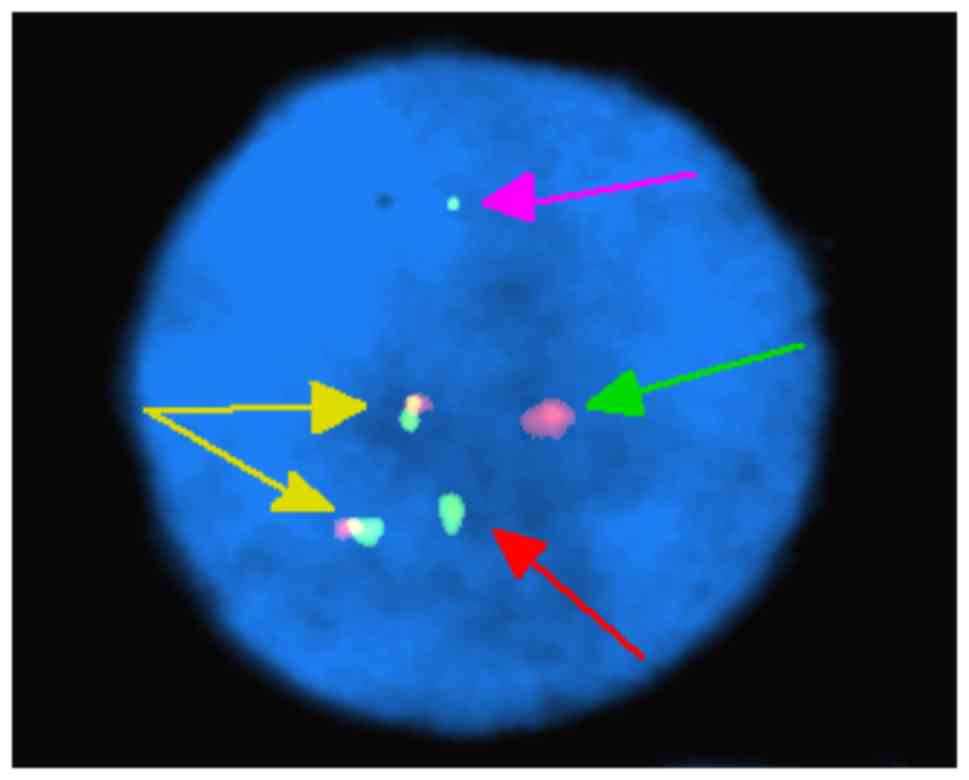
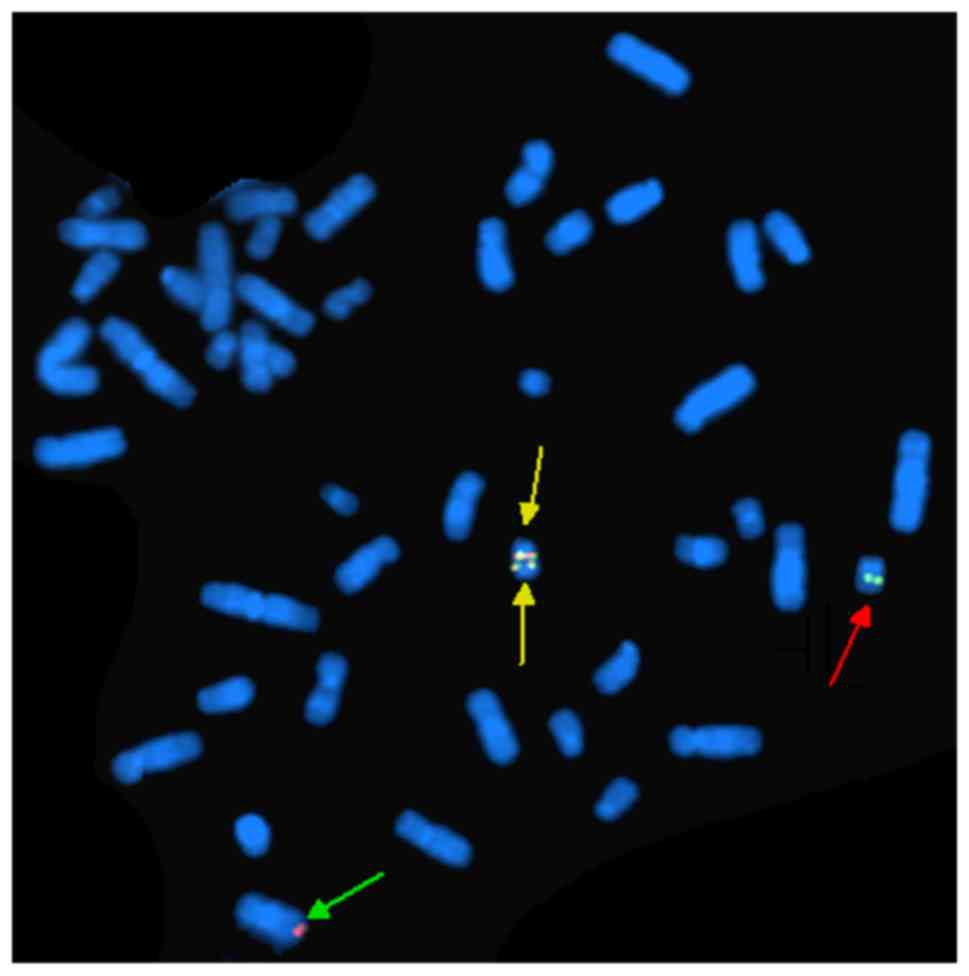
FISH analysis of nuclei of 200 interphase cells
using the probes ES-BCR-ABL and DF-BCR-ABL revealed two
non-overlapping fusion signals in 90% of cells (Fig. 4). These results indicate that the
idic(Ph) chromosome contained two BCR-ABL fusions. FISH analysis of
metaphase chromosomes using the same two probes revealed the
presence of two copies of the BCR-ABL fusion on one idic(Ph)
chromosome and the deletion of ABL gene (Fig. 5), as well as the major breakpoint of
BCR (Fig. 6). FISH analysis of
interphase and metaphase chromosomes using the probe CSP16/22
revealed three copies of the BCR-ABL fusion on normal chromosome 22
and idic(Ph) (data not shown). FISH analysis using the EWSR1
(22q12) probe, which covers the downstream region of the BCR
breakpoint, demonstrated two copies of the fusion on normal
chromosome 22 and chromosome 3p-(data not shown). This result
implied that the BCR-ABL fusion translocated to chromosome 3p-, and
that the ABL and ASS genes were deleted. In addition, karyotyping
analysis demonstrated that 3p21 exhibited deep R-banding originated
from 22q11-ter, and 9q34 exhibited longer shallow R-banding than
itself, which came from 3p21-pter (Fig.
3).
Furthermore, the BCL6 (3q27) gene was present on
normal chromosome 3 and chromosome 3p-, confirming that 3p21
contained the breakpoint for formation of the variant Ph
translocation in the CML patient (data not shown). The present
results are consistent with three-way Ph translocation (P210
pattern).
Discussion
In rare cases, CML is associated with three-way Ph
variant translocation involving chromosomes 9 and 22 (11,12). This
event occurs even more rarely in acute leukemia (13). Two main mechanisms have been proposed
for three-way translocations: A one-step mechanism in which
chromosome breakage occurs simultaneously on 3 chromosomes, which
undergo 3-way translocation; and a two-step mechanism in which a
standard t(9;22) translocation is followed by a second
translocation involving additional chromosomes (4,14,15). The two-step mechanism may be
associated with clonal evolution and poorer prognosis (14,15). The
FISH pattern of the patient analyzed in the present study indicated
one ABL copy (native), two BCR copies (one native and one on
chromosome 3), and two copies of the BCR-ABL fusion on the idic(Ph)
chromosome. This pattern is consistent with a two-step
mechanism.
The present results provide evidence that the
cytogenetic origins of CML may affect response to imatinib therapy
and therefore patient prognosis. Prior to imatinib therapy becoming
widespread, patients with variant translocations were considered to
be at risk of poorer prognosis than those with the standard
translocation (5,15–17). For
example, the proportion of patients in the accelerated phase of CML
was higher among those with variant translocations (56%) compared
with those with classic translocations (38%) (16). Today, however, this prognostic
distinction is considered controversial; for example, the European
LeukemiaNet recommendations do not mention higher risk of poor
prognosis for patients with CML with variant translocations
(17). The present results suggest
that the three-way translocation t(3;9;22)(p21;q34;q11) may be
associated with poor prognosis of patients with CML treated with
imatinib (18).
The implication of 3p21 in the present patient's
three-way translocation may help explain the onset of CML. A total
of 37 reports on CML patients with three-way Ph variant
translocations involving chromosome 3 were identified in the
Mitelman database (https://cgap.nci.nih.gov/Chromosomes/Mitelman/) and
the recent literature (18,19), and the breakpoint in these patients
occurs most often at 3p21. This region contains tumor suppressor
genes (H37/Luca15/RBM5, RASSFIA) as well as tumor susceptibility
genes (hMLH1) (20,21). Overexpression of H37/Luca15/RBM5 has
been demonstrated to result in cell cycle arrest and apoptosis in
human lung carcinoma (22). Deletions
or translocations involving 3p21 have been linked to acute
leukemia, myelodysplastic syndrome and solid tumor types, including
small cell lung and renal cell carcinomas (21,23,24).
In addition to the three-way translocation
t(3;9;22)(p21;q34;q11), the present patient possessed the idic(Ph)
chromosome. First reported in 1973 (25), idic(Ph) is a rare cytogenetic
aberration in which two identical Ph chromosomes fuse while
retaining their centromeres. In the Mitelman database and the
recent literature, there have been reports of 11 patients with CML
and one patient with acute lymphoblastic leukemia that possess
idic(Ph) chromosomes (26–31). In nearly all cases, these idic(Ph)
chromosomes formed by fusion at the satellite region in 22p13
(27–29). In the present case, two previous cases
of CML (30,31) and the single case with acute
lymphoblastic leukemia (26),
idic(Ph) chromosomes formed by fusion at 22q11. However, the
causative factor of the formation of idic(Ph) chromosomes remains
unknown. Isodicentric chromosomes may lead to breakage and reunion
cycles during mitosis, potentially forming ring chromosomes and
thus leading to genomic instability and heterogeneity in the cell
population.
The presence of the idic(Ph) chromosome in the
present patient may also explain his poor response to imatinib.
Often observed at later stages of CML, or in the accelerated phase
of the disease, idic(Ph) chromosomes can be associated with
resistance to standard chemotherapy and poor prognosis (28–30). In
particular, higher copy numbers of idic(Ph) chromosomes result in
amplification of the BCR-ABL gene, which is a key contributor to
imatinib resistance (32,33). Indeed, the presence of a double Ph
chromosome in patients with CML has been associated with poor
response to imatinib but good response to dasatinib (34,35). This
is consistent the present study, where the patient responded well
to dasatinib.
In conclusion, the present study described for the
first time the presence of a three-way Ph variant
t(3;9;22)(p21;q34;q11) and isodicentric Ph chromosomes in a CML
patient. These results may help define a new therapeutic standard
for treating CML involving isodicentric Ph chromosomes.
References
|
1
|
Baccarani M, Deininger MW, Rosti G,
Hochhausd A, Soverini S, Apperley JF, Cervantes F, Clark RE, Cortes
JE, Guilhot F, et al: European leukemiaNet recommendations for the
management of chronic myeloid leukemia: 2013. Blood. 122:872–884.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Czechowska A, Poplawski T, Drzewoski J and
Blasiak J: Imatinib (STI571) induces DNA damage in
BCR/ABL-expressing leukemia cells but not in normal lymphocytes.
Chem Biol Interact. 152:139–150. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jain P, Kantarjian H, Alattar ML, Jabbour
E, Sasaki K, Nogueras Gonzalez G, Dellasala S, Pierce S, Verstovsek
S, Wierda W, et al: Long-term molecular and cytogenetic response
and survival outcomes with imatinib 400 mg, imatinib 800 mg,
dasatinib and nilotinib in patients with chronic-phase chronic
myeloid leukaemia: Retrospective analysis of patient data from five
clinical trials. Lancet Haematol. 2:e118–e128. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gorusu M, Benn P, Li Z and Fang M: On the
genesis and prognosis of variant translocations in chronic myeloid
leukemia. Cancer genet Cytogenet. 173:97–106. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marzocchi G, Castagnetti F, Luatti S,
Baldazzi C, Stacchini M, Gugliotta G, Amabile M, Specchia G,
Sessarego M, Giussani U, et al: Variant Philadelphia
translocations: Molecular-cytogenetic characterization and
prognostic influence on frontline imatinib therapy, a GIMEMA
Working Party on CML analysis. Blood. 117:6793–6800. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Melo JV and Barnes DJ: Chronic myeloid
leukaemia as a model of disease evolution in human cancer. Nat Rev
Cancer. 7:441–453. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hehlmann R, Hochhaus A and Baccarani M:
Chronic myeloid leukaemia. Lancet. 370:342–350. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Skorski T: BCR/ABL, DNA damage and DNA
repair: Implications for new treatment concepts. Leuk Lymphoma.
49:610–614. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brothman AR, Persons DL and Shaffer LG:
Nomenclature evolution: Changes in the ISCN from the 2005 to the
2009 edition. Cytogenet Genome Res. 127:1–4. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Al-Achkar W, Wafa A, Ali BY, Manvelyan M
and Liehr T: A rare chronic myeloid leukemia case with Philadelphia
chromosome, BCR-ABL e13a3 transcript and complex translocation
involving four different chromosomes. Oncol Lett. 1:797–800. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Al-Achkar W, Wafa A, Ikhtiar A and Liehr
T: Three-way Philadelphia translocation t(9;10;22)(q34;p11.2;q11.2)
as a secondary abnormality in an imatinib mesylate-resistant
chronic myeloid leukemia patient. Oncol Lett. 5:1656–1658. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee J, Kim DS, Lee HS, Choi SI and Cho YG:
A novel t(9;22;11) translocation involving 11q24 in a patient with
chronic myeloid leukemia: A case report. Oncol Lett. 13:1711–1713.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cho SY, Kim SY, Jeon YL, Oh SH, Cho EH,
Lee WI, Cho KS and Park TS: A novel three-way Ph variant t(8;9;22)
in adult acute lymphoblastic leukemia. Ann Clin Lab Sci. 41:71–78.
2011.PubMed/NCBI
|
|
14
|
Huret JL: Complex translocations, simple
variant translocations and Ph-negative cases in chronic myelogenous
leukaemia. Hum Genet. 85:565–568. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Richebourg S, Eclache V, Perot C, Portnoi
MF, Van den Akker J, Terré C, Maareck O, Soenen V, Viguié F, Laï
JL, et al: Mechanisms of genesis of variant translocation in
chronic myeloid leukemia are not correlated with ABL1 or BCR
deletion status or response to imatinib therapy. Cancer Genet
Cytogenet. 182:95–102. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
El-Zimaity MM, Kantarjian H, Talpaz M,
O'Brien S, Giles F, Garcia-Manero G, Verstovsek S, Thomas D,
Ferrajoli A, Hayes K, et al: Results of imatinib mesylate therapy
in chronic myelogenous leukaemia with variant Philadelphia
chromosome. Br J Haematol. 125:187–195. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Baccarani M, Cortes J, Pane F,
Niederwieser D, Saglio G, Apperley J, Cervantes F, Deininger M,
Gratwohl A, Guilhot F, et al: Chronic myeloid leukemia: An update
of concepts and management recommendations of European Leukemia
Net. J Clin Oncol. 27:6041–6051. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tan J, Cang S, Seiter K, Primanneni S,
Ahmed N, Mathews T and Liu D: t(3;9;22) 3-way chromosome
translocation in chronic myeloid leukemia is associated with poor
prognosis. Cancer Invest. 27:718–722. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Buda G, Orciuolo E, Galimberti S,
Benedetti E, Caracciolo F, Cervetti G, Carulli G, Papineschi F and
Petrini M: Complex translocation t(3;9;22)(q21;q34;q11) at
diagnosis is a negative prognostic index in chronic myeloid
leukemia. Leuk Res. 32:192–194. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Morrissey C, Martinez A, Zatyka M,
Agathanggelou A, Honorio S, Astuti D, Morgan NV, Moch H, Richards
FM, Kishida T, et al: Epigenetic inactivation of the RASSF1A 3p21.3
tumor suppressor gene in both clear cell and papillary renal cell
carcinoma. Cancer Res. 61:7277–7281. 2001.PubMed/NCBI
|
|
21
|
Hemminki A, Peltomäki P, Mecklin J,
Järvinen H, Salovaara R, Nyström-Lahti M, de la Chapelle A and
Aaltonen LA: Loss of the wild-type MLH1 gene is a feature of
hereditary nonpolyposis colorectal cancer. Nat Genet. 8:405–410.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Oh JJ, Razfar A, Delgado I, Reed RA,
Malkina A, Boctor B and Slamon DJ: 3p21.3 tumor suppressor gene
H37/Luca15/RBM5 inhibits growth of human lung cancer cells through
cell cycle arrest and apoptosis. Cancer Res. 66:3419–3427. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shi G, Weh HJ, Martensen S, Seeger D and
Hossfeld DK: 3p21 is a recurrent treatment-related breakpoint in
myelodysplastic syndrome and acute myeloid leukemia. Cytogenet Cell
Genet. 74:295–299. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Johansson B, Billström R, Kristoffersson
U, Akerman M, Garwicz S, Ahlgren T, Malm C and Mitelman F: Deletion
of chromosome arm 3p in hematologic malignancies. Leukemia.
11:1207–1213. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Whang-peng J, Knutsen TA and Lee EC:
Dicentric Ph1 chromosome. J Natl Cancer Inst. 51:2009–2012. 1973.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yamamoto K, Nagata K, Morita Y, Inagaki K
and Hamaguchi H: Isodicentric Philadelphia chromosome in acute
lymphoblastic leukemia with der (7;12)(q10;q10). Leuk Res.
31:713–718. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Becher R, Ohl S, Schaefer UW, Wendehorst
E, Quiskamp F, Mahmoud HK, Schüning F and Schmidt CG: Clonal
evolution with isodicentric Ph1 chromosome in Ph1-positive CML:
Karyotypic conversion after bone marrow transplantation. Blut.
48:247–250. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kovacs G, Georgii A and Mainzer K: Three
isodicentric Philadelphia chromosomes in acute phase of chronic
myeloid leukemia: A case report. Cancer Genet Cytogenet. 20:29–33.
1986. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pernice F, Squadrito G, Saitta A, Mazza G
and Musolino C: Isodicentric Philadelphia chromosome in accelerated
phase of chronic myeloid leukemia. Cancer Genet Cytogenet.
66:113–116. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Szych CM, Liesveld JL, Iqbal MA, Li L,
Siebert S, Asmus C, O'Malley J, Lee A and Wang N: Isodicentric
Philadelphia chromosomes in imatinib mesylate (Gleevec)-resistant
patients. Cancer Genet Cytogenet. 174:132–137. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Ming, Chua C, Tan YY, Chua SP, Ma HB,
Koay E, Li Min, Poon M, Liu TC and Gole L: Multiple copies of a
rare rearrangement of Philadelphia chromosome in a chronic myeloid
leukemia patient: A case report. Cancer Genet Cytogenet. 199:66–68.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hochhaus A and Hughes T: Clinical
resistance to imatinib: Mechanisms and implications. Hematol Oncol
Clin North Am. 18:641–656. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Campbell LJ, Patsouris C, Rayeroux KC,
Somana K, Januszewicz EH and Szer J: BCR/ABL amplification in
chronic myelocytic leukemia blast crisis following imatinib
mesylate administration. Cancer Genet Cytogent. 139:30–33. 2002.
View Article : Google Scholar
|
|
34
|
Ikeda K, Harada-Shirado K, Matsumoto H,
Noji H, Ogawa K and Takeishi Y: Molecular response of e19a2
BCR-ABL1 Chronic myeloid leukemia with double Philadelphia
chromosome to Dasatinib. J Clin Oncol. 34:e130–e133. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Martin SE, Sausen M, Joseph A and Kingham
BF: Chronic myeloid leukemia with e19a2 atypical transcript: Early
imatinib resistance and complete response to dasatinib. Cancer
Genet Cytogenet. 201:133–134. 2010. View Article : Google Scholar : PubMed/NCBI
|