Introduction
Lung cancer has remained the most common type of
cancer since 1985, with 1.8 million novel cases reported in 2012
(1). Despite decades of research and
prolific cancer preventive strategies, the 5-year survival rate for
patients with lung cancer remains poor, and lung cancer remains the
primary cause of cancer-associated mortality (2,3). Globally,
1.6 million individuals die from lung cancer annually, accounting
for 20% of all cancer mortalities (4). The poor prognosis of lung cancer is
primarily due to late clinical diagnosis, metastases, multiple drug
resistance and comorbidity (5). Lung
cancer consists of two common forms: Small cell lung cancer (SCLC;
20%) and non-small cell lung cancer (NSCLC; 80%). NSCLC can be
further divided into adenocarcinoma (40–45%), squamous cell
carcinoma (20–25%) and large cell carcinoma (10–15%) subtypes
(6). The primary molecular cause of
NSCLC is not fully understood. Therefore, there is an urgent
requirement to clarify the pathogenesis of these tumors, and
identify a reliable and effective treatment strategy.
Cancer research in the area of endogenous microRNAs
(miRNAs/miRs) has markedly increased. miRNAs are endogenous small
(~22 nucleotides) non-coding RNAs that are able to suppress gene
expression or protein translation by interacting with the
3′-untranslated region (3′-UTR) of the target mRNAs. In addition,
miRNAs are also able to promote the expression of target genes
(7). Single miRNAs do not target a
single mRNA and the majority of mRNAs may be modulated by numerous
miRNAs; this results in complex regulation of gene expression
(8). Therefore, miRNAs are able to
participate in various fundamental biological mechanisms (9). It has been demonstrated that miRNAs
serve a pivotal role in lung cancer, with specific roles of miRNAs
as tumor suppressors (let-7, miR-126, −145, −200 and −34) and
oncogenes (miR-17, −92, −21, −31, −221 and −222) identified
(10). Although the number of
verified human miRNAs has increased in recent years, few have been
functionally described (11). Among
numerous miRNAs, miR-569 has been revealed to be a novel
cancer-associated miRNA which is increased partially owing to
amplification of 3q26.2. A previous in vivo and vitro
study demonstrated that miR-569 contributes to ovarian and breast
cancer cell survival and proliferation (12). Thus, miR-569 may be a feasible
biomarker and target for these types of cancer.
Genetic aberrations, including mutations and copy
number aberrations (CNAs), are signs of oncogenesis. CNAs have been
identified to be associated with the survival time of patients with
lung cancer (13,14). Non-coding miRNAs may be affected by
CNAs and serve as drivers in oncogenesis (15). Amplification of 3q26.2 is prevalent in
lung cancer (16). Accordingly,
investigation of the expression of miR-569 at 3q26.2 in lung cancer
cell (LCC) and its functional roles as well as the underlying
molecular mechanisms may possess clinical value.
The results of the present study demonstrated that
miR-569 was significantly decreased in LCC. Additionally,
functional experiments indicated that miR-569 may be able to
regulate cell proliferation, apoptosis and migration in LCC.
Furthermore, the present study identified that FOS and high
mobility group A2 (HMGA2) were potential targets of miR-569. The
data from the present study expands the current understanding of
the specific roles and the underlying molecular mechanisms of
miR-569 in LCC.
Materials and methods
Cell culture
The human lung cancer cell lines A549, H1299, HCC827
and 95D, and the normal human bronchial epithelial cell line HBE,
were acquired from the Regenerative Medicine Center of The First
Affiliated Hospital of Dalian Medical University (Dalian, China).
All cell lines used were cultured in RPMI-1640 medium (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
penicillin (100 U/ml), streptomycin (100 mg/ml) (Beijing Solarbio
Science and Technology Co., Ltd., Beijing, China) and 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). All
cells were incubated at 37°C in a humidified atmosphere containing
5% CO2.
Target identification
Potential targets for miR-569 were identified using
miRanda (www.microrna.org) and TargetScan
(www.targetscan.org/vert_71).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from the cells using RNAiso
Plus reagent (Takara Bio, Inc., Otsu, Japan), according to the
manufacturer's protocol. The purity and concentration of RNA was
measured using a NanoDrop 2000 instrument (Thermo Fisher
Scientific, Inc.). The RNA was transcribed into cDNA using a
microRNA Stem-Loop Reverse Transcription kit (GenePharma, Shanghai,
China), according to the manufacturer's protocol. To evaluate c-FOS
and HMGA2 expression, corresponding RNA was reverse-transcribed
into cDNA using the PrimeScript™ RT Reagent kit with genomic DNA
Eraser (Takara Bio, Inc.) 48 h after transfection in A549 cells.
qPCR was performed with a TransStart Top Green qPCR SuperMix kit
(TransGene Biotech Co., Ltd, Beijing, China) using the StepOne Plus
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling settings were as follows: 30 sec at 94°C, then 40
cycles of 5 sec at 94°C and 30 sec at 60°C. Small nuclear RNA(U6)
was used as an internal marker of miRNA. For the analysis of c-FOS
and HMGA2 expression, β-actin was used for normalization. All
reactions were performed in triplicate. The 2−∆∆Cq
method was used for relative quantification of gene expression
(17). The primers used are listed in
Table I.
 | Table I.Reverse transcription-polymerase chain
reaction primer sequences. |
Table I.
Reverse transcription-polymerase chain
reaction primer sequences.
| Name | Primer direction | Sequence (5′-3′) |
|---|
| hsa-miR-569 | F |
AGACTGCTGAGTTAATGAATCCTG |
|
| R |
TATGGTTGTTCACGACTCCTTC |
| U6 | F |
CTCGCTTCGGCAGCACATATACT |
|
| R |
ACGCTTCACGAATTTGCGTGTC |
| High mobility group
A2 | F |
CCAGGAAGCAGCAGCAAGA |
|
| R |
CCAGGCAAGGCAACATTGAC |
| c-FOS | F |
TACTACCACTCACCCGCAGAC |
|
| R |
GAATGAAGTTGGCACTGGAGA |
| β-actin | F |
ATCATGTTTGAGACCTTCAACA |
|
| R |
CATCTCTTGCTCGAAGTCCA |
Transfection
The miR-569 mimic (5′-AGUUAAUGAAUCCUGGAAAGU-3′,
5′-UUUCCAGGAUUCAUUAACUUU-3′) and corresponding negative control
(miR-NC; 5′-CAGUACUUUUGUGUAGUACAA-3′, 5′-ACGUGACACGUUCGGAGAATT-3′)
were synthesized by GenePharma. A549 cells were transfected with a
final oligonucleotide concentration of 50 nM using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Transfection efficiency was observed using
an inverted fluorescence microscope (Olympus Corporation, Tokyo,
Japan) and flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA)
6 h after transfection with a fluorescein miR negative control
(miR-FAM-NC). To further measure transfection efficiency, RT-qPCR
was performed on each experimental sample 48 h after
transfection.
Cell proliferation
A total of 8,000 cells were cultured per well in
96-well plates prior to the addition of the miR-569 mimic or miR-NC
into the well 1 day after seeding. Then a Cell Counting Kit-8
(CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was
used to measure cell proliferation at 24, 48 and 72 h after
transfection, and the absorbance of each well at 450 nm was
determined using a microplate reader (BioTek Instruments, Inc.,
Winooski, VT, USA). A total of 5 replicates were used for each
group in one experiment.
Cell cycle distribution and analysis
of cellular apoptosis
Cell cycle distribution and apoptosis were examined
using flow cytometry 48 h post-transfection. For the cell cycle
assay, 1×106 transfected cells were fixed in 75%
pre-cooled ethanol overnight, rinsed twice with PBS and stained
with propyl iodide dye solution (Beyotime Institute of
Biotechnology, Haimen, China) at room temperature for 30 min in the
dark prior to analysis by flow cytometry (BD Biosciences).
To analyze the effects of miR-569 on cell apoptosis,
the transfected cells were trypsinized, centrifuged at 179 × g for
5 min at 4°C and washed with ice-cold PBS, prior to being suspended
in 500 µl matched binding buffer containing 5 µl annexin
V-fluorescein isothiocyanate (FITC) and 5 µl propidium iodide (PI)
from an Annexin V-FITC Apoptosis Detection kit (Vazyme, Piscataway,
NJ, USA). Cells were incubated at room temperature for 15 min in
the dark prior to analysis by flow cytometry using the Apoptosis
Detection kit according to the manufacturer's protocol.
Wound-healing assay
Straight uniform lines were marked on the back of
the 6-well plates with a ruler. When the transfected cells reached
~80% confluence, the unilaminar cells were scratched across each
well using 10-µl pipette tips in order to evaluate cell migration
by observing the ability of the cell to migrate into the wounded
area. The wounds were recorded with images captured at0 and 24 h;
thereafter, the scratch region was examined using ImageJ software
version 1.38 (National Institutes of Health, Bethesda, MD,
USA).
Western blot analysis
Transfected cells were lysed in
radioimmunoprecipitation assay buffer (Beijing Solarbio Science and
Technology Co., Ltd.). The whole cell protein concentration was
determined using a bicinchoninic acid protein assay kit (Beijing
Solarbio Science and Technology Co., Ltd.), according to the
manufacturer's protocol. A total of 80 µg protein was loaded per
lane for 12% SDS-PAGE. Separated proteins were electrotransferred
onto polyvinylidene difluoride membranes and membranes were blocked
with 5% skimmed milk powder in Tris Buffered Saline-Tween 20 for 1
h at room temperature. The membranes were sealed in micro-plastic
bags containing polyclonal rabbit anti-human c-FOS (cat. no.
BM4864; dilution, 1:200; Wuhan Boster Biological Technology, Ltd.,
Wuhan, China), HMGA2 (cat. no. 20795-1-AP; dilution, 1:1,000; Wuhan
Sanying Biotechnology, Wuhan, China) or β-actin (cat. no. bs-0061R;
dilution, 1:1,500; BIOSS, Beijing, China) primary antibody.
Following incubation with the primary antibody overnight at 4°C,
the blots were then incubated with the corresponding horseradish
peroxidase-labeled goat anti-rabbit secondary antibody (cat. no.
bs-0295G-HRP; dilution, 1:5,000; Wuhan Boster Biological
Technology) at room temperature for 1 h. Protein bands were
visualized using enhanced chemiluminescence (Beijing Solarbio
Science and Technology Co., Ltd.) with FluorChemFC3 gel imaging
software (version 3.4; Protein Simple; Bio-Teche, Minneapolis, MN,
USA).
Statistical analysis
At least three repeats were performed for all
experiments. The data are presented as the mean ± standard
deviation. Statistical difference was determined by analysis of
variance followed by Fisher's least significant difference or
Student-Newman-Keuls post hoc test, or two-tailed Student's t-test
using SPSS (version 21.0; IBM Corp., Armonk, NY, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
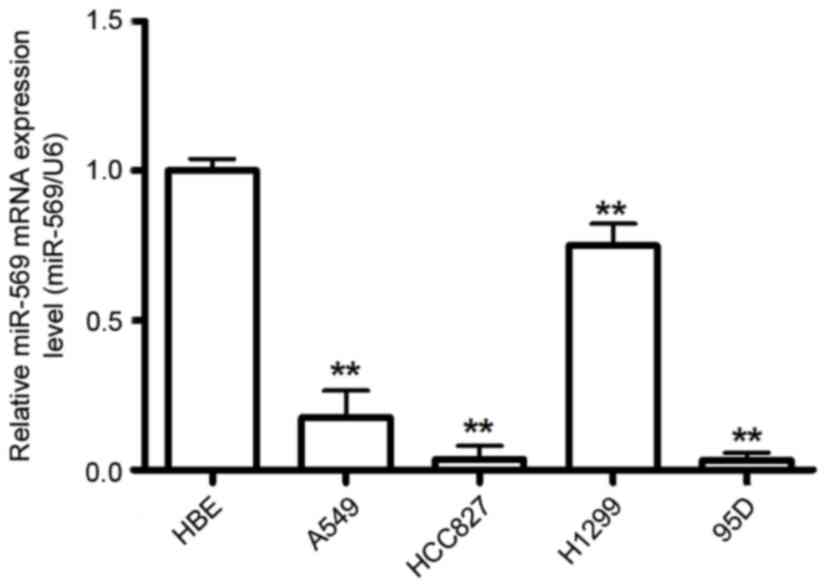
miR-569 is significantly decreased in
human LCC
To investigate the potential role of miR-569 in LCC
carcinogenesis, the present study examined miR-569 expression in a
group of human LCC using RT-qPCR. The results demonstrated that
miR-569 expression level was downregulated in the LCC compared with
the normal cell line HBE (P<0.05; Fig.
1).
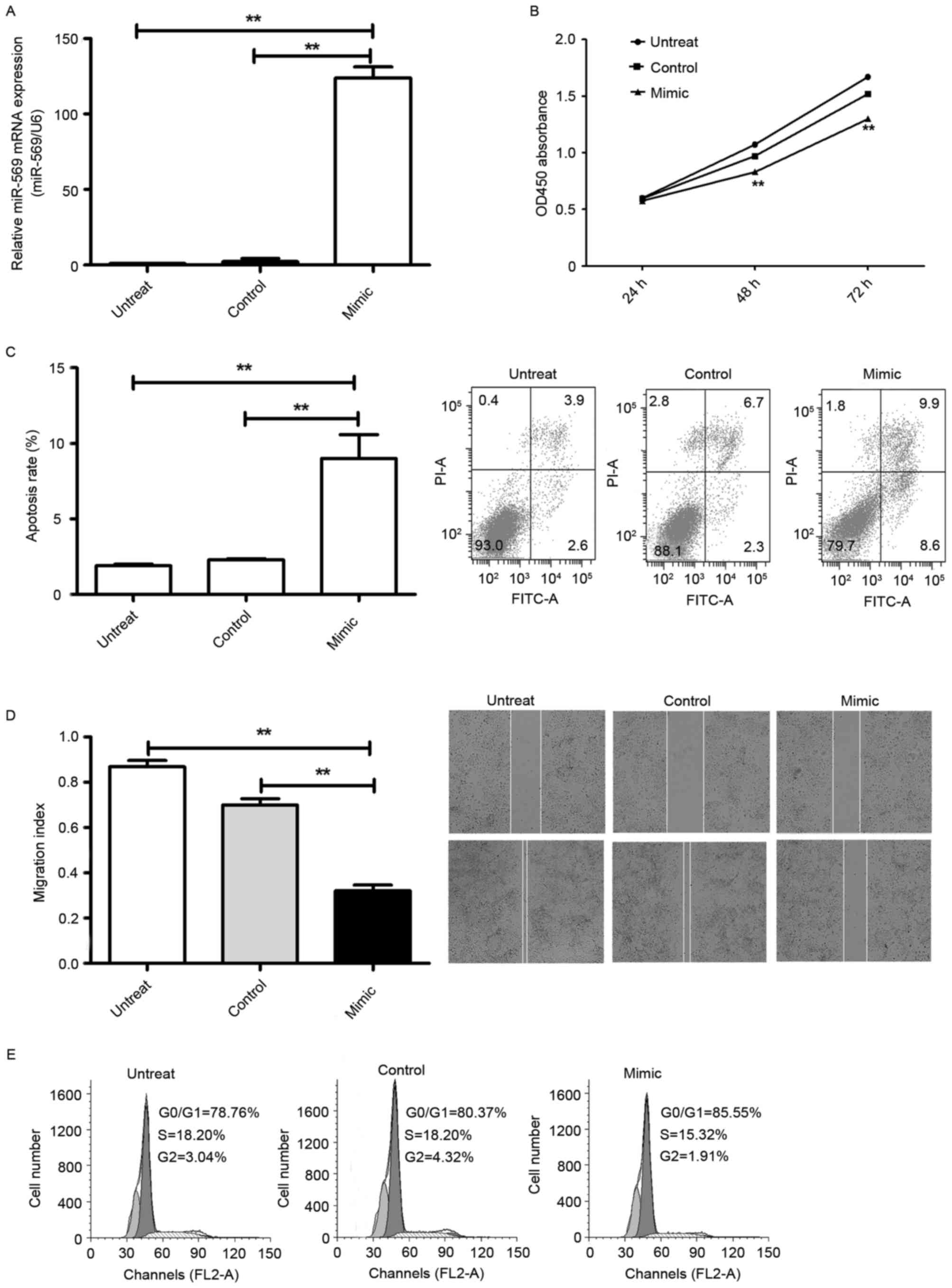
Ectopic overexpression of miR-569 is
associated with adverse effects on LCC
To explore the cellular functions of miR-569 in LCC,
first A549 cells were transfected with the fluorescently labelled
negative control (miR-FAM-NC), which exhibited high transfection
efficiency revealed using an inverted fluorescence microscope and
flow cytometry. Additionally, RT-qPCR reflected the successful
endogenic overexpression of miR-569: It was markedly increased
following transfection with the miR-569 mimic compared with the
miR-NC (P<0.05; Fig. 2A). CCK-8
and flow cytometry demonstrated that overexpression of miR-569
markedly prevented LCC proliferation (P<0.05; Fig. 2B) and induced cell apoptosis
(P<0.05; Fig. 2C). In addition, as
suggested by the flow cytometry, overexpression of miR-569 markedly
induced G1 phase cell cycle arrest in lung adenocarcinoma A549
cells (P<0.05; Fig. 2D). A
wound-healing assay was performed which revealed that
overexpression of miR-569 significantly suppressed migration
(P<0.05; Fig. 2E) in A549
cells.
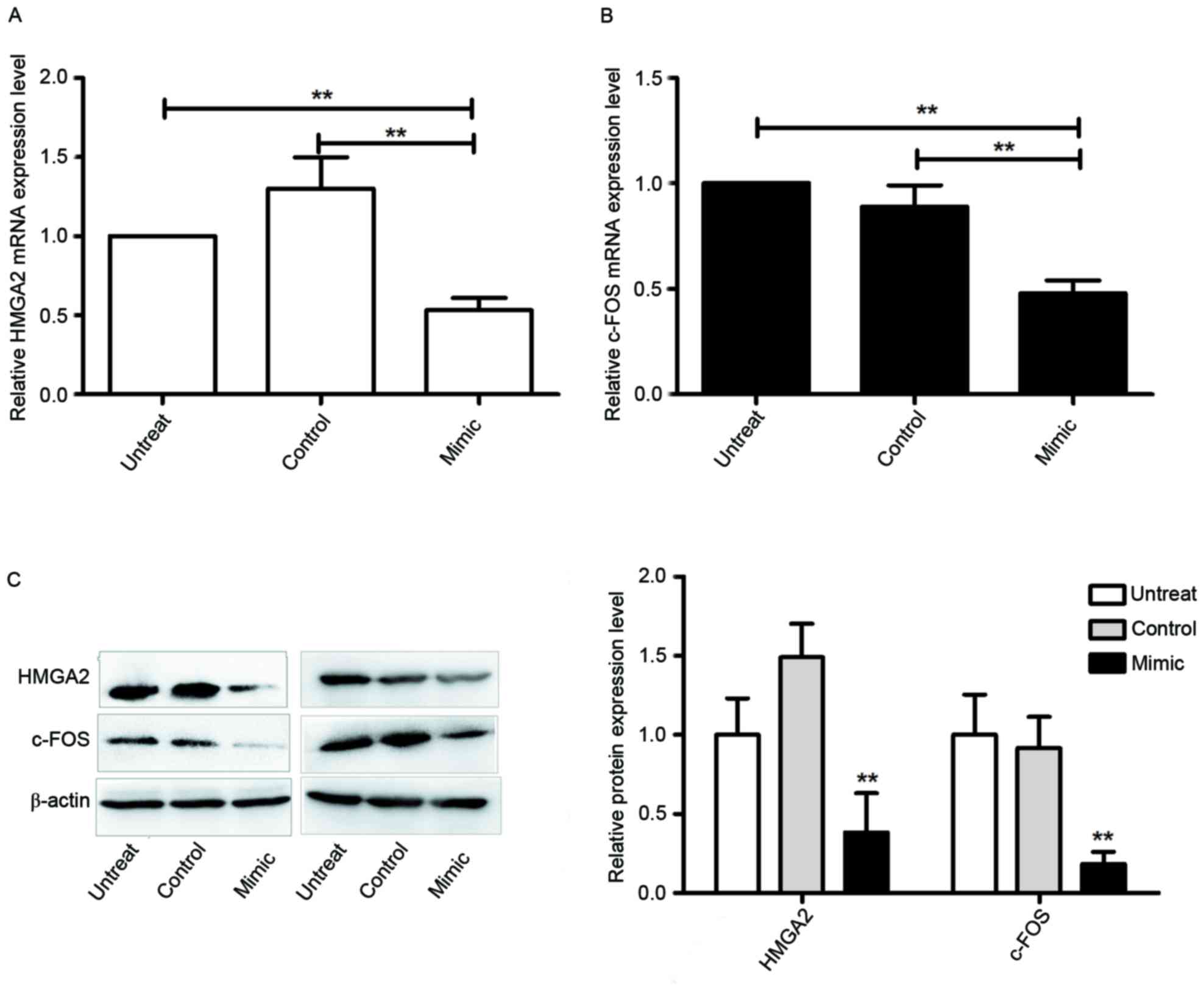
miR-569 inhibits c-FOS and HMGA2
expression
miRanda and TargetScan identified that miR-569 may
be associated with c-FOS and HMGA2 mRNA 3′-UTR binding sites. In
contrast with the upregulated expression of c-FOS and HMGA2
identified in previous studies (18–21), it
was observed in the present study that miR-569 expression was
decreased in LCC. This suggested that c-FOS and HMGA2 were
potential targets of miR-569. This was investigated in the present
study by detecting endogenous c-FOS and HMGA2 expression of the
miR-569 mimic-transfected cells in comparison with
miR-NC-transfected cells. It was revealed that the miR-569 mimic
markedly decreased the expression levels of HMGA2 and c-FOS mRNA
(P<0.05; Fig. 3A and B) and
protein (P<0.05; Fig. 3C). Taken
together, the results of the present study suggest that c-FOS and
HMGA2 are potential miR-569 targets in A549 cells.
Discussion
Previous studies have demonstrated that miRNAs may
serve key roles in cancer either as onco-miRNAs or tumor
suppressive miRNAs in various types of cancer (22–24),
including lung cancer (10). Despite
this, the underlying molecular mechanisms between miRNAs in cancer
remain somewhat unknown. A novel family of small single-stranded
non-coding RNAs, miRNAs be affected by CNAs (25). Additionally, changes in DNA copy
number, particularly at the 3q26.2 amplicon, have been identified
as predictors of lung cancer (26).
Thus, characterization of miR-569 associated with CNAs may expand
current knowledge of the etiology underlying lung cancer, and
provide new insights into molecular markers and targets for lung
cancer therapy.
HMGA2 is a crucial regulator of tumorigenesis and
embryogenesis (27). As a
transcription factor, HMGA2 is usually upregulated and serves
important roles in various biological processes of numerous
malignant tumors including LCC (28–30). The
3′-UTR of HMGA2 is >3 kb, allowing it to bind to numerous
miRNAs. It has previously been demonstrated that the let-7 miRNA
family may prevent early lung cancer progression by specifically
inhibit HMGA2 expression (31). In
addition, other miRNAs, including miR-26 a and miR-98, are able to
regulate HMGA2 expression to modulate cisplatin resistance of human
NSCLC (32,33). c-FOS is the human homolog of the
retroviral oncogene v-FOS (34). In
combination with their Jun partners, FOS forms the activator
protein 1 transcription factor complex protein, which has a
leucine-zipper region to bind DNA, giving rise to changes of gene
expression (35). As a
proto-oncogene, c-FOS regulates diverse cellular functions in a
variety of types of cancer including LCC (36,21).
To the best of our knowledge, the results of the
present study demonstrate for the first time that miR-569
expression levels are markedly downregulated in LCC cells. Of note,
miR-569 overexpression prevented cell proliferation and migration,
and inversely induced cell apoptosis. In addition, upregulation of
miR-569 inhibited HMGA2 and c-FOS mRNA and protein expression.
Accordingly, the results of the present study demonstrated that
miR-569 functions as a tumor suppressor in LCC, at least in part
due to targeting HMGA2 and c-FOS, and contributed to the
progression and metastasis of LCC.
To the best of our knowledge, the present study
revealed for the first time that miR-569 functioned as a potential
tumor suppressor gene by targeting HMGA2 and c-FOS in lung cancer.
Which may provide a new breakthrough to explore the pathogenesis of
lung cancer. However, the present study lacked tissue sample and
in vivo murine experiments to provide further evidence of
the role miR-569 serves in LCC. Therefore, further in-depth studies
are required to support and expand the results from the present
study.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wink KC, Roelofs E, Solberg T, Lin L,
Simone CB II, Jakobi A, Richter C, Lambin P and Troost EG: Particle
therapy for non-small cell lung tumors: Where do we stand? A
systematic review of the literature. Front Oncol. 4:2922014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bray F, Jemal A, Grey N, Ferlay J and
Forman D: Global cancer transitions according to the Human
Development Index (2008–2030): A population-based study. Lancet
Oncol. 13:790–801. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Grose D, Morrison DS, Devereux G, Jones R,
Sharma D, Selby C, Docherty K, McIntosh D, Nicolson M, McMillan DC,
et al: The impact of comorbidity upon determinants of outcome in
patients with lung cancer. Lung Cancer. 87:186–192. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mehta A, Dobersch S, Romero-Olmedo AJ and
Barreto G: Epigenetics in lung cancer diagnosis and therapy. Cancer
Metastasis Rev. 34:229–241. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vasudevan S, Tong Y and Steitz JA:
Switching from repression to activation: microRNAs can up-regulate
translation. Science. 318:1931–1934. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
van de Vrie M, Heymans S and Schroen B:
MicroRNA involvement in immune activation during heart failure.
Cardiovasc Drugs Ther. 25:161–170. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Garzon R, Marcucci G and Croce CM:
Targeting microRNAs in cancer: Rationale, strategies and
challenges. Nat Rev Drug Discov. 9:775–789. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Joshi P, Middleton J, Jeon YJ and Garofalo
M: MicroRNAs in lung cancer. World J Methodol. 4:59–72. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li F, Liu J and Li S: MicroRNA 106b ~25
cluster and gastric cancer. Surg Oncol. 22:e7–10. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chaluvally-Raghavan P, Zhang F, Pradeep S,
Hamilton MP, Zhao X, Rupaimoole R, Moss T, Lu Y, Yu S, Pecot CV, et
al: Copy number gain of hsa-miR-569 at 3q26.2 leads to loss of
TP53INP1 and aggressiveness of epithelial cancers. Cancer Cell.
26:863–879. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kawano O, Sasaki H, Okuda K, Yukiue H,
Yokoyama T, Yano M and Fujii Y: PIK3CA gene amplification in
Japanese non-small cell lung cancer. Lung Cancer. 58:159–160. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Go H, Jeon YK, Park HJ, Sung SW, Seo JW
and Chung DH: High MET gene copy number leads to shorter survival
in patients with non-small cell lung cancer. J Thorac Oncol.
5:305–313. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qian J and Massion PP: Role of Chromosome
3q amplification in lung cancer. J Thorac Oncol. 3:212–215. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Di Cello F, Hillion J, Hristov A, Wood LJ,
Mukherjee M, Schuldenfrei A, Kowalski J, Bhattacharya R, Ashfaq R
and Resar LM: HMGA2 participates in transformation in human lung
cancer. Mol Cancer Res. 6:743–750. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu Y, Song Y and Liu H: Expression and its
clinical significance of HMGA2 in the patients with non-small cell
lung cancer. Zhongguo Fei Ai Za Zhi. 11:377–381. 2008.(In Chinese).
PubMed/NCBI
|
|
20
|
Meyer B, Loeschke S, Schultze A, Weigel T,
Sandkamp M, Goldmann T, Vollmer E and Bullerdiek J: HMGA2
overexpression in non-small cell lung cancer. Mol Carcinog.
46:503–511. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Volm M, Drings P and Wodrich W: Prognostic
significance of the expression of c-fos, c-jun and c-erbB-1
oncogene products in human squamous cell lung carcinomas. J Cancer
Res Clin Oncol. 119:507–510. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gartel AL and Kandel ES: miRNAs: Little
known mediators of oncogenesis. Semin Cancer Biol. 18:103–110.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Garzon R and Marcucci G: Potential of
microRNAs for cancer diagnostics, prognostication and therapy. Curr
Opin Oncol. 24:655–659. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Holland AJ and Cleveland DW: Boveri
revisited: Chromosomal instability, aneuploidy and tumorigenesis.
Nat Rev Mol Cell Biol. 10:478–487. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang Z, Zhuan B, Yan Y, Jiang S and Wang
T: Integrated analyses of copy number variations and gene
differential expression in lung squamous-cell carcinoma. Biol Res.
48:472015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu J and Wei JJ: HMGA2 and high-grade
serous ovarian carcinoma. J Mol Med (Berl). 91:1155–1165. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Di Cello F, Hillion J, Hristov A, Wood LJ,
Mukherjee M, Schuldenfrei A, Kowalski J, Bhattacharya R, Ashfaq R
and Resar LM: HMGA2 participates in transformation in human lung
cancer. Mol Cancer Res. 6:743–750. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu Y, Song Y and Liu H: Expression and its
clinical significance of HMGA2 in the patients with non-small cell
lung cancer. Zhongguo Fei Ai Za Zhi. 11:377–381. 2008.(In Chinese).
PubMed/NCBI
|
|
30
|
Meyer B, Loeschke S, Schultze A, Weigel T,
Sandkamp M, Goldmann T, Vollmer E and Bullerdiek J: HMGA2
overexpression in non-small cell lung cancer. Mol Carcinog.
46:503–511. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Park SM, Shell S, Radjabi AR, Schickel R,
Feig C, Boyerinas B, Dinulescu DM, Lengyel E and Peter ME: Let-7
prevents early cancer progression by suppressing expression of the
embryonic gene HMGA2. Cell Cycle. 6:2585–2590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang Y, Zhang P, Zhao Y, Yang J, Jiang G
and Fan J: Decreased MicroRNA-26a expression causes cisplatin
resistance in human non-small cell lung cancer. Cancer Biol Ther.
17:515–525. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xiang Q, Tang H, Yu J, Yin J, Yang X and
Lei X: MicroRNA-98 sensitizes cisplatin-resistant human lung
adenocarcinoma cells by up-regulation of HMGA2. Pharmazie.
68:274–281. 2013.PubMed/NCBI
|
|
34
|
Milde-Langosch K: The fos family of
transcription factors and their role in tumourigenesis. Eur J
Cancer. 41:2449–2461. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chiu R, Boyle WJ, Meek J, Smeal T, Hunter
T and Karin M: The c-Fos protein interacts with c-Jun/AP-1 to
stimulate transcription of AP-1 responsive genes. Cell. 54:541–552.
1988. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Taniguchi S, Kawano T, Mitsudomi T, Kimura
G and Baba T: Fos oncogene transfer to a transformed rat fibroblast
cell line enhances spontaneous lung metastasis in rat. Jpn J Cancer
Res. 77:1193–1197. 1986.PubMed/NCBI
|