Introduction
Cervical cancer (CC) is one of the most common types
of gynecological malignant tumor (1).
The main histological type is cervical squamous cell carcinoma
(CSCC), which accounts for 75–80% of cases; adenocarcinoma accounts
for 10–15% and other histological subtypes represent 10–15%
(2,3).
Although there has been a declining trend over the past few
decades, CC remains a major health problem for Chinese women,
particularly for those living in rural areas (4). CC originating from cervical
intraepithelial neoplasia (CIN) is often caused by high risk-human
papilloma virus (HR-HPV) infection (5). CIN is a group of cervical lesions that
are associated with CC and are divided into three grades (I, II and
III) (6,7). The majority of low-grade CIN cases
naturally subside, but high-grade lesions continue to develop and
break through the sub-epithelial basement membrane, at which point
they are referred to as cervical invasive carcinoma (7,8). Histology
is currently the basis for the diagnosis and classification of CIN
(6).
The early detection of lesions by screening remains
the primary prevention method for CC. CIN often occurs in women
aged 25–35 years (6) and the highest
incidence of CC occurs at ~47 years of age (9), suggesting a slow evolution from
precancerous lesions to CC. Additionally, HPV vaccination for the
prevention of CC is offered in numerous regions around the world,
but it is not widely applied in a number of countries, including
mainland China (4,10). Finally, since the 1950s, due to the
widespread use of cervical cytology screening, cervical
precancerous and cancerous lesions may be identified and treated
early, resulting in a significant decrease in the incidence, and
mortality of CC (11,12). Therefore, early detection of lesions
by screening is effective for CC prevention. In addition, screening
and detection of high-grade CIN lesions and early CC, and providing
timely treatments may represent effective measures for increasing
the rate of cure in these diseases.
The Papanicolaou (Pap) test is currently the main
screening method for cervical precancerous changes and CC (13,14).
However, the sensitivity of the Pap test varies greatly, ranging
between 30–87%, and sometimes being as low as 20% (14,15). In
addition, the infrastructure used for Pap screening is expensive
and is difficult to implement in developing countries (16). Although the thin-layer liquid-based
cervical cytology technique has improved recently, the sensitivity
of the test has led to an important number of equivocal cytological
results that require confirmation (9,17). Due to
the fact that persistent HR-HPV infection is a well-established
cause of cervical neoplasia, HPV nucleotide detection is an
attractive method for the detection of cervical lesions (18–20), but
only a small proportion of the HR-HPV infected individuals exhibit
cervical lesions, and the HPV test demonstrates limited specificity
in diagnosing CC, particularly in young women (21,22).
DNA methylation involves intensive epigenetic
modifications that serve important roles in gene expression or
silencing in normal mammalian cells (10,23). DNA
methylation-induced alteration of C-phosphate-G (CpG) islands in
tumor suppressor gene promoter regions is often observed in human
cancer (24–27). It is currently acknowledged that
hyper- or hypo-methylation of tumor suppressor gene promoter
regions may contribute to cell transformation and thus, that the
DNA methylation status is a promising biomarker for the detection
of cancer (28). For CC, HPV viral
DNA methylation acts as a potential biomarker for early cancer
detection. For example, methylation of multiple genes, including
paired box gene 1 (PAX1) (29), LIM
homeobox transcription factor 1α (LMX1A), NK6 transcription
factor-related locus 1 (30), SRY-box
1, Wilms tumor 1 and one cut homeobox 1 (31), have demonstrated varying degrees of
sensitivity, specificity, and accuracy for the detection of CIN
grades III and above. Nevertheless, the diagnostic accuracy of
these genes requires further evaluation.
The purpose of the present study was to explore the
diagnostic accuracy of the quantitative methylation analysis of two
genes, PAX1 and LMX1A, in a full spectrum of cervical lesions in an
Eastern Chinese population.
Patients and methods
Study design
This single-center prospective clinical study was
approved by the Ethics Committee of the Central Hospital of Minhang
District (Shanghai, China). Between July 2013 and September 2014,
121 subjects with cervical cancer were recruited, and tested for
PAX1 and LMX1A methylation genes prior to undergoing pathological
examination at the gynecological department of the Central Hospital
of Minhang District. The tissue histopathology of cervical tissue
was used for diagnosis and the diagnostic accuracy of CIN and CC
was assessed using the gene methylation test.
Patients
The study recruited adult women from a population
undergoing routine health examination and those clinically
diagnosed with CC (age range 21–57; mean age, 37.15±8.6). Potential
participants were screened for eligibility using a structured
questionnaire and detailed clinical assessment. The inclusion
criteria were as follows: i) Patients and healthy women receiving
cytological examination, using the thin-layer liquid-based
technique (32), of the cervical
exfoliated cells and quantitative detection of HR-HPV DNA; ii)
women aged 21–57 years with a history of sexual activity; and iii)
women who provided written informed consent to participate in the
study. The exclusion criteria were: i) Refusal to undergo further
colposcopy, cervical biopsy or cervical loop electrosurgical
excision and hysterectomy; ii) history of other malignant tumors;
iii) treatment for other cervical diseases during the study; or iv)
histopathological diagnosis of cervical adenocarcinoma.
Examination and grouping
All the participants received a histological
examination and underwent a colposcopic cervical biopsy by a
trained medical doctor. Cervical exfoliated cell specimens were
taken by a nurse using a Cervex-Brush (Rovers Medical Devices, Oss,
The Netherlands) and smeared onto a slide for cytological
examination. The hospital team comprised 10 pathologists, each with
>20 years of experience. Biopsied tissues were fixed in 10%
neutral-buffered formalin for 24 h at 37°C. Paraffin-embedded
sections (4-µm thick) were cut for hematoxylin and eosin staining
(5–15 min at 37°C) and immunohistochemistry. To generate frozen
(−20°C) sections, fresh tissues were embedded in Tissue-Tek O.C.T
compound (Sakura Finetek Europe B.V., Flemingweg, The Netherlands)
immediately after removal. Frozen sections (7 µm) were used for
immunofloresence.
The participants were then grouped according to the
tissue biopsy results as NC (normal cervix), LSIL (low-grade
squamous intraepithelial lesion), HSIL (high-grade squamous
intraepithelial lesion) or CSCC (cervical squamous cell carcinoma).
Since 60% of the grade I CIN lesions naturally fade and require no
treatment (only follow-up if the disease does not progress within 2
years), CIN1 lesions were classified as LSIL. Approximately 20% of
CIN2 lesions progress to CIN3 and 5% of these lesions eventually
become invasive cancer; therefore, CIN2 and CIN3 lesions were
classified as HSIL (33).
Immunohistochemistry
For immunohistochemistry, all antibodies and
reagents were purchased from Fuzhou Maixin Biotechnology
Development Co., Ltd. (Fuzhou, China) and immunohistochemistry was
performed according to the UltraSensitive™ SP (Mouse/Rabbit) IHC
kit (cat no. KIT-9710; Fuzhou Maixin Biotechnology Development Co.,
Ltd.) manufacturer's protocol. The first step was to bake the
paraffin section at 60°C for 2 h, followed by xylene dewaxing and
alcohol hydration for 3 h at 60°C, following these procedures:
xylene I for 60 min, xylene II for 30 min, 100% alcohol for 30 min,
95% alcohol for 15 min, 75% alcohol for 15 min, 50% alcohol for 15
min and washing with distilled water for 15 min. In order to block
inactivated endogenous peroxidase, cells were incubated with 3%
H2O2 at 37°C for 10 min, followed by washing
with phosphate-buffered saline (PBS) three times for 5 min. Then,
for antigen repair, 0.01 M citric acid tissue antigen repair
solution (pH 6.0) was used for boiling (at 95°C, for 15 to 20 min),
and then the cells were allowed to cool naturally for 20 min. Then,
the cylinder was rinsed with cold water and cooling was accelerated
to room temperature, followed by washing with PBS three times for 5
min. Normal sheep serum was enclosed with the cells and incubated
at 37°C for 20 min, followed by removing the normal sheep serum
without washing. The primary antibodies p16 (cat no. MAB-0673;
Fuzhou Maixin Biotechnology Development Co., Ltd.) at 1:100
dilution and ki67 (cat no. MAB-0672; Fuzhou Maixin Biotechnology
Development Co., Ltd) at 1:200 dilution were added respectively and
refrigerated at 4°C overnight, and washed with PBS three times for
5 min (with PBS buffer used as a negative control). A total of 50
µl biotinylated goat anti-mouse/rabbit IgG [Buffer C from the
UltraSensitive SP (Mouse/Rabbit) IHC kit; cat no. KIT-9710] was
performed at 37°C for a 30-min incubation, followed by washing with
PBS three times for 5 min. A total of 50 µl horseradish
peroxidase-labeled streptomycin avidin working liquid [Buffer D
from the UltraSensitive SP (Mouse/Rabbit) IHC kit, product number:
KIT-9710], for the specific recognition of the biotin-labeled
secondary antibody, was then added for a 30-min incubation at 37°C,
followed by washing with PBS three times for 5 min. This method
ensures a higher sensitivity (34).
Then 3,3′-Diaminobenzidine/H2O2 reaction
staining at 37°C for 3–10 min was performed. After washing with the
tap water for 5 times within 15 min, the haematin was used for
re-dying at 37°C for 1 min, followed by normal alcohol dehydration
at 37°C for 5–10 min, treated with xylene for 4–6 min at 37°C to
increase the transmittance of the specimen, and the specimen was
covered with a square coverslip by adding the neutral gum to seal,
followed by drying naturally at room temperature for later
observation.
Immunofluorescence
Frozen sections (7 µm thick) were fixed with 2%
paraformaldehyde in PBS at room temperature for 10 min followed by
extraction using 0.5% Triton X-100 in PBS for 5 min, at room
temperature. Blocking was then performed using 0.01 M PBS (pH 7.4)
containing 10% normal goat serum and 0.3% Trixton X-100 for 1 h at
room temperature, followed by the addition of the primary
antibodies as follows: Human Anti-CDKN2A mouse monoclonal antibody
(cat no. D199930; Sangon Biotech Co., Ltd., Shanghai, China) at a
1:100 dilution, diluted with with 0.01 M PBS (pH 7.4) containing 1%
BSA and 0.3% Triton X-100 and human Ki-67 (D3B5) Rabbit mAb (Alexa
Fluor® 647 Conjugate; cat no. 12075; Cell Signaling
Technology) at a 1:50 dilution, diluted with 0.0 1 M PBS (pH 7.4)
containing 1% BSA and 0.3% Triton X-100, and incubated with these
antibodies at 4°C overnight. This was followed by washing with PBS
three times for 10 min in the dark. Then, incubation with the
secondary antibody FITC-conjugated Donkey Anti-Mouse IgG (cat no.
D110081; Sangon Biotech Co., Ltd.) was performed at a 1:100
dilution, diluted with 0.01 M PBS (pH 7.4) containing 1% BSA and
0.3% Triton X-100 and incubated for 1 h at room temperature,
followed by washing with PBS three times for 15 min. Fluorescence
microscopy was performed following sealing. DNA was visualized
using ~1.5 g/ml Hoechst 33342 (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). Fluorescence microscopy was performed using a
60× Plan Apo (NA 1.40) oil immersion objective lens and a Nikon
TE2000-U inverted microscope equipped with a SPOT-RT CCD
system.
DNA methylation: Cervical DNA
extraction, bisulfite treatment and modification
DNA was extracted from the obtained exfoliated cells
using an Omniscript RT kit (Qiagen, Inc., Valencia, CA, USA),
according to the manufacturer's protocols. The extracted DNA was
subjected to bisulfite treatment, DNA was tested for purity and
concentration using Nanodrop™ 2000 (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). 500 ng DNA was placed into a polymerase chain
reaction (PCR) tube, followed by denaturation at 95°C for 5 min,
and refolding at 60°C for three rounds (for 25, 85 and then 175
min) of reciprocation on a PCR instrument using an EpiTect
Bisulfite kit (Qiagen GmbH, Hilden, Germany). The reaction solution
was transferred to a 1.5 ml Eppendorf tube and was then treated
with Buffer BL, Buffer BW and Buffer BD (part of the EpiTect
Bisulfite kit), respectively. Subsequently, the Eppendorf tube was
centrifuged at 95°C for 1 min at a speed of 14,100 × g in the
EpiTect spin columns (Qiagen GmbH), and finally the sulfite-treated
product was eluted with 20 µl Buffer EB, according to the
manufacturer's protocols. Polymerase chain reaction (PCR) was used
to amplify the target fragments. The primer sequences used were as
follows: PCR primer forward, 5′-TATTTTGGGTTTGGGGTCGC-3′ and
reverse, 5′-CCCGAAAACCGAAAACCG-3′; sequencing primer,
5′-TTTTTGTTTTAGAGAGGTTAGTAAT-3′. The primers were all synthesized
by Sangon Biotech Co., Ltd. (Shanghai, China). PCR was performed as
follows: i) Initial denaturation at 95°C for 5 min; ii)
denaturation at 94°C for 30 sec, annealing at 64°C for 30 sec and
elongation at 72°C for 40 sec, for a total of 40 cycles; and iii)
final elongation at 72°C for 10 min. The products were separated
using a 2% agarose gel.
Quantitative bisulfite pyrosequencing
analysis
Quantitative detection of methylation was performed
for the suppressor genes in the cervical tissues. According to the
methylation sequencing kit (Qiagen GmbH, Hilden, Germany), a
reaction solution was prepared, containing 0.1 mol/l Tris Ac buffer
(pH 7.7), 2 mmol/l EDTA, 10 mmol/l Mg(Ac) 2, 0.1% bovine serum
albumin, 1 mmol/l dithiothreitol, 3 µmol/l 5′-phosphorylated
adenosine sulfate, 0.4 µg/l polyvidone, 0.4 mmol/l D luciferin,
2×10-4 U/l ATP sulfurylase, 2×10−3 U/l dual phosphatase
ATP and 18×10−3 U/l Klenow DNA polymerase (without
exonuclease activity and containing 14.6 mg/l luciferase; New
England Biolabs, Ipswich, MA, USA). Next, the methylation of the
samples was measured and the average value was calculated based on
the degree of methylation of the nine loci (according to the
criteria (35) of methylation
grouping that represents the degree of methylation for each
sample).
Statistical analysis
One-way analysis of variance with Tukey's test used
for post hoc analysis was used to evaluate differences in the
percentage of methylation reference (PMR) among the groups.
Continuous data are presented as the mean ± standard deviation.
Categorical data were analyzed using the χ2 test.
Receiver operating characteristic (ROC) curves were generated to
confirm the accuracy of diagnosis for each gene, and the
sensitivity and specificity were calculated. P<0.05 was
considered to indicate a statistically significant difference.
Statistical analysis was performed using SPSS 20.0 (IBM Corp.,
Armonk, NY, USA).
Results
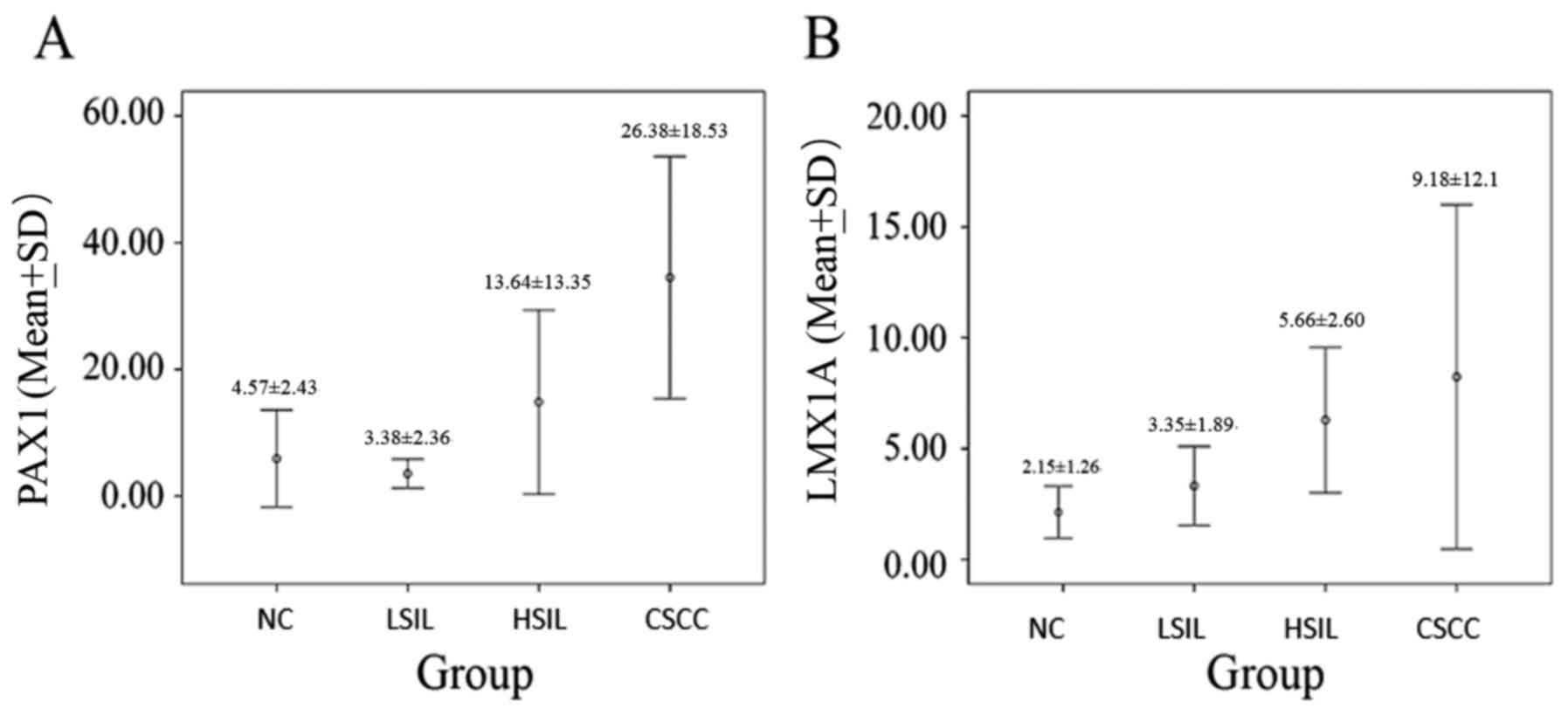
The patient distribution was n=28 for the NC group,
n=32 for the LSIL, n=34 for the HSIL and n=27 for the CSCC group
(Table I). The mean patient age
increased with disease severity (P<0.006). The proportion of
HPV-negative samples was significantly lower in the NC group
compared with in the CIN and CC groups (P<0.05). The proportions
of positive PAX1 and LMX1A methylated genes were higher in cervical
tissues and exfoliated cells in the HSIL and CSCC groups compared
with those in the LSIL and NC groups, but no significant difference
was observed between the NC and LSIL groups (Table I; Fig.
1).
 | Table I.Characteristics of participants and
gene methylation. |
Table I.
Characteristics of participants and
gene methylation.
| Variable | NC (n=28) | LSIL (n=32) | HSIL (n=34) | CSCC (n=27) | P-value |
|---|
| Age, years | 36.2±7.7 | 28.3±7.6 | 39.7±10.5 | 44.4±8.8 | 0.006 |
| HPV-negative | 18 | 6 | 3 | 2 | <0.001 |
| High-risk HPV | 10 | 26 | 31 | 25 |
| HPV DNA | 89.85±95.77 | 480.23±702.79 | 630.28±623.75 |
1650.80±4595.88 | 0.23 |
| PAX1 in tissue | 4.92±4.45 | 5.55±5.05 | 10.21±14.39 | 41.97±23.02 | <0.001 |
| PAX1 in exfoliated
cell | 4.57±2.43 | 3.38±2.36 | 13.64±13.35 | 26.38±18.53 | <0.001 |
| LMX1A in
tissue | 4.53±3.76 | 5.05±3.06 | 4.70±5.12 | 14.36±18.31 | <0.001 |
| LMX1A in exfoliated
cell | 2.15±1.26 | 3.35±1.89 | 5.66±2.60 | 9.18±12.1 | <0.001 |
| TCT, n |
|
|
|
| <0.001 |
| Normal (NC) | 20 | 7 | 4 | 5 |
|
| Low (CIN1) | 1 | 6 | 2 | 1 |
|
| High (CIN2-3) | 1 | 1 | 17 | 18 |
|
| ASC-US | 6 | 18 | 11 | 1 |
|
| CSCC | 0 | 0 | 0 | 2 |
|
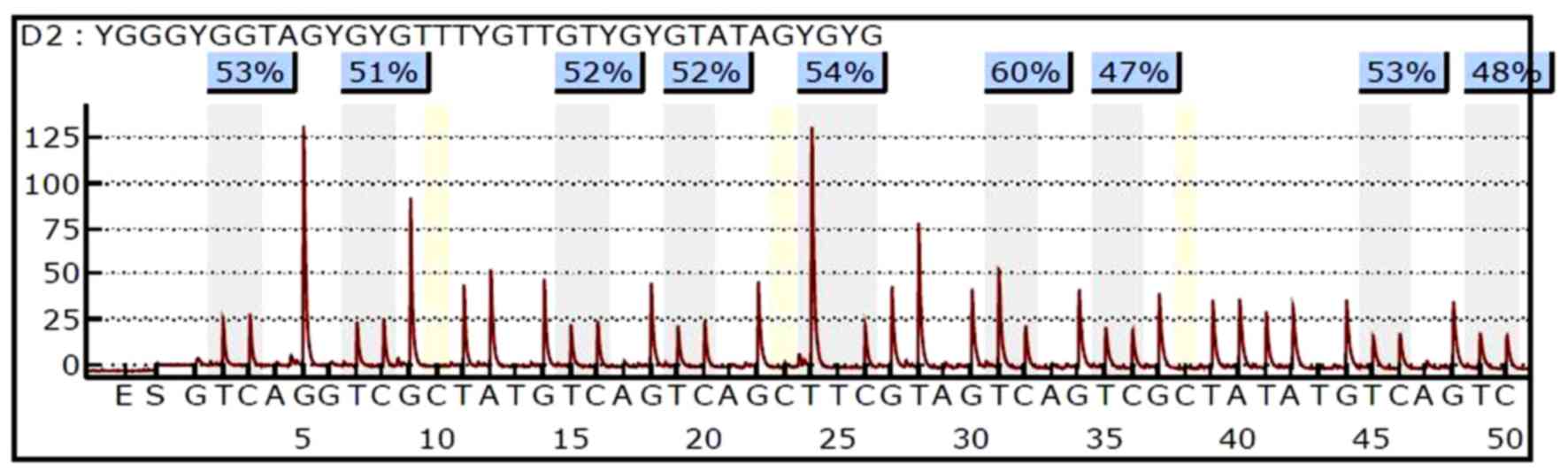
The methylation levels of PAX1 and LMX1A were
quantitatively detected by pyrosequencing. An example of the PAX1
pyrosequencing results of a single specimen was demonstrated in
Fig. 2.
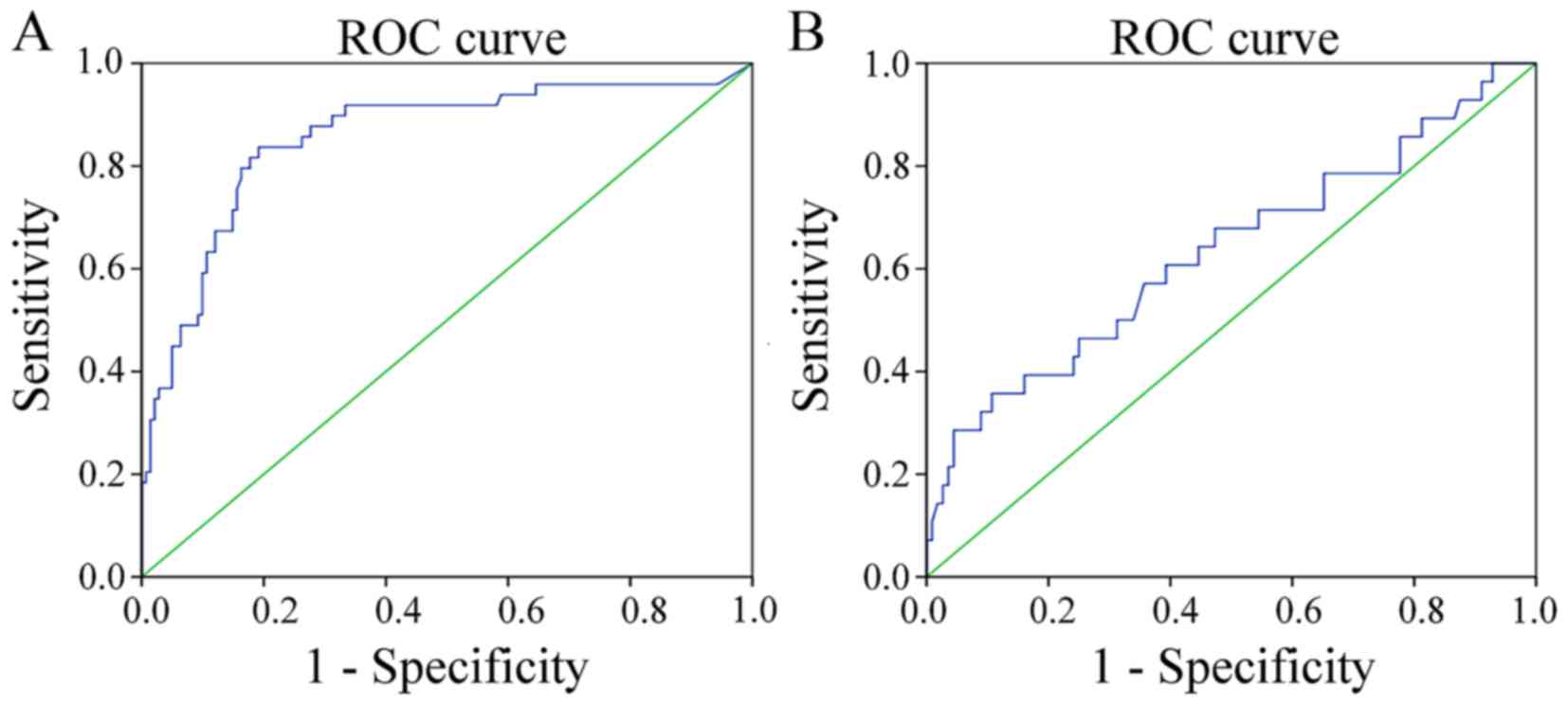
To evaluate the diagnostic potential of the two
genes, respective ROC curves were produced (Figs. 3 and 4).
Methylated PAX1 demonstrated a greater ability to detect cancer
compared with LMX1A (the sensitivities at the cut-off points were
0.837 and 0.357, respectively; P<0.001), although the
specificity values for the two genes were similar and high (0.809
and 0.893, respectively; Table II).
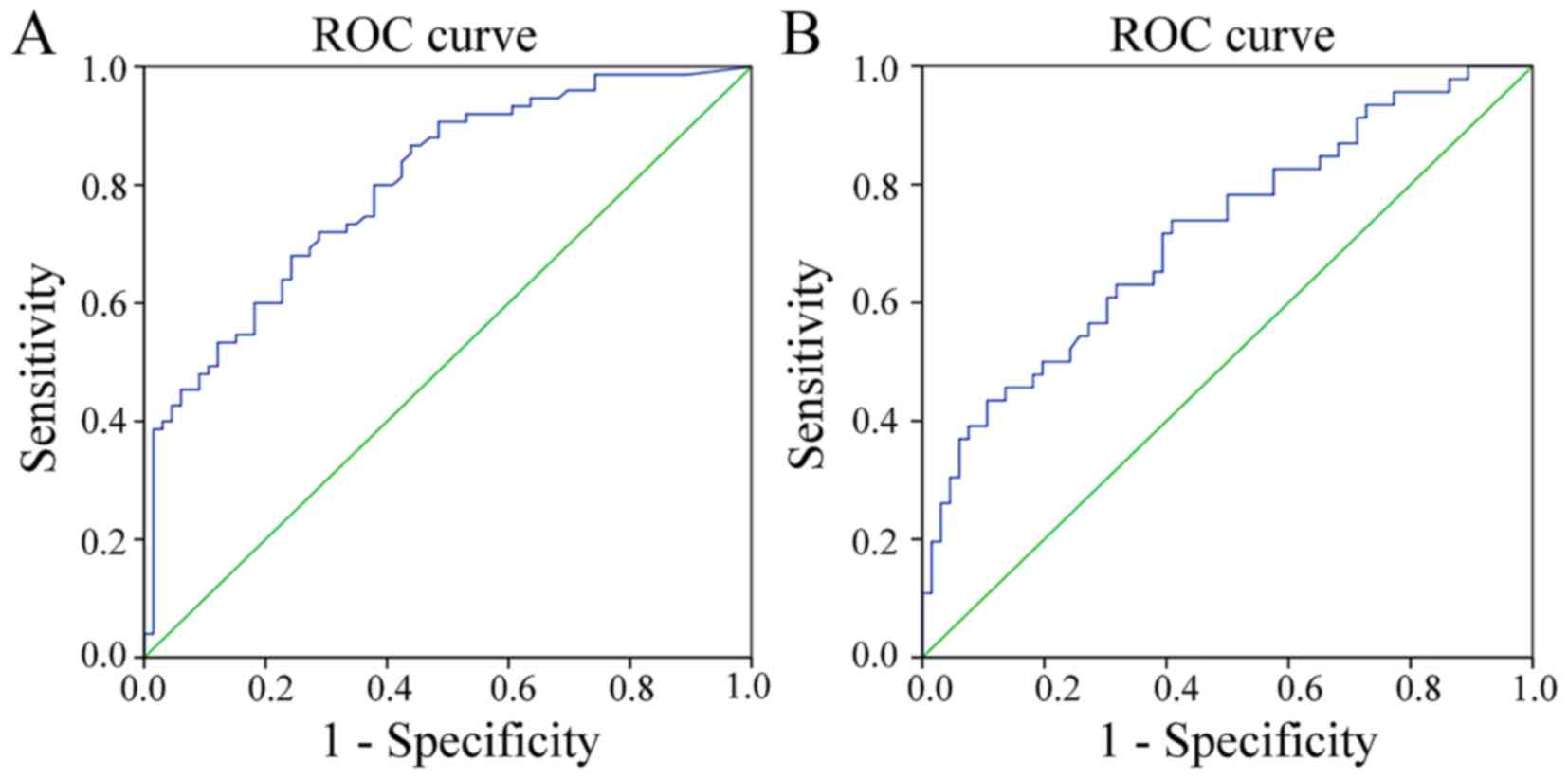
In addition, when comparing the ability to distinguish HSIL lesions
from normal tissues and LSIL, the sensitivities at the cut-off
values were similar, but not high (0.680 and 0.739, respectively),
while the specificities of the two genes differed significantly
(0.758 vs. 0.591; P<0.001; Table
III).
 | Table II.Sensitivity and specificity of PAX1
and LMX1A for distinguishing CSCC from HSIL, LSIL and NC (CSCC vs.
HSIL+LSIL+NC). |
Table II.
Sensitivity and specificity of PAX1
and LMX1A for distinguishing CSCC from HSIL, LSIL and NC (CSCC vs.
HSIL+LSIL+NC).
| Gene | Cut-off | AUC | Sensitivity, % | Specificity, % | P-value | OR (95% CI) |
|---|
| PAX1 | 11.78 | 0.790 | 0.837 | 0.809 | <0.001 | 0.788–0.923 |
| LMX1A | 7.185 | 0.633 | 0.357 | 0.893 | 0.029 | 0.508–0.758 |
 | Table III.Sensitivity and specificity of PAX1
and LMX1A for distinguishing HSIL from LSIL+NC (HSIL vs.
LSIL+NC). |
Table III.
Sensitivity and specificity of PAX1
and LMX1A for distinguishing HSIL from LSIL+NC (HSIL vs.
LSIL+NC).
| Gene | Cut-off | AUC | Sensitivity, % | Specificity, % | P-value | OR (95% CI) |
|---|
| PAX1 | 5.405 | 0.799 | 0.680 | 0.758 | <0.001 | 0.727–0.871 |
| LMX1A | 4.730 | 0.716 | 0.739 | 0.591 | <0.001 | 0.619–0.813 |
Discussion
DNA methylation serves an important role in the
regulation of gene expression or silencing in normal mammalian
cells and has been proposed as a potential biomarker for the
detection of cervical cancer (10,23). DNA
methylation-induced alteration of CpG islands in the tumor
suppressor gene promoter regions are often observed in human
cancer. The results of the present study revealed that the PMR of
the two genes were significantly higher in the HSIL and CSCC groups
compared with those in the NC, and LSIL groups. ROC curve analysis
demonstrated that the sensitivity, specificity and accuracy for
detecting CSCC of 0.790, 0.837 and 0.809, respectively using LMX1A
and 0.633, 0.357 and 0.893, respectively using PAX1. Previous
studies focused on detecting cervical cancer were hindered by
inconsistent results of quantitative DNA methylation analysis, and
moderate sensitivities and specificities using the available genes
(10,23).
Genes of the paired box (PAX) family serve important
roles in embryonic development and organogenesis, and may be
expressed persistently in stem cells and mature cells (36,37).
Specific PAX proteins are able to maintain stem cell properties,
and are involved in the development and progression of solid tumors
and hematologic cancer (38). As a
downstream product of the gene, the PAX protein may be expressed in
tissue-specific stem cells (39).
Previous studies have suggested that the anti-apoptotic function of
PAX proteins may encourage tumor cells to continuously grow without
undergoing apoptosis (38,40). In breast cancer cells, PAX2 and
estrogen receptor complexes regulate the expression of human
epidermal growth factor 2, which determines the response of tumor
cells to tamoxifen (41). PAX3 and
PAX7, as well as forkhead box protein O1, participate in
rhabdomyosarcoma formation through chromosomal rearrangement
(42,43). In hepatocellular carcinoma, PAX5
inhibits tumor formation by mediating P53-associated signaling
pathways (44). However, to the best
of our knowledge, there is no current literature regarding the
mechanisms of the PAX1 protein in malignant tumor development.
PAX1, as a member of the PAX family, serves an
important regulatory role in the early development of an embryo,
and is involved in the formation of bone, thymus and parathyroid
glands (38,45,46).
Inactivation of PAX1 has been observed in patients with CC and is
considered to be associated with the methylation of the promoter
region (47,48). In a hospital-based study on CC
detection, Huang et al (49)
observed that the quantitative measurement of PAX1 hypermethylation
in cervical samples was highly sensitive and more specific compared
with the Hybrid Capture 2 HPV test (0 vs. 5.9% in normal tissue).
In the past, the study of PAX1 gene methylation in the cervix was
found to be used for the differential diagnosis of invasive
carcinoma (50). In the present
study, the pyrosequencing quantitative methylation method confirmed
the methylation analysis levels of PAX1, which identified CSCC or
HSIL to a certain degree, as the AUCs were 0.790 (95% CI,
0.788–0.923) and 0.799 (95% CI, 0.727–0.871), respectively. When
PAX1 methylation was detected for differentiating CSCC, the
sensitivity and specificity were 0.837 and 0.809, respectively.
However, these indices decreased to 0.680 and 0.758 when HSIL was
screened for. These data revealed that the detection of PAX1
methylation has clinical diagnostic value in differentiating
invasive CC, but may not be sufficient alone in screening for HSIL.
A number of studies have indicated the potential value of PAX1 for
the screening and detection of CC (49,51,52), in
line with the findings of the present study, but the association
between PAX1 and tumors requires further investigation. The present
study used methylation-specific PCR to demonstrate that the PAX1
gene is abnormally methylated in cervical cancer specimens, with
methylation rates as high as 87.5%, which is significantly
different to those in normal cervical tissues and cervical
precancerous lesions (49).
Furthermore, the diagnostic sensitivity was twice that of the
HPV-HC2 assay (53).
LMX1A is an important homeobox transcription factor
in the process of cell development; it binds to AT-rich sequences
in the insulin promoter and stimulates the transcription of insulin
(54). In a previous study, LMX1A
methylation testing demonstrated great potential for cervical
lesion screening with a sensitivity, specificity and accuracy of
0.77, 0.88 and 0.90, respectively (30). In the present study, the
pyrosequencing method was used to confirm the methylation of the
LMX1A gene in cervical epithelial malignant transformation, but it
was hardly methylated in LSIL. Therefore, LMX1A methylation in
cervical tissue detection may provide valuable information
regarding the differentiation of invasive cancer, HSIL and LSIL.
ROC analysis revealed a sensitivity and specificity of 0.357 and
0.893, respectively, for CSCC; while the specificity for HSIL was
0.591. These unsatisfactory data indicated that it may be necessary
to combine other detection methods to improve accuracy. LMX1A
methylation is dysregulated in gastric, bladder, breast, ovarian
and pancreatic cancer (55–58). A study on various types of cancer may
provide useful insights into the involvement of LMX1A methylation
in tumorigenesis.
The present study has certain limitations. In
addition to the small sample size, there was no combined analysis
of the two genes or combined analysis of either gene using another
test, as the preliminary results suggested that combined
examination of multiple indices may be a feasible approach to
improving the diagnostic accuracy of differentiating cervical
lesions. In addition, the association between the two gene
methylation statuses and disease prognosis was not analyzed due to
problems at follow-up. Further multicenter studies, with larger
sample sizes and strictly designed diagnostic criteria are required
to obtain definitive conclusions.
In conclusion, quantitative detection of PAX1
methylation exhibited good diagnostic value in differentiating HSIL
from CSCC in cervical tissues, while the efficiency of LMX1A
methylation as a diagnostic tool requires further
investigation.
Acknowledgements
The present study was supported by the National Wu
Jieping Foundation for Clinical Scientific Research (grant no.
320.6750.13152) and the Project of Minhang Central Hospital (grant
no. 2016MHJC06).
References
|
1
|
Divine LM and Huh WK: Tertiary prevention
of cervical cancer. Clin Obstet Gynecol. 57:316–324. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vizcaino AP, Moreno V, Bosch FX, Muñoz N,
Barros-Dios XM, Borras J and Parkin DM: International trends in
incidence of cervical cancer: II. Squamous-cell carcinoma. Int J
Cancer. 86:429–435. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vizcaino AP, Moreno V, Bosch FX, Muñoz N,
Barros-Dios XM and Parkin DM: International trends in the incidence
of cervical cancer: I. Adenocarcinoma and adenosquamous cell
carcinomas. Int J Cancer. 75:536–545. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li J, Kang LN and Qiao YL: Review of the
cervical cancer disease burden in mainland China. Asian Pac J
Cancer Prev. 12:1149–1153. 2011.PubMed/NCBI
|
|
5
|
Burd EM: Human papillomavirus and cervical
cancer. Clin Microbiol Rev. 16:1–17. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kumar V, Abbas AK, Fausto N and Mitchell
RN: Robbins Basic Pathology. Saunders Elsevier; Dublin: 2007
|
|
7
|
Waxman AG, Chelmow D, Darragh TM, Lawson H
and Moscicki AB: Revised terminology for cervical histopathology
and its implications for management of high-grade squamous
intraepithelial lesions of the cervix. Obstet Gynecol.
120:1465–1471. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Agorastos T, Miliaras D, Lambropoulos AF,
Chrisafi S, Kotsis A, Manthos A and Bontis J: Detection and typing
of human papillomavirus DNA in uterine cervices with coexistent
grade I and grade III intraepithelial neoplasia: Biologic
progression or independent lesions? Eur J Obstet Gynecol Reprod
Biol. 121:99–103. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Leda G, Akila NV, Carlos AP, William PT
and Sharmila M: Cervical Cancer. http://www.cancernetwork.com/cancer-management/cervicalNov
01–2015
|
|
10
|
Jin B, Li Y and Robertson KD: DNA
methylation: Superior or subordinate in the epigenetic hierarchy?
Genes Cancer. 2:607–617. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee JY and Soong SJ: Cancer mortality in
the South, 1950 to 1980. South Med J. 83:185–190. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Joshi S, Kulkarni V, Darak T, Mahajan U,
Srivastava Y, Gupta S, Krishnan S, Mandolkar M and Bharti AC:
Cervical cancer screening and treatment of cervical intraepithelial
neoplasia in female sex workers using ‘screen and treat’ approach.
Int J Womens Health. 7:477–483. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pak SC, Martens M, Bekkers R, Crandon AJ,
Land R, Nicklin JL, Perrin LC and Obermair A: Pap smear screening
history of women with squamous cell carcinoma and adenocarcinoma of
the cervix. Aust N Z J Obstet Gynaecol. 47:504–507. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Geldenhuys L and Murray ML: Sensitivity
and specificity of the Pap smear for glandular lesions of the
cervix and endometrium. Acta Cytol. 51:47–50. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nanda K, McCrory DC, Myers ER, Bastian LA,
Hasselblad V, Hickey JD and Matchar DB: Accuracy of the
Papanicolaou test in screening for and follow-up of cervical
cytologic abnormalities: A systematic review. Ann Intern Med.
132:810–819. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lazcano-Ponce E, Alonso P, Ruiz-Moreno JA
and Hernández-Avila M: Recommendations for cervical cancer
screening programs in developing countries. The need for equity and
technological development. Salud Publica Mex. 45 Suppl 3:S449–S462.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nuovo J, Melnikow J and Howell LP: New
tests for cervical cancer screening. Am Fam Physician. 64:780–786.
2001.PubMed/NCBI
|
|
18
|
Flores-Miramontes MG, Torres-Reyes LA,
Alvarado-Ruíz L, Romero-Martínez SA, Ramírez-Rodríguez V,
Balderas-Peña LM, Vallejo-Ruíz V, Piña-Sánchez P, Cortés-Gutiérrez
EI, Jave-Suárez LF and Aguilar-Lemarroy A: Human papillomavirus
genotyping by linear array and next-generation Sequencing in
cervical samples from Western Mexico. Virol J. 12:1612015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Othman N and Othman NH: Detection of human
papillomavirus DNA in routine cervical scraping samples: Use for a
national cervical cancer screening program in a developing nation.
Asian Pac J Cancer Prev. 15:2245–2249. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Natter C, Polterauer S, Rahhal-Schupp J,
Cacsire Castillo-Tong D, Pils S, Speiser P, Zeillinger R, Heinze G
and Grimm C: Association of TAP gene polymorphisms and risk of
cervical intraepithelial neoplasia. Dis Markers. 35:79–84. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cuzick J, Clavel C, Petry KU, Meijer CJ,
Hoyer H, Ratnam S, Szarewski A, Birembaut P, Kulasingam S, Sasieni
P and Iftner T: Overview of the European and North American studies
on HPV testing in primary cervical cancer screening. Int J Cancer.
119:1095–1101. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Whitlock EP, Vesco KK, Eder M, Lin JS,
Senger CA and Burda BU: Liquid-based cytology and human
papillomavirus testing to screen for cervical cancer: A systematic
review for the U.S. Preventive Services Task Force. Ann Intern Med.
155:687–697, W214-W215. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Loscalzo J and Handy DE: Epigenetic
modifications: Basic mechanisms and role in cardiovascular disease
(2013 Grover Conference series). Pulm Circ. 4:169–174. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sakamoto A, Akiyama Y, Shimada S, Zhu WG,
Yuasa Y and Tanaka S: DNA Methylation in the Exon 1 region and
complex regulation of Twist1 expression in gastric cancer cells.
PLoS One. 10:e01456302015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fan H, Zhang H, Pascuzzi PE and Andrisani
O: Hepatitis B virus X protein induces EpCAM expression via active
DNA demethylation directed by RelA in complex with EZH2 and TET2.
Oncogene. 35:715–726. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schmid CA, Robinson MD, Scheifinger NA,
Müller S, Cogliatti S, Tzankov A and Müller A: DUSP4 deficiency
caused by promoter hypermethylation drives JNK signaling and tumor
cell survival in diffuse large B cell lymphoma. J Exp Med.
212:775–792. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sood S and Srinivasan R: Alterations in
gene promoter methylation and transcript expression induced by
cisplatin in comparison to 5-Azacytidine in HeLa and SiHa cervical
cancer cell lines. Mol Cell Biochem. 404:181–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kulis M and Esteller M: DNA methylation
and cancer. Adv Genet. 70:27–56. 2010.PubMed/NCBI
|
|
29
|
Chao TK, Ke FY, Liao YP, Wang HC, Yu CP
and Lai HC: Triage of cervical cytological diagnoses of atypical
squamous cells by DNA methylation of paired boxed gene 1 (PAX1).
Diagn Cytopathol. 41:41–46. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lai HC, Lin YW, Huang RL, Chung MT, Wang
HC, Liao YP, Su PH, Liu YL and Yu MH: Quantitative DNA methylation
analysis detects cervical intraepithelial neoplasms type 3 and
worse. Cancer. 116:4266–4274. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lai HC, Lin YW, Huang TH, Yan P, Huang RL,
Wang HC, Liu J, Chan MW, Chu TY, Sun CA, et al: Identification of
novel DNA methylation markers in cervical cancer. Int J Cancer.
123:161–167. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hammou JC, Bertino B, Blancheri A, Kon Man
P and Patoz L: Pap test: Liquid-based-thin-layer. A new method:
Results. Gynecol Obstet Fertil. 31:833–840. 2003.(In French).
|
|
33
|
WHO, . An introduction to cervical
intraepithelial neoplasia (CIN). http://screening.iarc.fr/colpochap.php?chap=2Aug
15–2015
|
|
34
|
Elias JM, Margiotta M and Gaborc D:
Sensitivity and detection efficiency of the peroxidase
antiperoxidase (PAP), avidin-biotin peroxidase complex (ABC), and
peroxidase-labeled avidin-biotin (LAB) methods. Am J Clin Pathol.
92:62–67. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu J, Xu L, Yang BH, Wang LF, Lin X and Tu
H: Assessing methylation status of PAX1 in cervical scrapings, as a
novel diagnostic and predictive biomarker, was closely related to
screen cervical cancer. Int J Clin Exp Pathol. 8:1674–1681.
2015.PubMed/NCBI
|
|
36
|
Wehr R and Gruss P: Pax and vertebrate
development. Int J Dev Biol. 40:369–377. 1996.PubMed/NCBI
|
|
37
|
Feiner N, Meyer A and Kuraku S: Evolution
of the vertebrate Pax4/6 class of genes with focus on its novel
member, the Pax10 gene. Genome Biol Evol. 6:1635–1651. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lang D, Powell SK, Plummer RS, Young KP
and Ruggeri BA: PAX genes: Roles in development, pathophysiology,
and cancer. Biochem Pharmacol. 73:1–14. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lang D, Lu MM, Huang L, Engleka KA, Zhang
M, Chu EY, Lipner S, Skoultchi A, Millar SE and Epstein JA: Pax3
functions at a nodal point in melanocyte stem cell differentiation.
Nature. 433:884–887. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sharma R, Sanchez-Ferras O and Bouchard M:
Pax genes in renal development, disease and regeneration. Semin
Cell Dev Biol. 44:97–106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hurtado A, Holmes KA, Geistlinger TR,
Hutcheson IR, Nicholson RI, Brown M, Jiang J, Howat WJ, Ali S and
Carroll JS: Regulation of ERBB2 by oestrogen receptor-PAX2
determines response to tamoxifen. Nature. 456:663–666. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Galili N, Davis RJ, Fredericks WJ,
Mukhopadhyay S, Rauscher FJ III, Emanuel BS, Rovera G and Barr FG:
Fusion of a fork head domain gene to PAX3 in the solid tumour
alveolar rhabdomyosarcoma. Nat Genet. 5:230–235. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bennicelli JL, Advani S, Schäfer BW and
Barr FG: PAX3 and PAX7 exhibit conserved cis-acting transcription
repression domains and utilize a common gain of function mechanism
in alveolar rhabdomyosarcoma. Oncogene. 18:4348–4356. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu W, Li X, Chu ES, Go MY, Xu L, Zhao G,
Li L, Dai N, Si J, Tao Q, et al: Paired box gene 5 is a novel tumor
suppressor in hepatocellular carcinoma through interaction with p53
signaling pathway. Hepatology. 53:843–853. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
McGaughran JM, Oates A, Donnai D, Read AP
and Tassabehji M: Mutations in PAX1 may be associated with
Klippel-Feil syndrome. Eur J Hum Genet. 11:468–474. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bannykh SI, Emery SC, Gerber JK, Jones KL,
Benirschke K and Masliah E: Aberrant Pax1 and Pax9 expression in
Jarcho-Levin syndrome: Report of two Caucasian siblings and
literature review. Am J Med Genet A. 120A:1–246. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schnittger S, Rao VV, Deutsch U, Gruss P,
Balling R and Hansmann I: Pax1, a member of the paired
box-containing class of developmental control genes, is mapped to
human chromosome 20p11.2 by in situ hybridization (ISH and FISH).
Genomics. 14:740–744. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang Y, Chen FQ, Sun YH, Zhou SY, Li TY
and Chen R: Effects of DNMT1 silencing on malignant phenotype and
methylated gene expression in cervical cancer cells. J Exp Clin
Cancer Res. 30:982011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang TH, Lai HC, Liu HW, Lin CJ, Wang KH,
Ding DC and Chu TY: Quantitative analysis of methylation status of
the PAX1 gene for detection of cervical cancer. Int J Gynecol
Cancer. 20:513–519. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen W, Yang HJ, Xu J and Zhu HP:
Quantitative analysis of LMX1A and PAX1 gene methylation in
cervical cancer and cervical intraepithelial neoplasia. China
Oncol. 25:19–24. 2015.
|
|
51
|
Kan YY, Liou YL, Wang HJ, Chen CY, Sung
LC, Chang CF and Liao CI: PAX1 methylation as a potential biomarker
for cervical cancer screening. Int J Gynecol Cancer. 24:928–934.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lai HC, Ou YC, Chen TC, Huang HJ, Cheng
YM, Chen CH, Chu TY, Hsu ST, Liu CB, Hung YC, et al: PAX1/SOX1 DNA
methylation and cervical neoplasia detection: A Taiwanese
Gynecologic Oncology Group (TGOG) study. Cancer Med. 3:1062–1074.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lim EH, Ng SL, Li JL, Chang AR, Ng J,
Ilancheran A, Low J, Quek SC and Tay EH: Cervical dysplasia:
Assessing methylation status (Methylight) of CCNA1, DAPK1, HS3ST2,
PAX1 and TFPI2 to improve diagnostic accuracy. Gynecol Oncol.
119:225–231. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
German MS, Wang J, Fernald AA, Espinosa R
III, Le Beau MM and Bell GI: Localization of the genes encoding two
transcription factors, LMX1 and CDX3, regulating insulin gene
expression to human chromosomes 1 and 13. Genomics. 24:403–404.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Dong W, Feng L, Xie Y, Zhang H and Wu Y:
Hypermethylation-mediated reduction of LMX1A expression in gastric
cancer. Cancer Sci. 102:361–366. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhao Y, Guo S, Sun J, Huang Z, Zhu T,
Zhang H, Gu J, He Y, Wang W, Ma K, et al: Methylcap-seq reveals
novel DNA methylation markers for the diagnosis and recurrence
prediction of bladder cancer in a Chinese population. PLoS One.
7:e351752012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Su HY, Lai HC, Lin YW, Chou YC, Liu CY and
Yu MH: An epigenetic marker panel for screening and prognostic
prediction of ovarian cancer. Int J Cancer. 124:387–393. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hagihara A, Miyamoto K, Furuta J, Hiraoka
N, Wakazono K, Seki S, Fukushima S, Tsao MS, Sugimura T and
Ushijima T: Identification of 27 5′ CpG islands aberrantly
methylated and 13 genes silenced in human pancreatic cancers.
Oncogene. 23:8705–8710. 2004. View Article : Google Scholar : PubMed/NCBI
|