Introduction
Dual specificity phosphatase 2 (DUSP2) is a member
of the dual-specificity phosphatases, which specifically
inactivates mitogen-activated protein kinase (MAPK) signaling by
the direct dephosphorylation of phosphothreonine and
phosphotyrosine residues (1,2). DUSP2 is expressed at high levels in
immune cells, particularly in patients with inflammatory arthritis,
including rheumatoid arthritis, and DUSP2-knockout mice present
with a significant reduction in inflammatory responses (3). DUSP2 is crucial in regulating the
tumor-relevant MAPK pathways. These pathways drive proliferation,
differentiation via the regulation of MAP kinases,
extracellular-signal regulated kinase (ERK)1/2, p38 and c-Jun
N-terminal kinase (JNK), and apoptosis (4). For example, DUSP2 is a transcription
target of p53 and E2F1 in signaling apoptosis and growth
suppression (5). It is reported that
DUSP2 is epigenetically silenced by promoter methylation in several
cancer cell lines, including skin and lung cancer (6). It is also reported that DUSP2 is
suppressed by hypoxia and the loss of function of DUSP2 not only
leads to the prolonged activation of ERK and tumorigenesis, but
also contributes to drug resistance (7,8). For
example, hypoxia induces lapatinib resistance in Erb-B2 receptor
tyrosine kinase 2-positive breast cancer cells via the regulation
of DUSP2 (9).
Several studies have reported that DUSP2 may act as
a tumor suppressor (5,10,11),
whereas others have reported that it may be involved in promoting
cancer progression (12). For
example, a high mRNA expression level of DUSP2 predicted
significantly poorer overall survival rates, compared with low
expression in serous ovarian carcinoma (12). Therefore, the role of DUSP2 appears to
vary with the type of malignancy. However, whether DUSP2 acts as a
tumor promoter or tumor suppressor is controversial.
The expression of DUSP2 is downregulated in several
types of human cancer, and the loss of DUSP2 promotes cancer
progression. However, the clinical value of DUSP2 in patients with
colorectal cancer (CRC) remains to be elucidated. The present
study, using a retrospective CRC patient cohort, aimed to examine
the biological function and clinical significance of DUSP2 in
CRC.
Materials and methods
Patients and tissue microarray
The samples examined in the present study consisted
of 96 patients with CRC, which were obtained from the tissue
specimen bank of Shanghai Biological Technology Co., Ltd.
(Shanghai, China). The patient surgery was performed between July
2006 and May 2007, and follow-up was continued until August 2014.
The clinicopathological classification was determined according to
the tumor-node-metastasis (TNM) classification of malignant tumors.
All the patients were pathologically diagnosed with colorectal
cancer without any pre-surgical treatment. There were 51 men and 45
women, with a median age of 55 years. Each study specimen of cancer
tissue was provided with adjacent-carcinoma tissue, which was sited
at a distance of 1.5 cm from the cancer tissue.
Immunochemical (IHC) staining
The tissue samples were processed as formalin-fixed,
paraffin-embedded tissue specimens according to standard
institutional procedures. Sections (4-µm thick) were cut from the
paraffin-embedded tissue specimens and used for the IHC. Sections
were heat-immobilized at 60°C for 30 min and deparaffinized in
xylene and rehydrated through a series of graded ethanol solutions
(100, 95, 90, 80 and 70%) at room temperature for 10 min. Antigen
retrieval was performed in a pressure cooker at 95°C for 2 min
using 0.01 M citrate buffer (pH 6.0). The samples were analyzed
under ×400 magnification using a BX51 light microscope (Olympus
Corporation, Tokyo, Japan). The IHC results were reviewed by two
expert pathologists. The specimens were then divided into four
grades, according to the degree of positivity as follows: Grade 0,
grade 1 (1–25% positive), grade 2 (26–50% positive) and grade 3
(51–100% positive). For the statistical analyses, grades 0 and 1
were defined as negative, and grades 2 and 3 were defined as
positive.
The cancer genome atlas (TCGA) data
analysis
The data files used to analyze DUSP2 expression were
initially downloaded from the TCGA (http://cancergenome.nih.gov/) data portal website, by
using the data matrix link to access RNASeq data and by using the
UNC (IlluminaHiSeq_RNAseqV2) data platform. DUSP2 mRNA expression
data from 461 CRC patients were obtained from the TCGA.
Immunofluorescence
For the immunofluorescence analyses, the cells were
cultured in 6-cm dishes. The cells were mounted with cytospin on
polylysine-coated glass slides and fixed with 4% paraformaldehyde
for 15 min, followed by the addition of 100% ice-cold acetone for
10 min at 4°C. To detect the protein expression of DUSP2,
immunofluorescence analysis was performed with DUSP2 antibody
(dilution, 1:100; cat. no. LS-B14289; LifeSpan BioScienes, Inc.),
incubated for 24 h at 4°C, followed by incubation with anti-IgG-PE
(Santa Cruz Biotechnology, Inc., Dallas, TX, USA) at 37°C for 30
min in the dark and mounting with DAPI mounting medium. The samples
were investigated by laser scanning confocal microscopy (OLS4100;
Olympus Corporation, Tokyo, Japan).
Cell lines
The colon cancer cell line, SW48, was obtained from
the Shanghai Cell Bank Chinese Academy of Sciences (Shanghai,
China). SW48 cells were cultured in RPMI-1640 medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
fetal bovine serum (Hyclone; GE Healthcare Life Sciences, Logan,
UT, USA), 100 U/ml penicillin and 100 U/ml streptomycin
(Invitrogen; Thermo Fisher Scientific, Inc.), at 37°C, 5%
CO2 and 95% humidity. The human colorectal carcinoma
cell lines, HCT116 and HCT15, were purchased from the Shanghai
Institutes for Biological Sciences, Chinese Academy of Sciences
(Shanghai, China), and were cultured in RPMI-1640 medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (Hyclone; GE Healthcare Life Sciences), 100 U/ml of
penicillin and 100 U/ml of streptomycin (Invitrogen; Thermo Fisher
Scientific, Inc.), at 37°C, 5% CO2 and 95% humidity.
SiRNA transfection
SW48 cells, cultured in RPMI-1640 medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (Hyclone; GE Healthcare Life Sciences), were transfected with
siRNAs targeting DUSP2 or with control siRNA using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Cells were transfected with siRNA at a
concentration of 50 nM. The transfection was performed at 37°C in a
humidified incubator with 5% CO2. The siRNAs were
designed and synthesized by Shanghai GenePharma Co., Ltd.
(Shanghai, China). The sequences of the siRNAs used in the present
study were as follows: siRNA-DUSP2 sense, 5-GCAUCACAGCCGUCCUCAATT-3
and anti-sense, 5-UUGAGGACGGCUGUGAUGCTT-3, and NC-siRNA sense,
5-GCAACACCGCUGUCUCCAATT-3 and anti-sense,
5-UUGGAGACAGCGGUGUUGCTT-3. When SW48 cells reached 80% confluence
6-well plates, transfection was conducted by mixing 5 µl siRNA with
5 µl Lipofectamine® 2000 in a final volume of 2,000 µl
medium. Cell morphology and transfection efficiency were evaluated
after 6 h transfection. Transfections were performed in triplicate
and each experiment was repeated ≥3 times.
Reverse transcription-quantitative
(RT-qPCR)
Total RNA was isolated using TRIzol reagent (Thermo
Fisher Scientific, Inc.), and reverse transcription of total RNA
was carried out using MMLV-RT, SPCL (Invitrogen; Thermo Fisher
Scientific, Inc.). Quantitative PCR was performed by using SYBR
Green I (Thermo Fisher Scientific, Inc.). The reaction mixture
consisted of 2.5 µl 10X PCR buffer, 2.0 µl of 2.5 mM each dNTP, 2.0
µl 25 mM MgCl2, 0.5 µl 10 pmol/µl each primer, 0.2 µl 5 U/µl Taq
polymerase, 0.5 µl cDNA template and distilled water for a total
volume of 25 µl. The primer sequences used were as follows, human
GAPDH, forward 5′-CCACCCATGGCAAATTCCATGGCA-3′ and reverse
5′-TCTAGACGGCAGGTCAGGTCCAC-3′; DUSP2, forward
5′-TTTGAGGGCCTTTTCCGCTACAAGAG-3′ and reverse
5′-GCCTCCGCTGTTCTTCACCCAGTC-3′ (6).
The thermocycling conditions were as follows: 94°C for 5 min; 35
cycles of 94°C for 30 sec, 56°C (GAPDH)/65°C (DUSP2) for 30 sec,
and 72°C for 60 sec, and a final extension at 72°C for 10 min.
Triplicate tests were performed for each sample, and all reactions
were repeated 3 times independently to ensure reproducibility of
results. The data were then viewed and analyzed using the
Rotor-Gene Real-Time Analysis Software (Corbett Rotor-Gene 6000;
Qiagen, Doncaster, Australia). For each sample, amplification plot
and corresponding dissociation curves were examined. To obtain
standardized quantitative results, external controls were
constructed consisting of cDNA plasmid standards (13).
hEGF treatment
HCT116 and HCT15 cells (3×105/well) were
seeded in 6-well plates. After 24 h, the cells were treated with
100 ng/ml hEGF for another 24 h. Then the protein was extracted and
subjected to western blot analysis.
Western blot analysis
The cells were harvested and centrifuged at 110 × g,
for 5 min at room temperature. The supernatant was removed, and the
cell pellet was washed twice with PBS (0.01 M; pH 7.2–7.3). Each
tube of cells was added into 100 µl radioimmunoprecipitation assay
lysis buffer with phenylmethane sulfonyl fluoride (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany), and protein concentrations were
using a bicinchoninic acid assay. The samples (20 µg) of the cell
lysate were subjected to 10% SDS-PAGE gel electrophoresis,
following which the resolved proteins were transferred onto
nitrocellulose membranes (GE Healthcare Life Sciences, Chalfont,
UK). The membranes were then blocked with 5% non-fat milk and 0.1%
Tween 20 in Tris-buffered saline, and probed with anti-DUSP2
antibody (dilution, 1:500; cat. no. LS-B14289; LifeSpan BioScienes,
Inc.), and a secondary antibody for 60 min at room temperature
[goat anti-rabbit monoclonal antibody (dilution; 1:8,000; cat. no.
SA00001-1; ProteinTech Group, Inc., Chicago, IL, USA)], following
which the blots were visualized using enhanced chemiluminescence
(GE Healthcare Life Sciences).
Evaluation of apoptosis
Apoptosis was detected via flow cytometric analysis
of Annexin V staining. The Annexin V-FITC/PI assay was performed as
previously reported (14). Briefly,
the adherent cells were harvested and suspended in the
Annexin-binding buffer (1х106 cells/ml). The cells were
then incubated with Annexin V-FITC and PI for 15 min at room
temperature in the dark, and immediately analyzed via
flow-cytometry. The data are presented as bi-parametric dot plots
showing Annexin V-FITC green fluorescence, vs. PI red
fluorescence.
Cell viability assay
The cells were seeded at a density of 3,000
cells/well in 96-well plates. When the cells had completely adhered
to the well, the culture medium was replaced with medium containing
10% FBS and a certain concentration of cetuximab (0, 1, 2.5, 5 or
10 µg/ml), and cultivated at 37°C, 5% CO2 for 48 h.
After 2 h, cell viability was measured using a Cell Counting Kit-8
(CCK-8), according to the manufacturer's protocol. Briefly, a
mixture of 10 µl CCK-8 (Dojindo Molecular Technologies, Inc.) and
190 µl of RPMI-1640 with 10% FBS was added to each well. An MRX II
microplate reader was used to measure the optical density at 450
nm. A background reading of the media was subtracted from each well
to standardize the results. Optical density (OD) was utilized as an
indicator of cell survival.
Statistical analysis
To investigate the associations between relapse-free
survival rate and various clinicopathological factors, survival
analysis was performed using the Kaplan-Meier method, and the
differences were evaluated using the log-rank test. A multivariate
survival analysis was performed using Cox's proportional-hazard
model. Hazard ratios and 95% confidence intervals were used to
measure associations. SPSS 13 software (SPSS, Inc., Chicago, IL,
USA) was used for all statistical analyses. P<0.05 was
considered to indicate a statistically significant difference.
Results
DUSP2 is differentially expressed
between left-sided colon carcinoma (LSCC) and right-sided colon
carcinoma (RSCC)
To determine the expression of DUSP2 in CRC, the
present study analyzed a tissue microarray containing primary CRC
and paired adjacent normal tissues using IHC analysis. Several
antibodies used for IHC are notoriously prone to false positive
signals, and antibodies, which are capable of resolving a single
positive band in western blots, can be found to stain non-specific
targets in cells and tissue sections by IHC. The solution to this
problem is to show that DUSP2 small interfering RNA attenuates or
eliminates the DUSP2 signal (15).
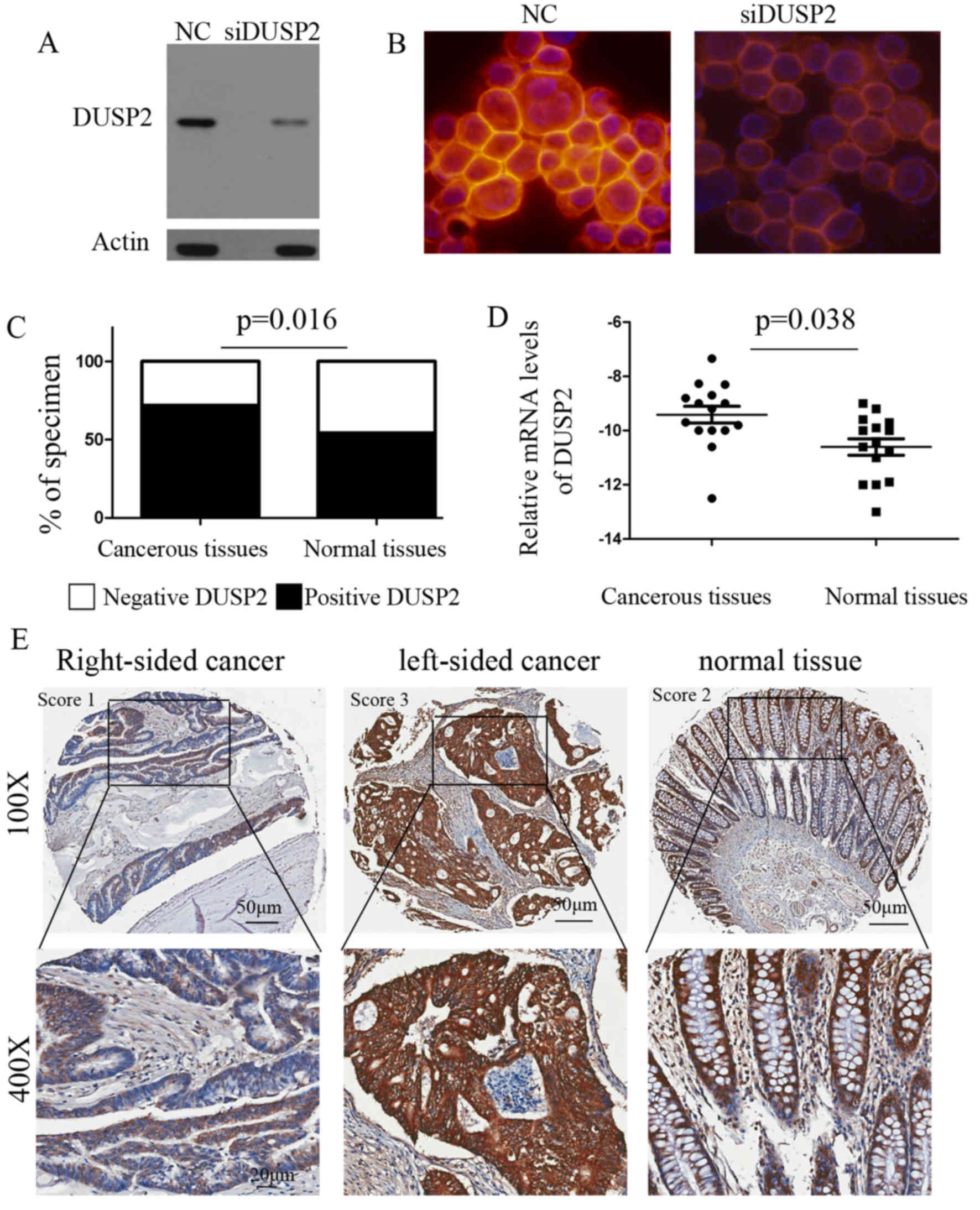
The silencing of DUSP2 was confirmed via western blot analysis in
the SW48 cell line (Fig. 1A). The
analysis was performed with DUSP2 antibody in SW48 control or
DUSP2-silenced cells and mounted with DAPI mounting medium. The
results indicated that the signals detected were those of DUSP2
(Fig. 1B). Overall, using a score to
semi-quantify immunoreactivity, it was found that the majority of
the CRC samples were graded as positive (69/96, 71.9%) and 34.27%
were graded as negative, whereas fewer normal tissues were graded
as positive (52/96, 54.2%; P=0.016; Fig.
1C). This was in contrast to previous findings that DUSP2 was
reduced in certain types of cancer (6). The expression of DUSP2 in human CRC
specimens was also analyzed using data of 461 CRC patients from The
Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov). Amongst these, matched
tumor and non-tumor specimens were available for 50 patients with
CRC. To further validate the results, the total expression of DUSP2
was determined in 15 paired normal and tumor tissues via RT-qPCR
analysis, which confirmed that DUSP2 was upregulated in tumor
tissues (Fig. 1D; P=0.038). The
correlations between the decreased expression of DUSP2 and the
clinicopathological features were also examined. The expression of
DUSP2 was not associated with age, gender, advanced TNM stage,
nodal metastasis or depth of tumor invasion (Table I). The decreased expression of DUSP2
was significantly associated with primary tumor site (P=0.008).
Compared with RSCC, the expression of DUSP2 was upregulated in
LSCC. Representative images of the protein expression of DUSP2 in
CRC and normal tissues are shown in Fig.
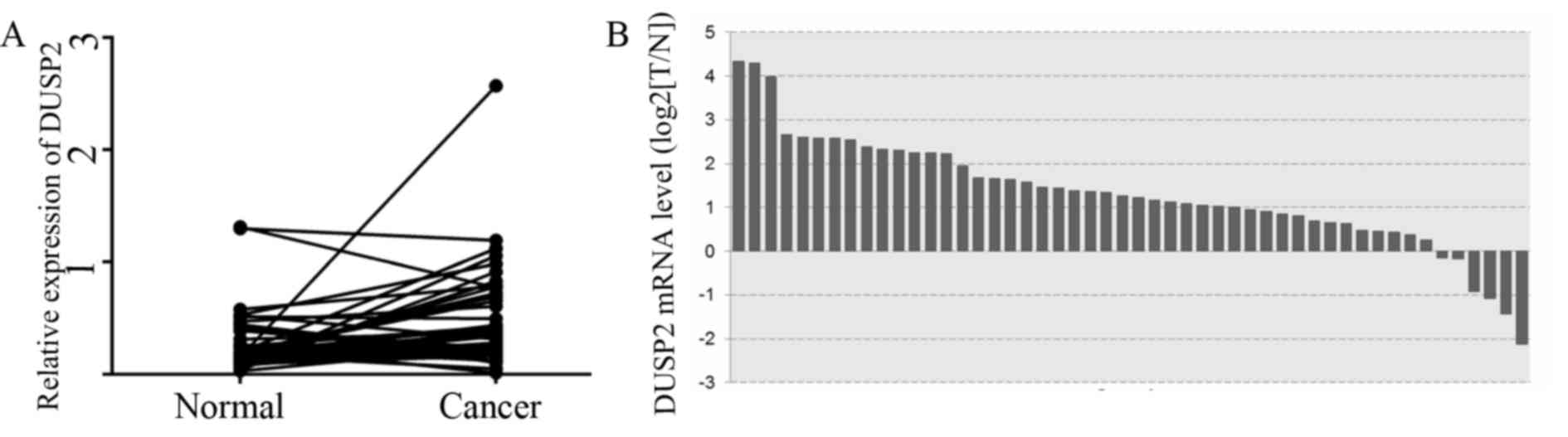
1E. Analysis of the relative gene expression of DUSP2 in all 50
patients with CRC from the TCGA gene expression datasets showed a
significant increase in the mRNA expression of DUSP2 in tumor
specimens, compared with that in non-tumor colorectal tissues
(Fig. 2A and B).
 | Table I.Association between the
clinicopathological features of patients with colorectal cancer and
the expression levels of DUSP2. |
Table I.
Association between the
clinicopathological features of patients with colorectal cancer and
the expression levels of DUSP2.
|
| Expression of
DUSP2 |
|
|---|
|
|
|
|
|---|
| Variable | Positive (n=69) | Negative (n=27) | P-value |
|---|
| Age (years) |
|
|
|
| ≤60 | 32 | 16 | 0.785 |
|
>60 | 37 | 11 |
|
| Gender |
|
|
|
| Male | 33 | 18 | 0.665 |
|
Female | 36 | 9 |
|
| Primary tumor
(T) |
|
|
|
|
T1/T2 | 31 | 12 | 0.572 |
|
T3/T4 | 38 | 15 |
|
| Nodal status (N) |
|
|
|
| N0 | 30 | 12 | 0.563 |
|
N1/N2/N3 | 39 | 15 |
|
| Stage |
|
|
|
| I–II | 37 | 15 | 0.671 |
|
III–IV | 32 | 12 |
|
| Primary tumor
site |
|
|
|
| RSCC | 26 | 20 | 0.008 |
|
LSCC | 43 | 7 |
|
Decreased expression of DUSP2 is
correlated with poor prognosis in patients with CRC
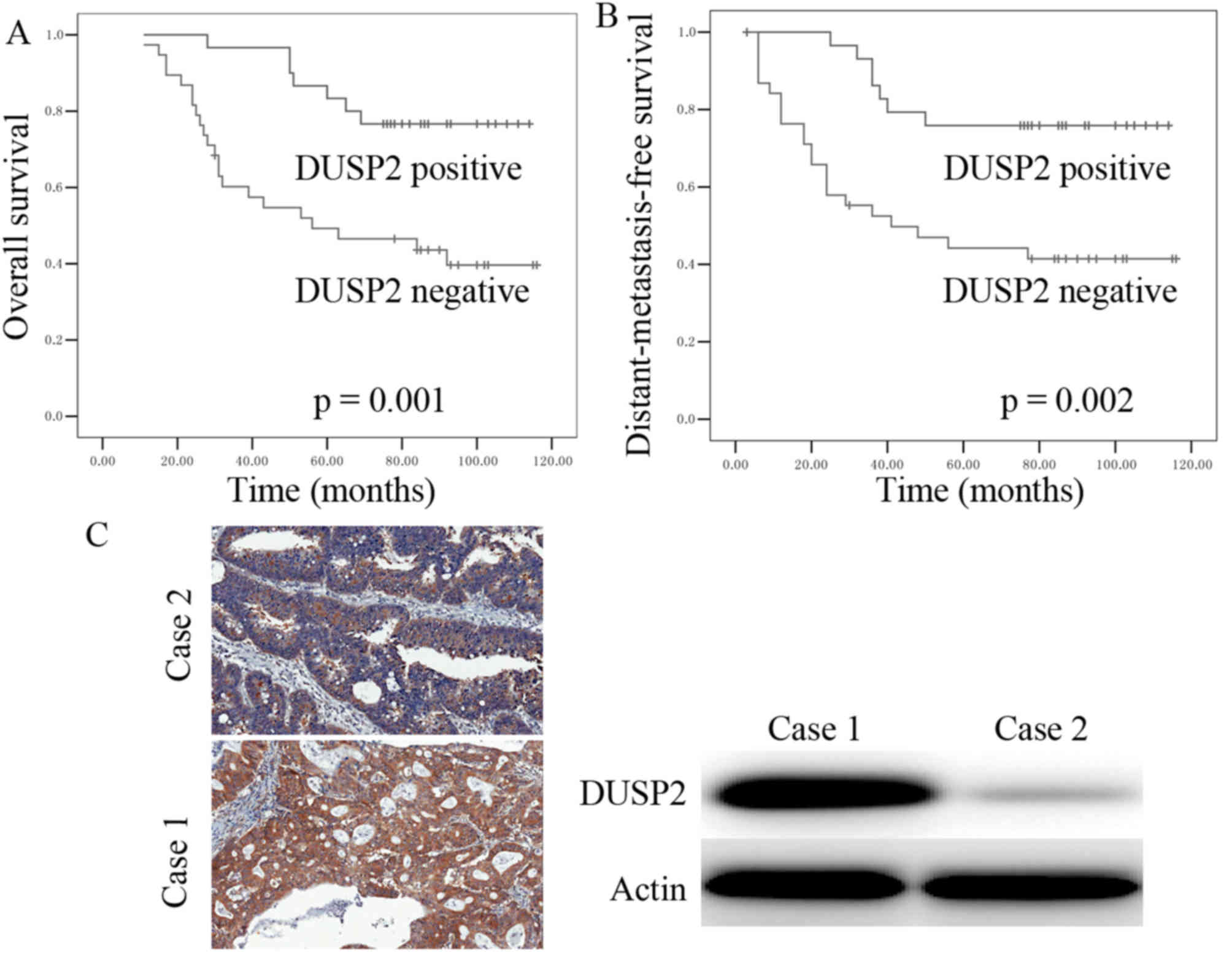
The log-rank test and Kaplan-Meier survival curve
were used to determine whether decreased expression of DUSP2 was
correlated with overall survival rates and distant-metastasis-free
survival (DMeFS). The results demonstrated that patients with CRC
and a decreased expression of DUSP2 had a poorer overall survival
rate, compared with patients with positive expression of DUSP2
(Fig. 3A). It was also found that a
decreased expression of DUSP2 was correlated with significantly
shorter DMeFS (Fig. 3B). To further
evaluate the prognostic value of the expression of DUSP2,
univariate Cox regression analysis was performed. The univariate
Cox regression analysis indicated that the decreased expression of
DUSP2 was an independent prognostic biomarker for CRC in patients
(HR 3.55, CI 1.092–9.896; P=0.002; Table
II). To further confirm that the signals detected were those of
DUSP2, the expression of DUSP2 was detected in tumor tissues using
western blot analysis. The results revealed that the signal of
DUSP2 in the IHC was consistent with the signal of DUSP2 obtained
from the western blot analysis (Fig.
3C).
 | Table II.Univariate analysis of the expression
of DUSP2 and overall survival in patients with colorectal
cancer. |
Table II.
Univariate analysis of the expression
of DUSP2 and overall survival in patients with colorectal
cancer.
|
| Univariate
analysis |
|
|---|
|
|
|
|
|---|
| Variable | Hazard ratio | 95% CI | P-value |
|---|
| Age (years) |
|
|
|
| ≥60,
vs. <60 | 1.251 | 0.602–2.602 | 0.548 |
| Gender |
|
|
|
| Male,
vs. female | 1.012 | 0.984–1.040 | 0.405 |
| Primary tumor
(T) |
|
|
|
| T1/T2,
vs. T3/T4 | 1.067 | 0.172–8.028 | 0.545 |
| Nodal status
(N) |
|
|
|
| N0, vs.
N1/N2/N3 | 2.071 | 0.473–6.404 | 0.126 |
| Stage |
|
|
|
| I–II,
vs. III–IV | 2.136 | 0.322–3.453 | 0.235 |
| DUSP2 |
|
|
|
|
Positive, vs. negative | 3.55 | 1.092–9.896 | 0.002 |
Upregulation of DUSP2 is a consequence
rather than a cause of tumor progression
DUSP2 specifically inactivates MAPK signaling by the
direct dephosphorylation of phosphothreonine and phosphotyrosine
residues of ERK and AKT. It is also reported that loss of function
of DUSP2 leads to prolonged ERK activation. The present study
hypothesized that the upregulation of DUSP2 may be a consequence
rather than a cause of tumor progression in CRC cells. To address
this question, the expression levels of DUSP2 and phosphorylated
(p)ERK were analyzed using IHC to determine whether the expression
of DUSP2 was positively correlated with pERK in CRC tissues. The
results showed no significant correlation between DUSP2 and pERK
(data not shown). It is well known that epidermal growth factor
(EGF) stimulation causes a significant increase in pERK, and this
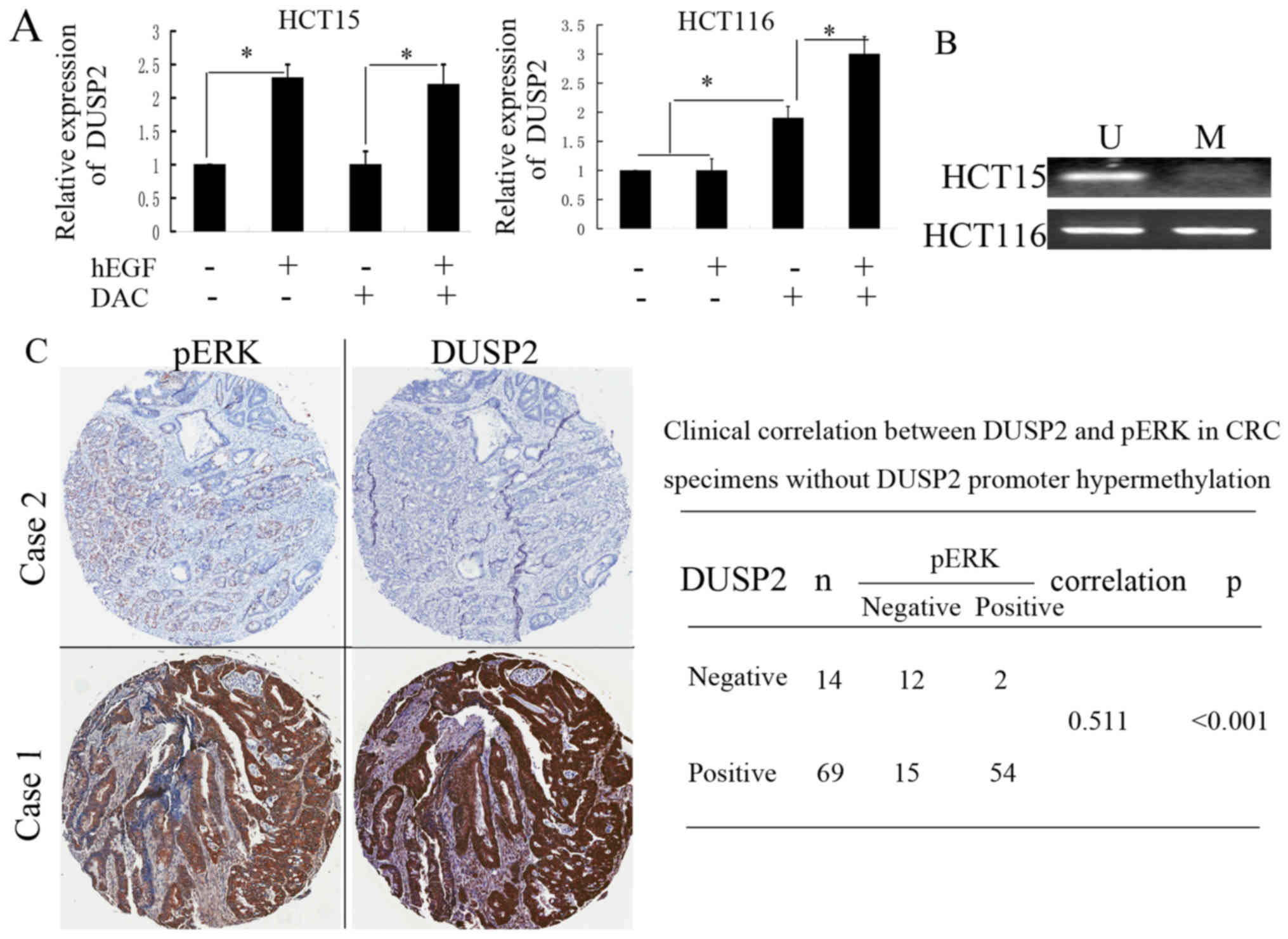
effect is observed even following EGF withdrawal (16). In the present study, when the HCT15
and HCT116 cells were treated with 100 ng/ml hEGF for 24 h, it was
found that the expression of DUSP2 was upregulated in the HCT15
cells. However, the treatment of HCT116 cells under the same
conditions had no effect on the expression of DUSP2 (Fig. 4A). It has been reported that DUSP2 is
epigenetically silenced in several cancer cells (6). The present study analyzed the promoter
methylation status of DUSP2 in HCT15 and HCT116 cells via
methylation-specific PCR (MSP). In accordance with the
above-mentioned result, the hypermethylation of DUSP2 was observed
in HCT116 cells, but not HCT15 cells (Fig. 4B). The treatment of HCT116 cells with
EGF in combination with DAC, a methyltransferase inhibitor,
markedly upregulated the expression of DUSP2 and exhibited a
synergetic effect. The DUSP2 methylation status in CRC tissues was
also detected by MSP. It was found that hypermethylation of DUSP2
was observed in 13 of 96 patients with CRC. The primary tumors of
these 13 patients originated in the RSCC. The expression of DUSP2
in the cancer tissues from these 13 patients was negative. There
was a significant correlation between promoter methylation and loss
of DUSP2 in the RSCC (P<0.001). When the expression levels of
DUSP2 and pERK were determined in CRC tissues without
hypermethylation of DUSP2, it was found that the expression of
DUSP2 was positively correlated with that of pERK in the CRC
tissues (Fig. 4C). These results
suggested that the hypermethylation of DUSP2 may inhibit the
upregulation of DUSP2 induced by the increase of pERK. Although it
is overexpressed in partial CRC tissues, the results suggested that
DUSP2 functions as a tumor suppressor.
Enforced expression of DUSP2
sensitizes CRC cells to cetuximab treatment
It is also reported that the loss of function of
DUSP2 leads not only to the prolonged activation of ERK and
tumorigenesis, but also contributes to drug resistance. Low
expression levels of DUSP5 and DUSP6 are involved in cetuximab
resistance in head and neck squamous cell carcinoma (17). The present study examined whether
DUSP2 is associated with cetuximab resistance in CRC. A plasmid
carrying cDNA of DUSP2 was transfected into HCT15 and HCT116 cells.
HCT15 and HCT116 cells harboring a KRAS mutation are primarily
resistant to cetuximab. After 48 h of cetuximab treatment in the
two cell lines, apoptosis was determined via flow cytometry using
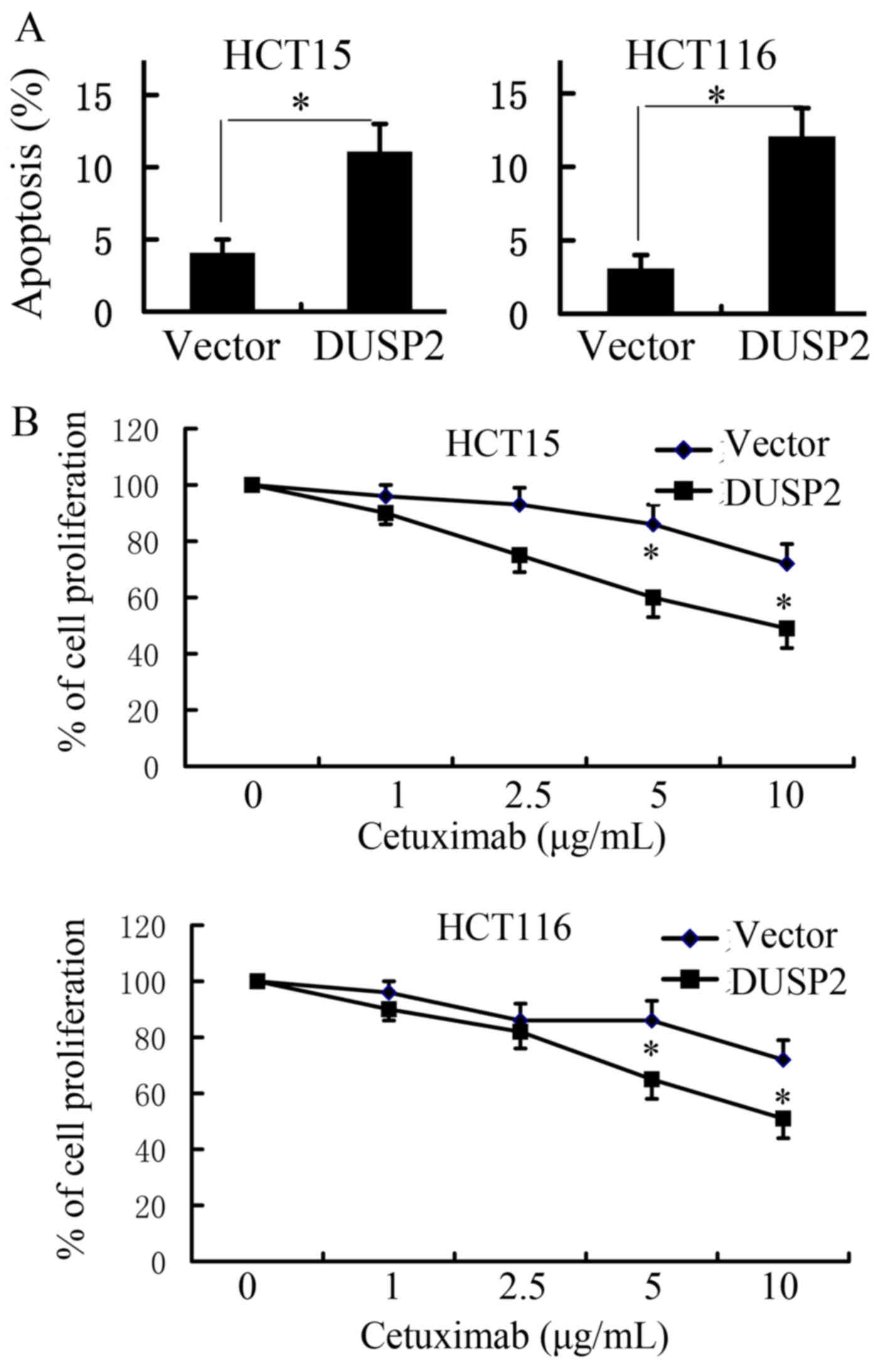
an Annexin V-FITC/PI staining kit. As shown in Fig. 5A, the overexpression of DUSP2
sensitized CRC cells to cetuximab, compared with the control
plasmid (P<0.05). To investigate the function of DUSP2 on the
sensitivity of CRC cell lines to cetuximab, the viability of HCT15
and HCT116 cells incubated in the presence of different cetuximab
concentrations was evaluated using a cell viability assay (CCK-8
assay). As shown in Fig. 5B, the
overexpression of DUSP2 enhanced the inhibitory effect of cetuximab
on HCT15 and HCT116 cells, compared with the control plasmid. The
present study also examined whether the inhibition of DUSP2 results
in resistance to cetuximab in CRC cells. This analysis was
performed in SW48 cells, which are sensitive to cetuximab. It was
found that the knockdown of DUSP2 significantly inhibited the
sensitivity of SW48 cells to cetuximab, compared with negative
control (data not shown).
Discussion
CRC is the most common type of malignancy with the
third highest incidence and mortality rates among all diagnosed
cases of cancer worldwide (18,19).
Cetuximab combined with chemotherapy has demonstrated therapeutic
efficacy in patients with metastatic CRC with all RAS wild-type
tumors (20). However, the efficacy
of cetuximab is limited by the development of resistance mechanisms
in cancer cells (21). Certain
patients with all RAS wild-type tumors are primarily resistant to
cetuximab (22). Increasing data has
shown that the location of the primary tumor can be prognostic and
predictive of responses to cetuximab in metastatic CRC, although
the exact reason remains to be elucidated (23). The results of previous clinical
studies have indicated that patients with left-sided RAS wild-type
metastatic CRC require preferential treatment with cetuximab
(24,25). The latest National Comprehensive
Cancer Network guideline for colon cancer recommends that cetuximab
combination therapy is only used for left-sided RAS wild-type
metastatic CRC (21). Significant
differences have been observed to exist between LSCC and RSCC, with
regard to epidemiological, biological and clinical data concerned
with carcinogenesis and survival (26). Zhu et al reported that 11
genes, including DUSP2, were found to be differentially expressed
in LSCC and RSCC by expression profiling with microarray analysis
(27). In addition, the loss of DUSP2
promotes angiogenesis and metastasis via the upregulation of
interleukin-8 in colon cancer (11).
In the present study, the expression of DUSP2 was
investigated using IHC in 96 patients with CRC. It was found that
the expression level of DUSP2 was significantly upregulated in CRC
tissue, compared with that in paired normal colon tissue. The IHC
analyses also demonstrated that the expression of DUSP2 in LSCC was
significantly higher, compared with that in RSCC. Low expression
levels of DUSP5 and DUSP6 are involved in cetuximab resistance in
head and neck squamous cell carcinoma, and decreased expression of
DUSP2 is associated with drug resistance in cells of several types
of cancer. It has also been reported that the loss of function of
DUSP2 leads to the prolonged activation of ERK (17). The EGFR-independent activation of the
RAS/RAF/MAPK kinase/MAPK pathway is one of the resistance
mechanisms to cetuximab (28). The
present study examined whether DUSP2 is associated with cetuximab
resistance in CRC. The results demonstrated that the overexpression
of DUSP2 increased the inhibitory effect of cetuximab in CRC.
It is reported that the expression of DUSP2 is
downregulated in several types of cancer in humans and that the
loss of DUSP2 promotes cancer progression (2). By contrast, the present study found that
the expression level of DUSP2 was significantly upregulated in CRC
tissues, compared with that in paired normal colon tissues. It was
hypothesized that the upregulation of DUSP2 may be a consequence
rather than a cause of tumor progression in CRC cells. To address
this question, the expression levels of DUSP2 and pERK were
analyzed using IHC to determine whether the expression of DUSP2 was
positively correlated with pERK in CRC tissues. It is known that
EGF stimulation causes a significant increase in pERK and that this
effect is observed even following EGF withdrawal. When the HCT15
and HCT116 cells were treated with hEGF to induce the expression of
pERK, it was found that the expression of DUSP2 was upregulated in
HCT15 cells. It was also found that DAC and hEGF synergistically
induced the expression of DUSP2, suggesting that the
hypermethylation of DUSP2 may inhibit the upregulation of DUSP2
induced by the increase in pERK in HCT116 cells. The
hypermethylation of DUSP2 was observed in 13/96 CRC tissues, and
the primary tumors of these 13 patients all originated on the right
side of the colon. When the expression levels of DUSP2 and pERK
were analyzed in CRC tissues without hypermethylation of DUSP2, it
was found that the expression of DUSP2 was positively correlated
with pERK in CRC tissues. It was hypothesized that the upregulation
of DUSP2 may function via negative feedback to balance the
activation of MAPKs, including ERK1/2, p38 MAPK and JNK, in CRC
cells. This process may be inhibited by the hypermethylation of
DUSP2 in RSCC.
In conclusion, the findings of the present study
revealed that DUSP2 was overexpressed in LSCC and epigenetically
silenced in RSCC. The overexpression of DUSP2 may be a consequence
rather than a cause of tumor progression in CRC cells. The
upregulation of DUSP2 may function via negative feedback to balance
the activation of MAPKs, including ERK1/2, p38 MAPK and JNK, in CRC
cells. Furthermore, the results demonstrated that the
overexpression of DUSP2 increased the inhibitory effect of
cetuximab in CRC, suggesting that DUSP2 may be a novel biomarker
and therapeutic target in CRC therapy.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81672424) and the
Youth Innovation Fund of the First Affiliated Hospital of Zhengzhou
University (grant no. 20150101).
References
|
1
|
Bermudez O, Pages G and Gimond C: The
dual-specificity MAP kinase phosphatases: Critical roles in
development and cancer. Am J Physiol Cell Physiol. 299:C189–C202.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wei W, Jiao Y, Postlethwaite A, Stuart JM,
Wang Y, Sun D and Gu W: Dual-specificity phosphatases 2: Surprising
positive effect at the molecular level and a potential biomarker of
diseases. Genes Immun. 14:1–6. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hamamura K, Nishimura A, Chen A, Takigawa
S, Sudo A and Yokota H: Salubrinal acts as a Dusp2 inhibitor and
suppresses inflammation in anti-collagen antibody-induced
arthritis. Cellular signalling. 27:828–835. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Perander M, Al-Mahdi R, Jensen TC, Nunn
JA, Kildalsen H, Johansen B, Gabrielsen M, Keyse SM and Seternes
OM: Regulation of atypical MAP kinases ERK3 and ERK4 by the
phosphatase DUSP2. Sci Rep. 7:434712017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yin Y, Liu YX, Jin YJ, Hall EJ and Barrett
JC: PAC1 phosphatase is a transcription target of p53 in signalling
apoptosis and growth suppression. Nature. 422:527–531. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Haag T, Richter AM, Schneider MB, Jimenez
AP and Dammann RH: The dual specificity phosphatase 2 gene is
hypermethylated in human cancer and regulated by epigenetic
mechanisms. BMC Cancer. 16:492016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lin SC, Chien CW, Lee JC, Yeh YC, Hsu KF,
Lai YY, Lin SC and Tsai SJ: Suppression of dual-specificity
phosphatase-2 by hypoxia increases chemoresistance and malignancy
in human cancer cells. J Clin Invest. 121:1905–1916. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee YH, Morrison BL and Bottaro DP:
Synergistic signaling of tumor cell invasiveness by hepatocyte
growth factor and hypoxia. J Biol Chem. 289:20448–20461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Karakashev SV and Reginato MJ:
Hypoxia/HIF1α induces lapatinib resistance in ERBB2-positive breast
cancer cells via regulation of DUSP2. Oncotarget. 6:1967–1980.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hou PC, Li YH, Lin SC, Lin SC, Lee JC, Lin
BW, Liou JP, Chang JY, Kuo CC, Liu YM, et al: Hypoxia-Induced
downregulation of DUSP-2 phosphatase drives colon cancer stemness.
Cancer Res. 77:4305–4316. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin SC, Hsiao KY, Chang N, Hou PC and Tsai
SJ: Loss of dual-specificity phosphatase-2 promotes angiogenesis
and metastasis via up-regulation of interleukin-8 in colon cancer.
J Pathol. 241:638–648. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Givant-Horwitz V, Davidson B, Goderstad
JM, Nesland JM, Tropé CG and Reich R: The PAC-1 dual specificity
phosphatase predicts poor outcome in serous ovarian carcinoma.
Gynecol Oncol. 93:517–523. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu W, Huang L, Xu X, Qian H and Xu W:
Anti-proliferation effect of BMI-1 in U937 cells with siRNA. Int J
Mol Med. 25:889–895. 2010.PubMed/NCBI
|
|
14
|
Wang XF, Zhao YB, Wu Q, Sun ZH and Li HJ:
Triptolide induces apoptosis in endometrial cancer via a p53
independent mitochondrial pathway. Mol Med Rep. 9:39–44. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Trastour C, Benizri E, Ettore F, Ramaioli
A, Chamorey E, Pouysségur J and Berra E: HIF-1alpha and CA IX
staining in invasive breast carcinomas: Prognosis and treatment
outcome. Int J Cancer. 120:1451–1458. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tashiro E, Henmi S, Odake H, Ino S and
Imoto M: Involvement of the MEK/ERK pathway in EGF-induced
E-cadherin down-regulation. Biochem Biophys Res Commun.
477:801–806. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Boeckx C, Op de Beeck K, Wouters A,
Deschoolmeester V, Limame R, Zwaenepoel K, Specenier P, Pauwels P,
Vermorken JB, Peeters M, et al: Overcoming cetuximab resistance in
HNSCC: The role of AURKB and DUSP proteins. Cancer Lett.
354:365–377. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen Y, Huang Y, Hou P, Zhang Z, Zhang Y,
Wang W, Sun G, Xu L, Zhou J, Bai J and Zheng J: ING4 suppresses
tumor angiogenesis and functions as a prognostic marker in human
colorectal cancer. Oncotarget. 7:79017–79031. 2016.PubMed/NCBI
|
|
19
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang X, Song Y, Song N, Zhang Y, Zhang L,
Wang Y, Wang Z, Qu X and Liu Y: RANKL/RANK pathway abrogates
cetuximab sensitivity in gastric cancer cells via activation of
EGFR and c-Src. Onco Targets Ther. 10:73–83. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Taieb J, Balogoun R, Le Malicot K,
Tabernero J, Mini E, Folprecht G, Van Laethem JL, Emile JF, Mulot
C, Fratté S, et al: Adjuvant FOLFOX +/−cetuximab in full RAS and
BRAF wildtype stage III colon cancer patients. Ann Oncol.
28:824–830. 2017.PubMed/NCBI
|
|
22
|
Queralt B, Cuyàs E, Bosch-Barrera J,
Massaguer A, de Llorens R, Martin-Castillo B, Brunet J, Salazar R
and Menendez JA: Synthetic lethal interaction of cetuximab with
MEK1/2 inhibition in NRAS-mutant metastatic colorectal cancer.
Oncotarget. 7:82185–82199. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang F, Bai L, Liu TS, Yu YY, He MM, Liu
KY, Luo HY, Zhang DS, Jin Y, Wang FH, et al: Right-sided colon
cancer and left-sided colorectal cancers respond differently to
cetuximab. Chin J Cancer. 34:384–393. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brule SY, Jonker DJ, Karapetis CS,
O'Callaghan CJ, Moore MJ, Wong R, Tebbutt NC, Underhill C, Yip D,
Zalcberg JR, et al: Location of colon cancer (right-sided versus
left-sided) as a prognostic factor and a predictor of benefit from
cetuximab in NCIC CO.17. Eur J Cancer. 51:1405–1414. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Moretto R, Cremolini C, Rossini D,
Pietrantonio F, Battaglin F, Mennitto A, Bergamo F, Loupakis F,
Marmorino F, Berenato R, et al: Location of primary tumor and
benefit from anti-epidermal growth factor receptor monoclonal
antibodies in patients with RAS and BRAF wild-type metastatic
colorectal cancer. Oncologist. 21:988–994. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dienstmann R, Vermeulen L, Guinney J,
Kopetz S, Tejpar S and Tabernero J: Consensus molecular subtypes
and the evolution of precision medicine in colorectal cancer. Nat
Rev. 17:79–92. 2017. View Article : Google Scholar
|
|
27
|
Zhu H, Wu TC, Chen WQ, Zhou LJ, Wu Y, Zeng
L and Pei HP: Screening for differentially expressed genes between
left- and right-sided colon carcinoma by microarray analysis. Oncol
Lett. 6:353–358. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Joo D, Woo JS, Cho KH, Han SH, Min TS,
Yang DC and Yun CH: Biphasic activation of extracellular
signal-regulated kinase (ERK)1/2 in epidermal growth factor
(EGF)-stimulated SW480 colorectal cancer cells. BMB Rep.
49:220–225. 2016. View Article : Google Scholar : PubMed/NCBI
|