Introduction
Receptor for advanced glycation end products (RAGE)
is a pattern recognition receptor that binds multiple ligands,
including AGE (1), S100 proteins
(2), lipopolysaccharides (3), phosphatidylserine (4), amyloid-β (Aβ) (5), and high mobility group box (HMGB)-1
(6). Interactions of these diverse
ligands with RAGE result in intracellular signaling, including
nuclear factor κ-B (NF-κB) activation, which results in pathogenic
processes such as diabetic complications (7), inflammatory diseases, Alzheimer's
disease (AD) (8) and cancer (9). Takeuchi et al (10) demonstrated that RAGE expression in
HT1080 human fibrosarcoma cells induced tumor cells to proliferate,
migrate, invade and metastasize. HMGB-1 was revealed to induce
RAS-related C3 botulinum toxin substrate (Rac)1 and cell division
control protein 42 homolog (Cdc42) functions in RAGE-expressing
HT1080 fibrosarcoma cells (10).
Epidemiological studies also demonstrated that RAGE expression was
associated with tumor malignancies of the stomach (11), colon and rectum (12–14),
prostate (15), breast (16) and bone (17). Therefore, these previous studies
suggested that RAGE represents a potential therapeutic target, and
that inhibiting RAGE may be useful to anticancer strategies.
Previously, Sakai et al (18) developed a novel drug design system,
involving the conversion of optimized binding peptide to
non-peptidic small molecules by structure-based virtual screening
(SBVS), followed by optimization of the small molecules using a
structure-based drug design system, namely conversion to small
molecules through optimized-peptide strategy (COSMOS). Using this
strategy, the most optimized binding peptide is first
computationally designed on a hot spot in the target protein.
Subsequently, the optimized binding peptide may be converted to
small molecules by SBVS based on its pharmacophore. Then, the
selected candidates are evaluated using in vitro assays.
Therefore, this strategy decreases the cost and time required to
search for effective lead compounds, for drug design and for
optimization (18). The present study
identified a RAGE inhibitor, papaverine, using this drug design
system. Papaverine is an opiate alkaloid, originally isolated from
the plant Papaver somniferum and now synthetically produced
as a direct-acting smooth muscle relaxant. Its mechanism of action
may be associated with non-selective inhibition of
phosphodiesterases and direct inhibition of calcium channels
(19,20).
The present study assessed whether papaverine
functioned as a RAGE inhibitor using in vitro cell culture
systems of RAGE- and dominant-negative (dn)RAGE-expressing HT1080
fibrosarcoma cells.
Materials and methods
Papaverine
Papaverine hydrochloride (molecular weight, 375.85
Da; product number, P0016) was purchased from Tokyo Chemical
Industry Co., Ltd. (Tokyo, Japan).
Cell lines
HT1080 human fibrosarcoma cells (American Type
Culture Collection, Manassas, VA, USA) were transfected with a
plasmid containing human full-length RAGE cDNA or cytoplasmic
domain-deleted dnRAGE cDNA, or with the vector alone, as previously
described (21,22). The cells were designated as
RAGE-expressing, dnRAGE-expressing or mock-transfected (mock)
HT1080 cells, respectively. Cells were maintained in RPMI-1640
medium (Nakarai Tesque, Kyoto, Japan) supplemented with 10% fetal
bovine serum (FBS; Nichirei Biosciences Inc., Tokyo, Japan), 100
U/ml penicillin and 100 µg/ml streptomycin in the presence of G418
(geneticin, 200 µg/ml; Roche Applied Science, Mannheim,
Germany).
NF-κB luciferase assay
Stably transfected rat C6 glioma cells that
expressed human RAGE and the NF-κB enhancer-luciferase system
(pNF-κB-Luc; Agilent Technologies, Inc., Santa Clara, CA, USA) were
used in this assay, as previously described (21). After a 4 h incubation at 37°C in a
humidified 5% CO2 atmosphere in Dulbecco's modified
Eagle's medium supplemented with 0.1% FBS, the cells were
stimulated with 1 µg/ml HMGB-1 (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) or with 100 µg/ml glyceraldehyde-derived
AGE-modified bovine serum albumin (BSA) (Sigma-Aldrich; Merck KGaA)
(21) with/without 10 or 20 µM
papaverine or 0.08% dimethy sulfoxide (DMSO) as a negative control
for 4 h at 37°C in a humidified 5% CO2 atmosphere.
Luciferase activity was determined using the luciferase assay
system (Promega Corporation, Madison, WI, USA) and an LB 941
Multimode Reader TriStar (Berthold Technologies GmbH & Co. KG,
Bad Wildbad, Germany). These experiments were repeated three
times.
Western blot analysis
RAGE- and dnRAGE-expressing HT1080 cells and mock
control cells which were incubated with 0, 10 and 20 µM papaverine
for 72 h at 37°C in a humidified 5% CO2 atmosphere, were
washed with ice cold PBS, scraped off in PBS and pelleted by
centrifugation at 300 × g for 5 min at 4°C. The cells were lysed
immediately by sonication in 1% Triton X-100 (Sigma-Aldrich; Merck
KGaA), 50 mM Tris-HCl, pH 7.5, 150 mM NaCl and protease inhibitor
cocktail (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), and then
centrifuged at 10,000 × g for 10 min at 4°C. The cell lysates (50
µg of protein) were separated via 12.5% SDS-PAGE and electroblotted
onto polyvinylidene fluoride membranes (EMD Millipore, Billerica,
MA, USA). The membranes were blocked at room temperature for 1 h
with 5% (w/v) non-fat dried milk in PBS, and then incubated at room
temperature for 1 h with either a rabbit anti-human RAGE-specific
polyclonal antibody (1:1,000) produced as described previously
(21) or a mouse anti β-actin
antibody (cat. no. A5441, 1:10,000; Sigma-Aldrich; Merck KGaA).
Goat anti-rabbit IRDye 680 (cat. no. P/N 926-32221) and goat
anti-mouse IRDye 800CW (P/N 925-32210) were diluted 10,000-fold and
used as the secondary antibodies. The antigen-antibody complex was
visualized using the Odyssey Infrared Imaging System version 3.0
(LI-COR Biosciences, Lincoln, NE, USA). These experiments were
repeated two times. Secondary antibody only was used as a negative
control.
Plate-binding assays
Competitive binding inhibition assay with papaverine
was performed using a 96-well AGE-BSA-coated plate as previously
described (21). Briefly, 50 ng/ml
soluble RAGE (sRAGE) was incubated with or without 10 or 20 µM of
papaverine or 0.08% DMSO as a negative control on an AGE-BSA-coated
plate at room temperature for 1 h. Following incubation and washed
three times with 0.01% Tween-20, 0.15 M NaCl, 20 mM Tris-HCl (pH
7.5), horseradish peroxidase (HRP)-labeled anti-RAGE antibody (in
human esRAGE ELISA kit; ready-to-use; cat. no. K1009-1; B-Bridge
International, Inc., Santa Clara, CA, USA) was added and the plate
was further incubated at room temperature for 1 h. The HRP-labeled
antibody-sRAGE-AGE complex was then detected by measuring the
absorbance at 450 nm using the microplate reader iMark (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). These experiments were
repeated three times.
Cell proliferation assay
RAGE- and dnRAGE-expressing HT1080 cells and mock
control cells were inoculated (1×103 cells/well) at 37°C
in a humidified 5% CO2 atmosphere in a 96-well plate (BD
Biosciences, Franklin Lakes, NJ, USA) containing RPMI-1640 medium
supplemented with 10% FBS and 0, 10 or 20 µM papaverine or 0.08%
DMSO as a negative control. Following inoculation, the total viable
cell number was counted using a hemocytometer using the dye
exclusion method with 0.2% Trypan blue at room temperature (Thermo
Fisher Scientific, Inc.) using an inverted light microscope
(Primovert, Zeiss, Carl Zeiss Industrielle Messtechnik GmbH,
Oberkochen, Germany) at magnification, ×4 objective at 0, 24, 48
and 72 h. These experiments were repeated three times.
Cell migration assay
Cell migration was evaluated using the monolayer
denudation assay as previously described (10). Briefly, RAGE- and dnRAGE-expressing
HT1080 and mock control cells were inoculated (2×105
cells/well) and were cultured to 100% confluence in a 12-well
plate. Cells were then wounded by denuding a strip of the monolayer
(width, ~1 mm) with a 200 µl pipette tip. Cells were washed twice
with serum-free RPMI-1640 medium, and then incubated for 20 h at
37°C under a humidified 5% CO2 atmosphere in RPMI-1640
containing 0.1% FBS with/without 10 or 20 µM papaverine or 0.08%
DMSO as negative control. The rate of wound closure was assessed in
four separate fields of view 20 h after denudation using a light
microscope (magnification, ×4 objective). These experiments were
repeated three times.
Cell invasion assay
A total of mg/ml of matrigel-coated porous filters
(pore size, 8 µm) in a 24-well format (BD Biosciences) were used as
barriers in Boyden chambers to assess the extent of invasion by
RAGE- and dnRAGE-expressing HT1080 cells and mock controls. Cells
were plated (2×105 cells) in the upper chambers with
RPMI-1640 containing 0.1% BSA in the presence or absence of 10 or
20 µM papaverine or 0.08% DMSO as negative control. The lower
chambers were filled with 750 µl RPMI-1640 containing 1% FBS in the
presence or absence of 10 or 20 µM papaverine. Following a 20-h
incubation at 37°C in a humidified 5% CO2 atmosphere,
membranes were cut and removed from the insert housings. The filter
membrane was fixed in 4% paraformaldehyde at room temperature for 2
min following the removal of non-invasive cells from the upper
surface using a cotton swab. The bottom surface with the invasive
cells was stained with 0.1% crystal violet at room temperature for
1 min, and the invasive cells were counted in six fields of view
using a light microscope (magnification, ×4 objective) as
previously described (10). These
experiments were repeated three times.
Statistical analysis
Statistical analysis was performed using one-way
analysis of variance with the Tukey-Kramer post-hoc test. These
tests were conducted using Ekuseru-Toukei 2015 (Social Survey
Research Information Co., Ltd., Tokyo, Japan). Data are presented
as mean ± standard error of the mean. P<0.05 was considered to
indicate a statistically significant difference.
Results
Inhibitory effects of papaverine on
RAGE-dependent NF-κB activity
The present study assessed the inhibitory effects of
papaverine on the RAGE-dependent NF-κB intracellular signaling
pathway. Using the C6 glioma system, which reflected RAGE-dependent
NF-κB activity (21), HMGB-1-induced
activation of NF-κB was evaluated in the presence or absence of
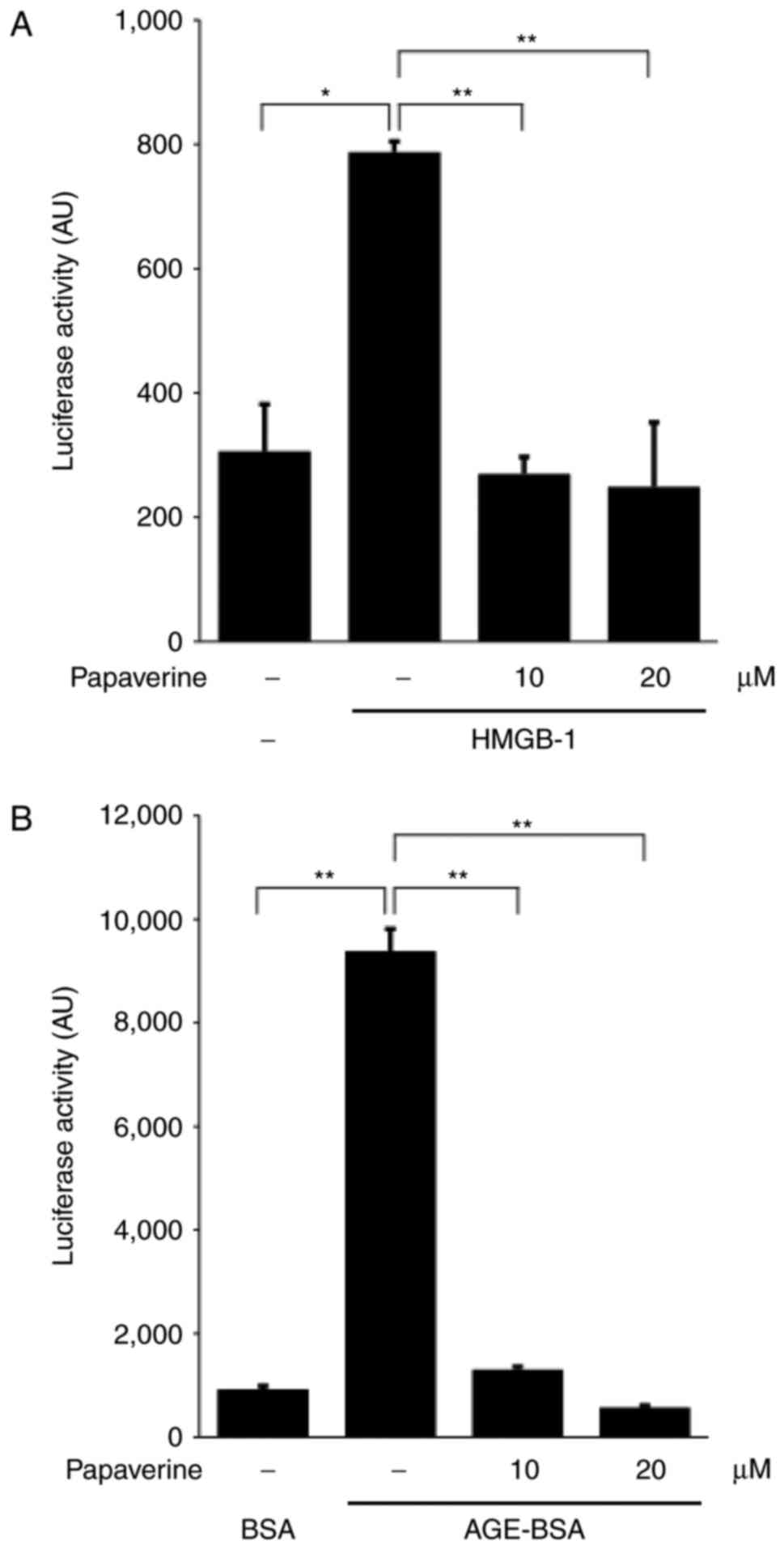
papaverine in the culture media. Adding HMGB-1 significantly
induced intracellular NF-κB activation (P<0.01); however, this
upregulation was inhibited by papaverine treatment (10 or 20 µM;
Fig. 1A). In addition, the increase
in NF-κB activity induced by glyceraldehyde-derived AGE-BSA,
another RAGE ligand, was significantly inhibited by papaverine
(Fig. 1B). The results of the present
study suggested that papaverine could inhibit the RAGE-dependent
intracellular signaling pathway. The present study subsequently
confirmed the papaverine-RAGE binding using a plate assay (21). Papaverine significantly and
dose-dependently competed for the binding between
glyceraldehyde-derived AGE-BSA and recombinant soluble RAGE
(Fig. 1C), which suggested that the
binding site of papaverine to RAGE may be shared with that of
AGE-BSA.
Inhibitory effects of papaverine on
RAGE-dependent cell proliferation, migration and invasion
To mimic RAGE-dependent tumor malignant behaviors
and assess the inhibitory effects of papaverine, the present study
used an established human fibrosarcoma cell line HT1080 that
expressed human RAGE and dnRAGE and the mock control cells. The
HT1080 cells expressed HMGB1 and secreted HMGB1 levels did not
differ among the mock-transfected, RAGE-expressing and
dnRAGE-expressing HT1080 cells (10).
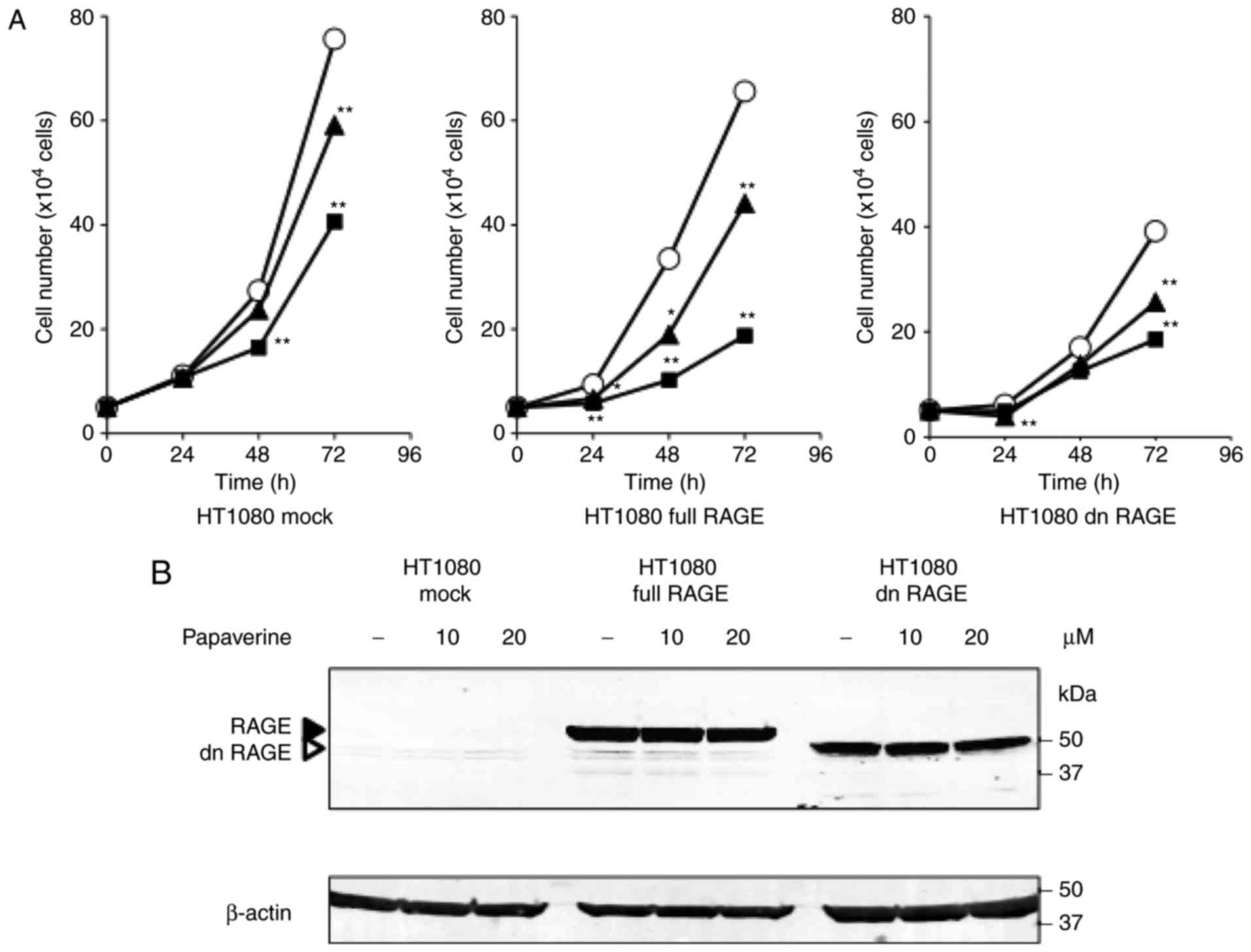
The present study assessed the effects of papaverine treatment on
HT1080 cell proliferation. Papaverine significantly and
dose-dependently decreased the proliferation rate of
RAGE-expressing HT1080 cells (P<0.05 vs. non-treated HT1080
cells; P<0.01 vs. non-treated HT1080 cells). In particular, at
20 µM it was most effective between treated and non-treated
conditions in RAGE-expressing HT1080 cells (65.5% reduction) at 48
h (Fig. 2A). A reduction (39.8%) was
also observed between treated and non-treated conditions in the
mock control cells at 48 h; however, its effectiveness was
decreased in dnRAGE-expressing HT1080 cells (26.2% reduction)
(Fig. 2A). Papaverine did not change
the protein expression levels of RAGE and dnRAGE in the
RAGE-expressing and dnRAGE-expressing HT1080 cells, respectively
(Fig. 2B). Whether papaverine could
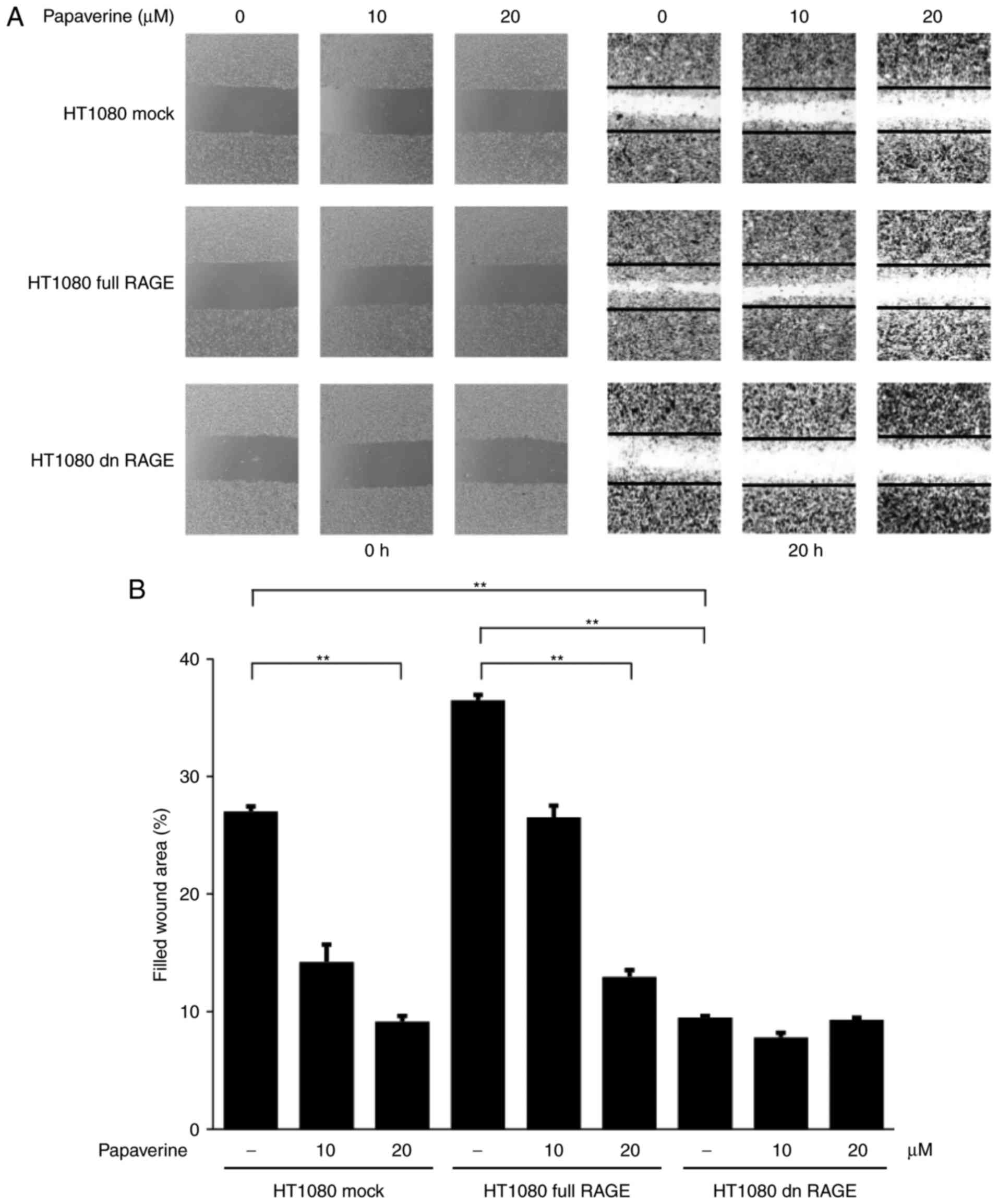
inhibit the RAGE-dependent migration and invasion of HT1080 cells
was also assessed in vitro. The inhibitory effects of
papaverine on cell migration were observed between treated and
non-treated RAGE-expressing HT1080 cells. Additionally, the
inhibitory effects were observed between treated and non-treated
conditions in mock control cells (Fig.
3). However, no inhibitory effects of papaverine on migration
were observed in dnRAGE-expressing HT1080 cells (Fig. 3), which indicated a selective
inhibitory action of papaverine against RAGE. Furthermore,
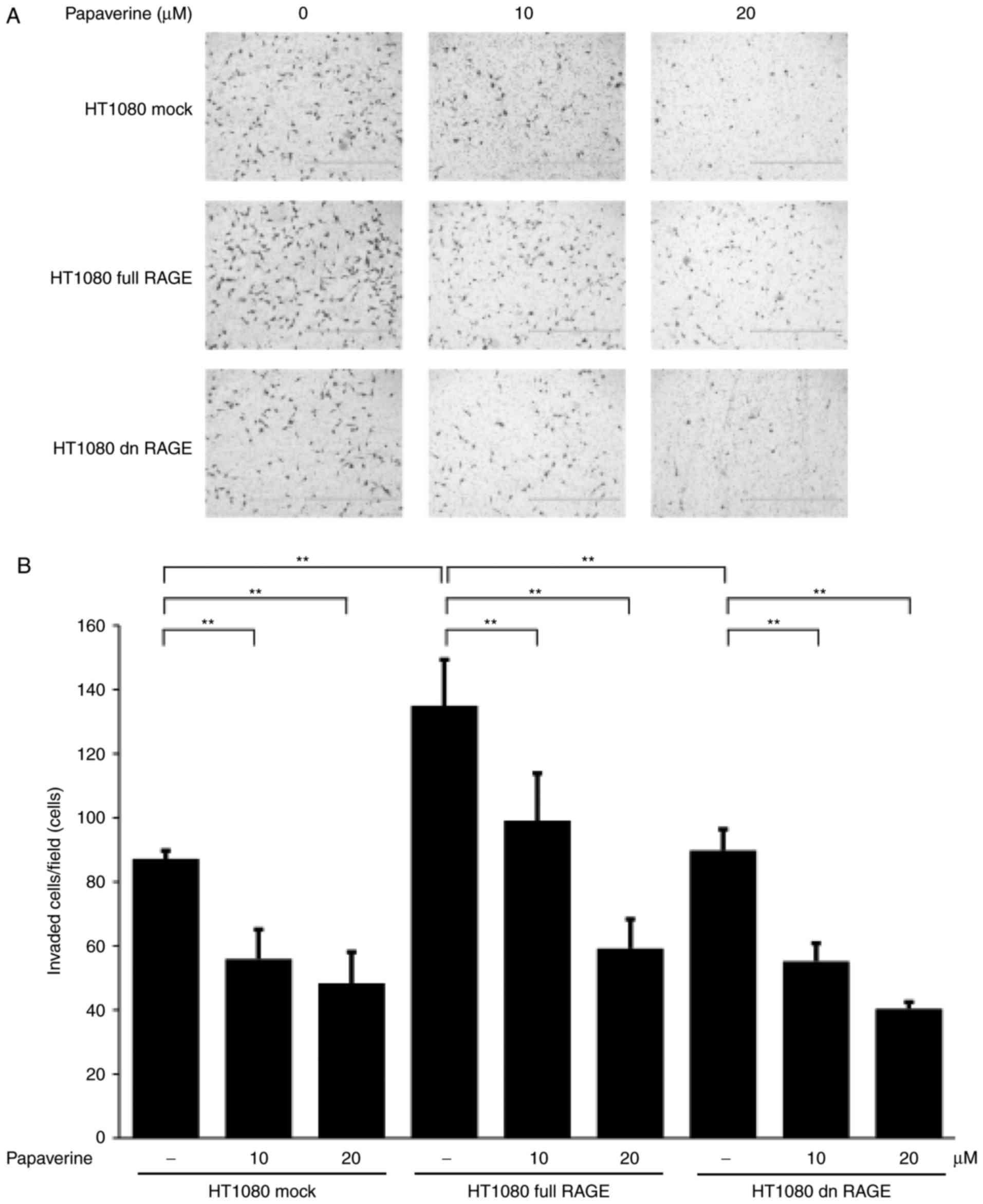
papaverine treatment significantly and dose-dependently inhibited
HT1080 cell invasion in the Matrigel assay (P<0.01; Fig. 4). A statistically significant
inhibitory effect of papaverine treatment was also identified
between treated and non-treated dnRAGE-expressing HT1080 cells
(P<0.01). The results of the present study demonstrated that
papaverine may inhibit RAGE-dependent and RAGE-independent
malignant phenotypes of cancer cells; the latter may include an
antitumor effect of papaverine via an elevation of intracellular
cyclic AMP by phosphodiesterase inhibition (8).
Discussion
RAGE is a multi-ligand, pattern recognition receptor
that has been implicated in the growth, progression and metastasis
of multiple types of human cancer (23,24). RAGE
ligands include S100/calgranulins (S100A4, S100A6, S100A7,
S100A8/9, S100A14, S100B and S100P) (2,25–29) and HMGB-1 protein, which have been
demonstrated to be upregulated in glioma, melanoma, bladder, liver,
pancreatic, prostate, colorectal, gastric and lung cancer (30–34).
Ligand-RAGE signaling pathways enhance the properties associated
with malignant tumor phenotypes (10,22,35–37),
including by activating members of the small GTPase family (Cdc42
and Rac1), members of the mitogen-activated protein kinase family
(extracellular signal-regulated kinase, p38 and stress-activated
protein kinases/c-Jun amino-terminal kinases), NF-κB and the formin
homology 1 domain (10,22,35–37).
Therefore, blocking RAGE and ligand-RAGE signaling could represent
a potential strategy for treating certain types of cancer. Previous
experimental studies have revealed that inhibiting RAGE suppressed
tumor growth, invasion and angiogenesis in multiple types of cancer
(10,16,38). The
therapeutic efficacy of blocking RAGE from interacting with HMGB-1
was initially demonstrated in glioma cells, in which this blockade
inhibited tumor growth and invasion (16). Subsequently, strategies and components
of RAGE inhibition have been reported in oncology and other fields,
including neurology (39,40). For example, RAGE-neutralizing
antibodies and sRAGE decreased the emergence of lung metastasis
following intracardiac injection of Lewis lung carcinoma cells
(39,40). In addition, endogenous secretory
receptor for advanced glycation end products, another soluble decoy
form of RAGE, was demonstrated to inhibit Aβ-42 uptake into mouse
brain; therefore, it may be effective in AD (41). Hong et al (42) evaluated the effects of the
RAGE-specific inhibitor FPS-ZM1 on Aβ metabolism, AGE-induced
inflammation and oxidative stress in rat hippocampus. In addition,
blocking RAGE signaling in tumor-associated macrophages has been
proposed as a potential anticancer strategy; the macrophages form
the tumor microenvironment, which could drive tumor angiogenesis
(30). FPS-ZM1 directly inhibited
primary tumor growth; in addition, it blocked RAGE signaling in
tumor-associated macrophages, inhibited tumor angiogenesis and
inflammatory cell recruitment and inhibited metastasis to the lungs
and liver (16). Another RAGE-binding
peptide, RP-1, was demonstrated to inhibit Aβ-induced cellular
stress in human neuroblastoma cells in vitro (43). Furthermore, Han et al (44) reported that
4,6-bisphenyl-2-(3-alkoxyanilino) pyrimidine inhibited the binding
of Aβ to RAGE.
The present study identified papaverine as a RAGE
inhibitor using the drug design system COSMOS, an example of drug
repositioning, the application of well-known existing drugs and
compounds for novel indications. As a structure from which
researchers could develop novel drugs, papaverine could represent a
potential precursor to a therapeutic RAGE inhibitor. RAGE has been
implicated in multiple pathogenic processes, in cancer and numerous
other diseases, including diabetes, atherosclerosis, inflammatory
and neurodegenerative diseases (7–9,45). Therefore, papaverine and its
derivatives could be useful in preventing and treating multiple
RAGE-associated diseases. To clarify our understanding of RAGE
inhibition by papaverine, in vivo animal models should be
used in future studies.
To conclude, the results of the present study
suggested papaverine could inhibit RAGE and provided novel insights
into the field of RAGE biology.
Acknowledgements
The authors would like to thank Ms. Yuko Niimura
(Kanazawa University) for her technical assistance. The Government
of Egypt funded the visit of Dr El-Far to Japan as a post-doctoral
fellow for 6 months in 2015. The present study was supported by the
Japan Society for the Promotion of Science (grant no.
26450152).
Glossary
Abbreviations
Abbreviations:
|
RAGE
|
receptor for advanced glycation
end-products
|
|
COSMOS
|
conversion to small molecules through
optimized-peptide strategy
|
|
NF-κB
|
nuclear factor κB
|
|
AGE
|
advanced glycation end-products
|
|
Aβ
|
amyloid-β
|
|
HMGB
|
high-mobility group box
|
|
Rac1
|
RAS-related C3 botulinum toxin
substrate 1
|
|
Cdc42
|
cell division control protein 42
homolog
|
|
SBVS
|
structure-based virtual screening
|
|
ERK
|
extracellular signal-regulated
kinase
|
References
|
1
|
Schmidt AM, Vianna M, Gerlach M, Brett J,
Ryan J, Kao J, Esposito C, Hegarty H, Hurley W, Clauss M, et al:
Isolation and characterization of two binding proteins for advanced
glycosylation end products from bovine lung which are present on
the endothelial cell surface. J Biol Chem. 267:14987–14997.
1992.PubMed/NCBI
|
|
2
|
Hofmann MA, Drury S, Fu C, Qu W, Taguchi
A, Lu Y, Avila C, Kambham N, Bierhaus A, Nawroth P, et al: RAGE
mediates a novel proinflammatory axis: A central cell surface
receptor for S100/calgranulin polypeptides. Cell. 97:889–901. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yamamoto Y, Harashima A, Saito H,
Tsuneyama K, Munesue S, Motoyoshi S, Han D, Watanabe T, Asano M,
Takasawa S, et al: Septic shock is associated with receptor for
advanced glycation endproducts (RAGE) ligation of LPS. J Immunol.
186:3248–3257. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He M, Kubo H, Morimoto K, Fujino N, Suzuki
T, Takahasi T, Yamada M, Yamaya M, Maekawa T, Yamamoto Y and
Yamamoto H: Receptor for advanced glycation end products binds to
phosphatidylserine and assists in the clearance of apoptotic cells.
EMBO Rep. 12:358–364. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yan SD, Chen X, Fu J, Chen M, Zhu H, Roher
A, Slattery T, Zhao L, Nagashima M, Morser J, et al: RAGE and
amyloid-beta peptide neurotoxicity in Alzheimer's disease. Nature.
382:685–691. 1996. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hori O, Brett J, Slattery T, Cao R, Zhang
J, Chen J, Nagashima M, Lundh ER, Vijay S, Nitecki D, et al: The
receptor for advanced glycation end products (RAGE) is a cellular
binding site for amphoterin: Mediation of neurite outgrowth and
co-expression of rage and amphoterin in the developing nervous
system. J Biol Chem. 270:25752–25761. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yamamoto Y, Kato I, Doi T, Yonekura H,
Ohashi S, Takeuchi M, Watanabe T, Yamagishi S, Sakurai S, Takasawa
S, et al: Development and prevention of advanced diabetic
nephropathy in RAGE-overexpressing mice. J Clin Invest.
108:261–268. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schmidt AM, Yan SD, Yan SF and Stern DM:
The multiligand receptor RAGE as a progression factor amplifying
immune and inflammatory responses. J Clin Invest. 108:949–955.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sims GP, Rowe DC, Rietdijk ST, Herbst R
and Coyle AJ: HMGB1 and RAGE in inflammation and cancer. Annu Rev
Immunol. 28:367–388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takeuchi A, Yamamoto Y, Munesue S,
Harashima A, Watanabe T, Yonekura H, Yamamoto H and Tsuchiya H: Low
molecular weight heparin suppresses receptor for advanced glycation
end products-mediated expression of malignant phenotype in human
fibrosarcoma cells. Cancer Sci. 104:740–749. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kuniyasu H, Oue N, Wakikawa A, Shigeishi
H, Matsutani N, Kuraoka K, Ito R, Yokozaki H and Yasui W:
Expression of receptors for advanced glycation end-products (RAGE)
is closely associated with the invasive and metastatic activity of
gastric cancer. J Pathol. 196:163–170. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fuentes MK, Nigavekar SS, Arumugam T,
Logsdon CD, Schmidt AM, Park JC and Huang EH: RAGE activation by
S100P in colon cancer stimulates growth, migration, and cell
signaling pathways. Dis Colon Rectum. 50:1230–1240. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Onyeagucha BC, Mercado-Pimentel ME,
Hutchison J, Flemington EK and Nelson MA: S100P/RAGE signaling
regulates microRNA-155 expression via AP-1 activation in colon
cancer. Exp Cell Res. 319:2081–2090. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mercado-Pimentel ME, Onyeagucha BC, Li Q,
Pimentel AC, Jandova J and Nelson MA: The S100P/RAGE signaling
pathway regulates expression of microRNA-21 in colon cancer cells.
FEBS Lett. 589:2388–2393. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ishiguro H, Nakaigawa N, Miyoshi Y,
Fujinami K, Kubota Y and Uemura H: Receptor for advanced glycation
end products (RAGE) and its ligand, amphoterin are overexpressed
and associated with prostate cancer development. Prostate.
64:92–100. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kwak T, Drews-Elger K, Ergonul A, Miller
PC, Braley A, Hwang GH, Zhao D, Besser A, Yamamoto Y, Yamamoto H,
et al: Targeting of RAGE-ligand signaling impairs breast cancer
cell invasion and metastasis. Oncogene. 36:1559–1572. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang Q, Jin Y, Zhao CF, Wang WJ and Liu
GY: Receptor for advanced glycation end-products (RAGE) is
overexpressed in human osteosarcoma and promotes the proliferation
of osteosarcoma U-2OS cells in vitro. Genet Mol Res. 15:2016.
|
|
18
|
Sakai J, Yoshimori A, Nose Y, Mizoroki A,
Okita N, Takasawa R and Tanuma S: Structure-based discovery of a
novel non-peptidic small molecular inhibitor of caspase-3. Bioorg
Med Chem. 16:4854–4859. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kukovetz WR and Pöch G: Inhibition of
cyclic-3′, 5′-nucleotide-phosphodiesterase as a possible mode of
action of papaverine and similarly acting drugs. Naunyn
Schmiedebergs Arch Pharmakol. 267:189–194. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Iguchi M, Nakajima T, Hisada T, Sugimoto T
and Kurachi Y: On the mechanism of papaverine inhibition of the
voltage-dependent Ca++ current in isolated smooth muscle
cells from the guinea pig trachea. J Pharmacol Exp Ther.
263:194–200. 1992.PubMed/NCBI
|
|
21
|
Myint KM, Yamamoto Y, Doi T, Kato I,
Harashima A, Yonekura H, Watanabe T, Shinohara H, Takeuchi M,
Tsuneyama K, et al: RAGE control of diabetic nephropathy in a mouse
model: Effects of RAGE gene disruption and administration of
low-molecular weight heparin. Diabetes. 55:2510–2522. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huttunen HJ, Fages C and Rauvala H:
Receptor for advanced glycation end products (RAGE)-mediated
neurite outgrowth and activation of NF-κB require the cytoplasmic
domain of the receptor but different down signaling pathways. J
Biol Chem. 274:19919–19924. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huttunen HJ, Fages C, Kuja-Panula J,
Ridley AJ and Rauvala H: Receptor for advanced glycation end
products-binding COOH-terminal motif of amphoterin inhibits
invasive migration and metastasis. Cancer Res. 62:4805–4811.
2002.PubMed/NCBI
|
|
24
|
Wajsman Z, Williams P and Murphy GP: A
study of the effect of papaverine in neuroblastoma using the
experimental C1300 murine system. Oncology. 35:1–4. 1978.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wolf S, Haase-Kohn C, Lenk J, Hoppmann S,
Bergmann R, Steinbach J and Pietzsch J: Expression, purification
and fluorine-18 radiolabeling of recombinant S100A4: A potential
probe for molecular imaging of receptor for advanced glycation
endproducts in vivo? Amino Acids. 41:809–820. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Leclerc E, Fritz G, Weibel M, Heizmann CW
and Galichet A: S100B and S100A6 differentially modulate cell
survival by interacting with distinct RAGE (receptor for advanced
glycation end products) immunoglobulin domains. J Biol Chem.
282:31317–31331. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wolf R, Howard OM, Dong HF, Voscopoulos C,
Boeshans K, Winston J, Divi R, Gunsior M, Goldsmith P, Ahvazi B, et
al: Chemotactic activity of S100A7 (Psoriasin) is mediated by the
receptor for advanced glycation end products and potentiates
inflammation with highly homologous but functionally distinct
S100A15. J Immunol. 181:1499–1506. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jin Q, Chen H, Luo A, Ding F and Liu Z:
S100A14 stimulates cell proliferation and induces cell apoptosis at
different concentrations via receptor for advanced glycation end
products (RAGE). PLoS One. 6:e193752011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Padilla L, Dakhel S and Hernández JL: S100
to receptor for advanced glycation end-products binding assay:
Looking for inhibitors. Biochem Biophys Res Commun. 446:404–409.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen X, Zhang L, Zhang IY, Liang J, Wang
H, Ouyang M, Wu S, da Fonseca AC, Weng L, Yamamoto Y, et al: RAGE
expression in tumor-associated macrophages promotes angiogenesis in
glioma. Cancer Res. 74:7285–7297. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Khorramdelazad H, Bagheri V, Hassanshahi
G, Karami H, Moogooei M, Zeinali M and Abedinzadeh M: S100A12 and
RAGE expression in human bladder transitional cell carcinoma: A
role for the ligand/RAGE axis in tumor progression? Asian Pac J
Cancer Prev. 16:2725–2729. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Meghnani V, Wagh A, Indurthi VS, Koladia
M, Vetter SW, Law B and Leclerc E: The receptor for advanced
glycation end products influences the expression of its S100
protein ligands in melanoma tumors. Int J Biochem Cell Biol.
57:54–62. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kostova N, Zlateva S, Ugrinova I and
Pasheva E: The expression of HMGB1 protein and its receptor RAGE in
human malignant tumors. Mol Cell Biochem. 337:251–258. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sparvero LJ, Asafu-Adjei D, Kang R, Tang
D, Amin N, Im J, Rutledge R, Lin B, Amoscato AA, Zeh HJ and Lotze
MT: RAGE (Receptor for Advanced Glycation Endproducts), RAGE
ligands, and their role in cancer and inflammation. J Transl Med.
7:172009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu R, Duan L, Cui F, Cao J, Xiang Y, Tang
Y and Zhou L: S100A9 promotes human hepatocellular carcinoma cell
growth and invasion through RAGE-mediated ERK1/2 and p38 MAPK
pathways. Exp Cell Res. 334:228–238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Iotzova-Weiss G, Dziunycz PJ, Freiberger
SN, Läuchli S, Hafner J, Vogl T, French LE and Hofbauer GF:
S100A8/A9 stimulates keratinocyte proliferation in the development
of squamous cell carcinoma of the skin via the receptor for
advanced glycation-end products. PLoS One. 10:e01209712015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hudson BI, Kalea AZ, Del Mar Arriero M,
Harja E, Boulanger E, D'Agati V and Schmidt AM: Interaction of the
RAGE cytoplasmic domain with diaphanous-1 is required for
ligand-stimulated cellular migration through activation of Rac1 and
Cdc42. J Biol Chem. 283:34457–34468. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kalea AZ, See F, Harja E, Arriero M,
Schmidt AM and Hudson BI: Alternatively spliced RAGEv1 inhibits
tumorigenesis through suppression of JNK signaling. Cancer Res.
70:5628–5638. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Taguchi A, Blood DC, del Toro G, Canet A,
Lee DC, Qu W, Tanji N, Lu Y, Lalla E, Fu C, et al: Blockade of
RAGE-amphoterin signalling suppresses tumour growth and metastases.
Nature. 405:354–360. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mizumoto S, Takahashi J and Sugahara K:
Receptor for advanced glycation end products (RAGE) functions as
receptor for specific sulfated glycosaminoglycans, and anti-RAGE
antibody or sulfated glycosaminoglycans delivered in vivo inhibit
pulmonary metastasis of tumor cells. J Biol Chem. 287:18985–18994.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sugihara T, Munesue S, Yamamoto Y, Sakurai
S, Akhter N, Kitamura Y, Shiba K, Watanabe T, Yonekura H, Hayashi
Y, et al: Endogenous secretory receptor for advanced glycation
end-products inhibits amyloid-β1-42 uptake into mouse brain. J
Alzheimers Dis. 28:709–720. 2012.PubMed/NCBI
|
|
42
|
Hong Y, Shen C, Yin Q, Sun M, Ma Y and Liu
X: Effects of RAGE-specific inhibitor FPS-ZM1 on amyloid-β
metabolism and AGEs-induced inflammation and oxidative stress in
rat hippocampus. Neurochem Res. 41:1192–1199. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cai C, Dai X, Zhu Y, Lian M, Xiao F, Dong
F, Zhang Q, Huang Y and Zheng Q: A specific RAGE-binding peptide
biopanning from phage display random peptide library that
ameliorates symptoms in amyloid β peptide-mediated neuronal
disorder. Appl Microbiol Biotechnol. 100:825–835. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Han YT, Kim K, Son D, An H, Kim H, Lee J,
Park HJ, Lee J and Suh YG: Fine tuning of
4,6-bisphenyl-2-(3-alkoxyanilino)pyrimidine focusing on the
activity-sensitive aminoalkoxy moiety for a therapeutically useful
inhibitor of receptor for advanced glycation end products (RAGE).
Bioorg Med Chem. 23:579–587. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kalea AZ, Schmidt AM and Hudson BI: RAGE:
A novel biological and genetic marker for vascular disease. Clin
Sci (Lond). 116:621–637. 2009. View Article : Google Scholar : PubMed/NCBI
|