Introduction
Breast cancer is among the most commonly diagnosed
types of malignant tumor in women worldwide (1). Bone is the most common site of distant
metastasis in patients with breast cancer and >70% of all
patients with breast cancer eventually develop bone metastases,
which presents clinical challenges (2). There is an increased risk of mortality
for patients with breast cancer once bone metastasis has occurred
(3). Therefore, elucidation of the
mechanism of breast cancer bone metastasis is required for the
identification of novel therapeutic targets for the prevention or
control of bone metastasis.
Metastasis is associated with the migratory ability
of tumor cells. Pseudopodia form through the rearrangement of the
cell cytoskeleton and serve key roles in cell migration (4). Microtubules are essential for
pseudopodia extension and regulation of cell movement, katanin is
an ATPase that causes microtubule degradation (5–7). Katanin
consists of p60 and p80 subunits (8);
p80 targets the p60 subunit to the centrosome, and promotes the
microtubule-severing activity of p60 (9). Research has demonstrated that leucine
zipper tumor suppressor 2 (LAPSER1) and katanin p80 co-localize in
the centrosome, and are involved in mitosis and cell movement
(10). However, the activity of the
p60 subunit remains poorly characterized and requires further
research.
p60 is a 60-kDa enzymatic subunit containing an
ATPases associated with diverse cellular activities (AAA) domain,
which is responsible for microtubule-severing activity (11) and directly regulates
microtubule-severing by phosphorylating katanin p60 at the Ser131
site (12). E3 ubiquitin ligases
participate in the degradation of katanin p60 (13,14). In
the context of disease, research into p60 function has primarily
focused on the role of p60 in neurogenesis (15–17). To
the best of our knowledge, the expression of katanin p60 in tumor
metastasis has only been reported in prostate cancer (18), and its role in breast cancer is
unknown. In the present study, the distribution and expression of
katanin p60 in clinical breast cancer specimens was investigated,
and it was determined whether katanin p60 was involved in breast
cancer cell proliferation or promotion of breast cancer bone
metastasis.
Materials and methods
Cell culture
The triple negative breast cancer cell line,
MDA-MB-231 and metastatic invasive ductal carcinoma cell line,
MCF-7 were purchased from the Type Culture Collection of the
Chinese Academy of Sciences (Shanghai, China). Cells were cultured
in DMEM (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal calf serum (FCS; Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 U/ml streptomycin
(Beyotime Institute of Biotechnology, Shanghai, China) at 37°C in
5% CO2.
Tissue samples and
immunohistochemistry
The primary breast cancer specimens and breast
cancer bone metastases specimens were divided into primary, and
metastasis groups, each containing 10 specimens. All tissues were
obtained from the Xiangyang Central Hospital between April 2013 and
November 2016; the mean age of the patients was 61.2±13.1 years
(37–83 years), written informed consent was obtained from all
patients. The present study was approved by the Xiangyang Central
Hospital Ethics Committee (Xiangyang, China). Paraffin tissue
sections of 5 µm thickness were dewaxed with xylene and rehydrated
with graded alcohol, antigen retrieval was performed using 10 mM
citrate buffer pH 6.0 (3 mg/ml trisodium citrate, 0.4 mg/ml citric
acid; Sinopharm Chemical Reagent Co., Ltd., Shanghai, China). Then
sections were treated with 3% hydrogen peroxide at room temperature
for 15 min and blocked with 5% sheep serum (Beyotime Institute of
Biotechnology) at 37°C for 30 min. Subsequent to an overnight
incubation at 4°C with anti-katanin p60 primary antibody (dilution
1:200; cat no. ab111881; Abcam, Cambridge, UK), an ABC kit (cat no.
SA1022; Boster Biological Technology, Pleasanton, CA, USA) was used
for protein visualization. ImageJ software (version 1.46; National
Institutes of Health, Bethesda, MD, USA) was used to evaluate the
mean optical density of the immunohistochemical staining for each
group.
Katanin p60 plasmids and
transfection
Katanin p60 is encoded by the gene katanin catalytic
subunit A1 (KATNA1; GenBank Accession no. 007044). The pcDNA3.1 and
pcDNA3.1/KATNA1 plasmids were designed and constructed by Chongqing
Weisiteng Biomedical Science and Technology Co., Ltd. (Chongqing,
China). MDA-MB-231 and MCF-7 cells were seeded in 6-well plates at
a density of 1.5×105 cells/well, and transfected with
pcDNA3.1 or pcDNA3.1/KATNA1 when the cell confluence reached
50–70%. Two cell lines were transfected under the same condition. A
total of 1800 µl Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.)
was added to each well, followed by a mixture of 2 µg plasmid DNA,
6 µl X-tremeGENE transfection reagent (Roche, Basel, Switzerland)
and 200 µl Opti-MEM, the control group was cultured only with
Opti-MEM. Following an incubation of 5 h at 37°C, cells were
collected at different time points for subsequent studies. The
transfection efficiency was detected by western blotting and
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR).
shRNA
The shRNAs were constructed and identified by
Chongqing Weisiteng Biomedical Science and Technology Co., Ltd.,
and the sequences were as follows: shRNA-KATNA1, forward,
5′-GGUUCAGAUGGAUGGUGUUTT-3′ and reverse,
5′-AACACCAUCCAUCUGAACCTT-3′); shRNA-negative control (NC), forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′). A total of 9 µl
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) diluted in Opti-MEM and 3 µg shRNA-NC or
shRNA-KATNA1 (0.5 µg/µl) diluted in Opti-MEM were mixed for 20 min,
then the mixture was added into MDA-MB-231 or MCF-7 cells (6-well
plates) for 6 h at 37°C, the transfection efficiency be detected by
western blotting analysis and RT-qPCR.
RNA isolation and RT-qPCR
Cells were collected after transfected with plasmids
or shRNAs for 48 h. At room temperature, total RNA was extracted
with TRIzol (Thermo Fisher Scientific, Inc.) for 5 min, reacted
with chloroform for 2–3 min, isopropanol for 10 min and 75% ethanol
for 1 min sequentially, then RNA was dissolved in 0.1% diethyl
pyrocarbonate (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The
concentration and purity of RNA were measured using a
spectrophotometer (GE Healthcare, Chicago, IL, USA). Reverse
transcription was performed using PrimeScript™ 1st
strand cDNA Synthesis Kit (cat no. 6610A; Takara Biomedical
Technology, Beijing, China), for 10 min at 25°C, 50 min at 42°C and
5 min at 85°C. qPCR analysis was performed using SYBR®
Premix DimerEraser™ (Perfect Real Time; cat no. RR091A;
Takara Biomedical Technology). The primer sequences were as
follows: Katanin p60, forward, 5′-TAAACTGGACAGCACTCCCTTG-3′ and
reverse, 5′-CCTGGTGAGGGTCTTCGTTC-3′; actin, forward,
5′TGACGTGGACATCCGCAAAG-3′ and reverse, 5′-CTGGAAGGTGGACAGCGAGG-3′.
The thermocycling parameters were as follows: 94°C for 4 min; 35
cycles of 94°C for 20 sec, 60°C for 30 sec and 72°C for 30 sec. The
relative expression of katanin p60 was obtained according to the
2−ΔΔCq method (19).
Western blot analysis
After transfected with shRNAs or plasmids for 48 h,
cells were lysed using radioimmunoprecipitation assay buffer
containing 1% Triton X-100, 1% sodium deoxycholate and 0.1% SDS
(0.1 ml/1×106 cells; Beyotime Institute of
Biotechnology). Total protein was extracted and its concentration
was measured using a BCA protein assay kit (Beyotime Institute of
Biotechnology). Extracted proteins (50 µg) were separated by
SDS-PAGE (5% stacking gels, 12% resolving gels) and transferred
into a nitrocellulose membrane. Subsequent to blocking with 5% skim
milk (GE Healthcare, Chicago, IL, USA) for 2 h at room temperature,
the membranes were washed and incubated with anti-katanin p60
antibody (dilution, 1:500; Abcam, Cambridge, UK) and anti-GADPH
antibody (dilution, 1:1,000; Abcam) overnight at 4°C. Then the
membranes were washed with 1X TBST buffer (pH 7.6; 2.42 g/l Tris, 8
g/l NaCl, 0.5 ml Tween-20) and incubated with the
peroxidase-conjugated anti-rabbit secondary antibody (dilution,
1:1,000; cat no. A0545; Sigma-Aldrich; Merck KGaA) for 90 min at
room temperature, an ECL chemiluminescence kit (Pierce; Thermo
Fisher Scientific, Inc.) was used to visualize the proteins.
Densitometry was performed using ImageJ software (version 1.46;
National Institutes of Health, Bethesda, MD, USA).
Cell proliferation assay
Un-transfected group, negative control and
shRNA-KATNA1 or pcDNA3.1/KATNA1-transfected cells were seeded into
96-well plates at 1×104 cells/well for 24, 48, 72 and 96
h. Cells were cultured with 10 µl MTT (5 mg/ml; Sigma-Aldrich;
Merck KGaA) for 4–6 h, then 150 µl dimethyl sulfoxide was added
(Amresco, LLC, Solon, OH, USA) and incubated for 10 min.
Subsequently, the absorbance was measured using a multimode reader
at 490 nm. Each experiment was repeated three times for
construction of the growth curve.
Cell migration assay
Un-transfected group, negative control and
shRNA-KATNA1 or pcDNA3.1/KATNA1-transfected cells were cultured for
24 h. Then, 2.5×104 cells were added to the upper
chamber of Transwell filters, a total of 200 µl DMEM supplemented
with 5% BSA (Gibco; Thermo Fisher Scientific, Inc.) was added to
each lower chamber, and incubated for 24 h at 37°C. The cells were
fixed with 70% formaldehyde for 30–60 min and stained with 0.1%
crystal violet for 30 min (Sigma-Aldrich; Merck KGaA) at room
temperature, and the number of migrated cells was counted using
ImageJ software (version 1.46; National Institutes of Health).
Statistical analysis
All data were analyzed by SPSS 17.0 software (SPSS,
Inc., Chicago, IL, USA) and expressed as the mean ± standard error.
Student's t-test was used to determine the statistical
significance. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression of katanin p60 in breast
cancer and bone metastasis
It has been demonstrated that the expression of
katanin p60 contributes to the progression of prostate cancer,
indicated that katanin p60 may also serve an important role in
breast cancer (18). To test this
hypothesis, immunohistochemical staining of tissues with an
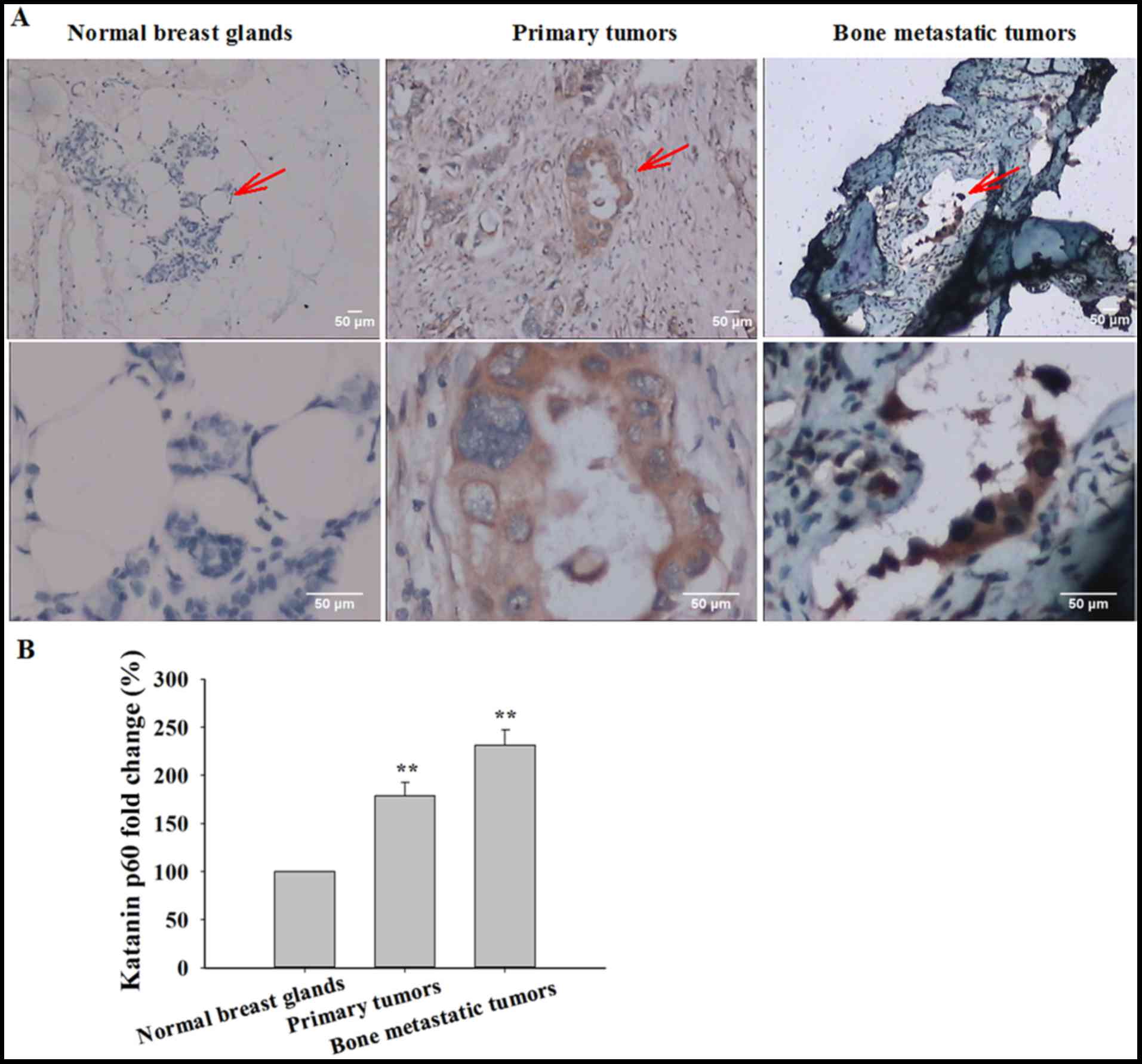
anti-katanin p60 antibody (Table I)
was performed. Low expression of katanin p60 was exhibited in
healthy breast tissue, and katanin p60 expression was primarily
identified in the cytoplasm (Fig.
1A). The expression of katanin p60 was significantly increased
in primary breast cancer tissue compared with healthy control
tissue (178.96±13.81%; P=0.001; Fig.
1B). In bone metastatic breast cancer, the expression of
katanin p60 was further increased when compared with healthy breast
tissue (231.48±16.00%; P=0.001), and with primary breast cancer
(P=0.023; Fig. 1B). Together, these
data indicate that katanin p60 may function in the regulation of
breast cancer cell proliferation and migration.
 | Table I.Immunohistochemical staining of
katanin p60. |
Table I.
Immunohistochemical staining of
katanin p60.
| Tissue type | No. of patients | Mean optical
density | Standard error | P-value (compared
with healthy breast) |
|---|
| Healthy breast | 10 | 0.1687 | 0.0074 |
|
| Primary breast
tumor | 10 | 0.3019 | 0.0233 | 0.001 |
| Bone metastatic
tumor | 10 | 0.3905 | 0.0270 | 0.001 |
Up- or downregulation of katanin
p60
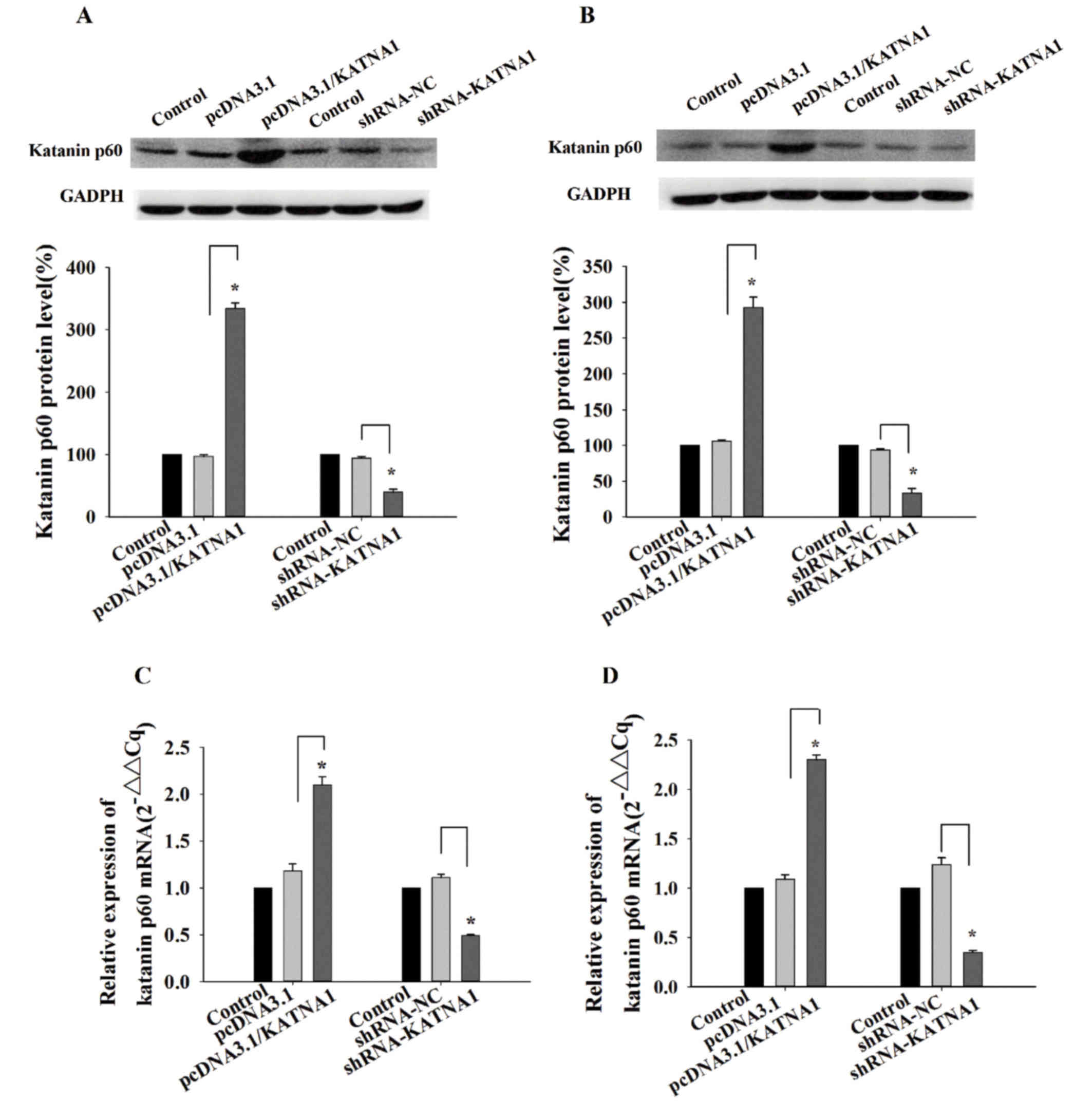
To determine the function of katanin p60 in breast
cancer cells, plasmids and shRNAs were utilized to up- or
downregulate its expression. RT-qPCR and western blotting were used
to detect the expression of katanin p60 at the mRNA, and protein
levels. In MDA-MB-231 cells and MCF-7 cells, the expression of
katanin p60 was significantly increased following transfection with
pcDNA3.1/KATNA1 (Fig. 2). In
addition, the protein and mRNA levels of katanin p60 were
significantly decreased following transfection with shRNA-KATNA1
(Fig. 2).
Function of katanin p60 on cell
proliferation
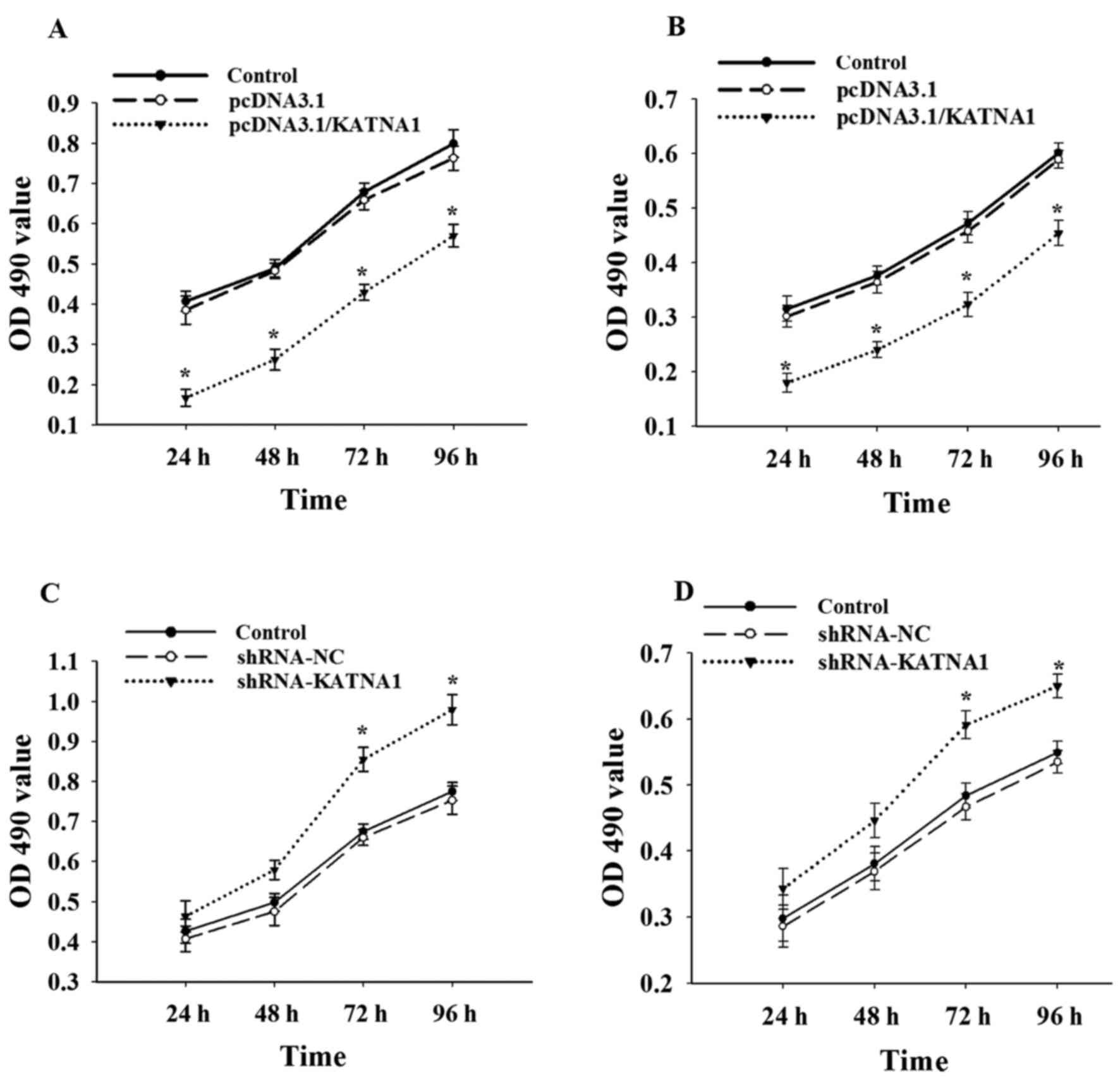
To further investigate whether katanin p60 was
involved in the regulation of cell proliferation, cells were
collected at 24, 48, 72 and 96 h after transfection with plasmids
or shRNAs (Fig. 3). The present study
demonstrated no significant difference in cell proliferation
between MDA-MB-231 and MCF-7 cells. Upregulation of katanin p60
expression with pcDNA3.1/KATNA1 reduced cell proliferation, the
percentage of proliferating cells at 24 h was 40.72±2.54% for
MDA-MB-231 cells and 56.82±1.34% for MCF-7 cells (Fig. 3A and B). No significant differences in
cell proliferation were identified compared with the shRNA-NC group
at 24 or 48 h following transfection with shRNA-KATNA1; however,
the percentage of proliferating cells at 72 h was significantly
increased, at 126.64±0.88% in MDA-MB-231 cells and 122.03±0.56% in
MCF-7 cells (Fig. 3C and D). This
indicates that katanin p60 may serve a role in breast cancer cell
proliferation.
Katanin p60 is necessary for cell
migration
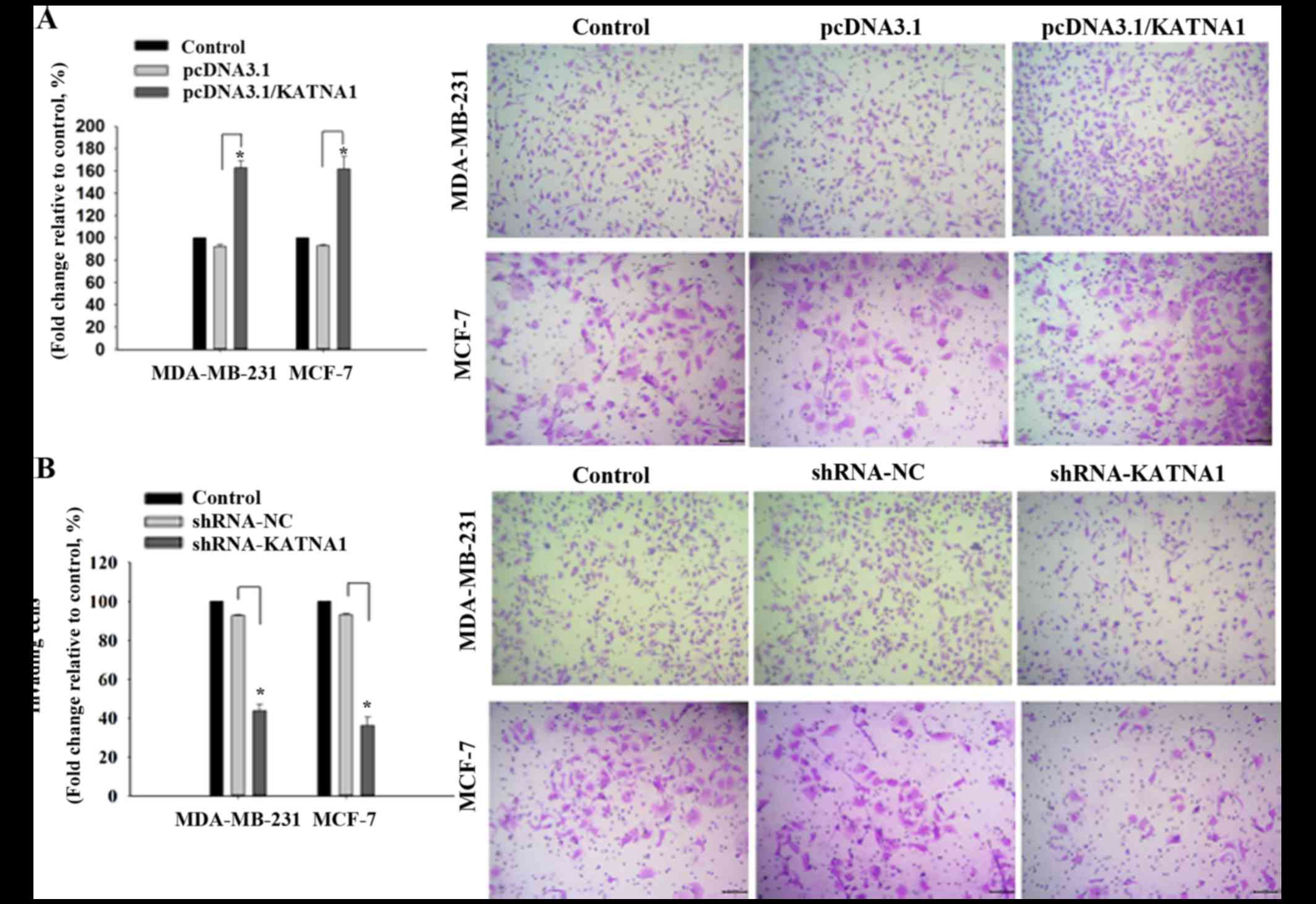
To investigate whether katanin p60 was involved in
cell migration, plasmids or shRNAs were transfected into cells to
achieve up- and downregulation of p60, respectively, and the
resultant effects on cell migration were analyzed. It was
demonstrated that upregulation of katanin p60 expression
accelerated cell migration in MDA-MB-231 and MCF-7 cells compared
with pcDNA3.1 cells, with fold changes of 162.95±6.20% and
161.84±11.23% (Fig. 4A).
Downregulated expression of katanin p60 reduced cell mobility in
MDA-MB-231 cells (43.78±3.28%) and MCF-7 cells (36.18±4.23%)
compared with shRNA-NC cells (Fig.
4B). No significant difference was identified in the metastatic
rate between the two cell lines. These results indicated that
katanin p60 may promote tumor cell spread to other sites, and are
supportive of the aforementioned results achieved in breast cancer
tissue specimens and bone metastasis tissue specimens.
Discussion
Katanin p60 contains an N-terminal domain that is
connected with microtubules, which are required for cell motility
and spindle formation (11). Previous
studies have reported that katanin p60 oligomerization increases
the affinity of katanin for microtubules, thus increasing
microtubule degradation (20,21). In the present study, it was
demonstrated that katanin p60, a member of the AAA ATPase family,
was expressed differently between healthy breast tissue specimens,
primary and bone metastatic breast cancer specimens. The expression
of katanin p60 in bone metastatic breast cancer was significantly
higher, which was positively associated with tumor metastasis,
indicating that katanin p60 may serve a role in breast cancer cell
proliferation and metastasis.
The present study demonstrates that the expression
levels of katanin p60 mRNA and protein increased significantly
subsequent to transfection with pcDNA3.1/KATNA1. Simultaneously,
the number of breast cancer cells decreased. Downregulation of
katanin p60 expression using shRNA resulted in an increased cell
number. These results indicate that katanin p60 expression and cell
proliferation are positively associated. Previous research has
reported an association between katanin p60 expression and cell
proliferation (18). Spindle length
is controlled by katanin in mitosis and meiosis, essential for
chromosome segregation and cytokinesis (22). Katanin p60 has also been demonstrated
to facilitate microtubule instability (23). Microtubule binding by the katanin p60
subunit is important for katanin in targeting the spindle poles
(24). These studies suggest that
katanin p60 may affect cell division by regulating spindle
length.
Cell migration is an essential process in tumor
metastasis. Previous studies have indicated that pseudopodial
protrusion is associated with tumor cell migration and invasion
(25,26). Drosophila katanin p60 is reported to
regulate the interactions of the cortical-microtubule plus-end,
which is involved in cell migration (27). The appropriate distribution and
content of katanin p60 has been demonstrated as critical for
neuronal migration (28). The present
study confirmed that overexpression of katanin p60 significantly
promoted cell migration, and reduced expression inhibited cell
migration in MDA-MB-231 and MCF-7 cell lines. Therefore, it is
hypothesized that katanin p60 may affect breast cancer cell
migration by regulating the formation of cellular pseudopodia.
Overall, the present study supports the concept that
katanin p60 functions in cell proliferation and migration. A
previous study reported that purine-type compounds interact with
katanin p60, which induce cell death of NSCLC cells (29). Together, these results suggest that
katanin p60 is required for breast cancer cell proliferation and
bone metastases, and, therefore, has exciting potential as a
therapeutic target. Future research by this group will focus on the
association between katanin p60, spindle and cellular pseudopodia
in breast cancer cells, in order to further elucidate the
regulatory mechanisms of katanin p60.
Acknowledgements
The current study was supported by the Hubei Health
and Family Planning Commission Programs (grant no. WJ2015MB185) and
the Hubei Nature Science Foundation (grant no. 2014CFC1077).
References
|
1
|
Kozlow W and Guise TA: Breast cancer
metastasis to bone: Mechanisms of osteolysis and implications for
therapy. J Mammary Gland Biol Neoplasia. 10:169–80. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mundy GR: Metastasis to bone: Causes,
consequences and therapeutic opportunities. Nat Rev Cancer.
2:584–593. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cetin K, Christiansen CF, Sværke C,
Jacobsen JB and Sørensen HT: Survival in patients with breast
cancer with bone metastasis: A Danish population-based cohort study
on the prognostic impact of initial stage of disease at breast
cancer diagnosis and length of the bone metastasis-free interval.
BMJ Open. 5:e0077022015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nabi IR: The polarization of the motile
cell. J Cell Sci. 112:1803–1811. 1999.PubMed/NCBI
|
|
5
|
Roll-Mecak A and McNally FJ:
Microtubule-severing enzymes. Curr Opin Cell Biol. 22:96–103. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hartman JJ, Mahr J, McNally K, Okawa K,
Iwamatsu A, Thomas S, Cheesman S, Heuser J, Vale RD and McNally FJ:
A microtubule-severing protein, is a novel AAA ATPase that targets
to the centrosome using a WD40-containing subunit. Cell.
93:277–287. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bershadsky AD and Vasiliev JM: Mechanisms
of regulation of pseudopodial activity by the microtubule system.
Symp Soc Exp Biol. 47:353–373. 1993.PubMed/NCBI
|
|
8
|
McNally FJ and Vale RD: Identification of
katanin, an ATPase that severs and disassembles stable
microtubules. Cell. 75:419–429. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu W, Solowska JM, Qiang L, Karabay A,
Baird D and Baas PW: Regulation of microtubule severing by katanin
subunits during neuronal development. J Neurosci. 25:5573–5583.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sudo H and Maru Y: LAPSER1 is a putative
cytokinetic tumor suppressor that shows the same centrosome and
midbody subcellular localization pattern as p80 katanin. FASEB J.
21:2086–2100. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Johjima A, Noi K, Nishikori S, Ogi H,
Esaki M and Ogura T: Microtubule severing by katanin p60 AAA+
ATPase requires the C-terminal acidic tails of both alpha- and
beta-tubulins and basic amino acid residues in the AAA+ ring pore.
J Biol Chem. 290:11762–11770. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Whitehead E, Heald R and Wilbur JD:
N-terminal phosphorylation of p60 katanin directly regulates
microtubule severing. J Mol Biol. 425:214–221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cummings CM, Bentley CA, Perdue SA, Baas
PW and Singer JD: The Cul3/Klhdc5 E3 ligase regulates p60/katanin
and is required for normal mitosis in mammalian cells. J Biol Chem.
284:11663–11675. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang SW, Oh KH, Park E, Chang HM, Park JM,
Seong MW, Ka SH, Song WK, Park DE, Baas PW, et al: USP47 and C
terminus of Hsp70-interacting protein (CHIP) antagonistically
regulate katanin-p60-mediated axonal growth. J Neurosci.
33:12728–12738. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu W, Qiang L, Solowska JM, Karabay A,
Korulu S and Baas PW: The microtubule-severing proteins spastin and
katanin participate differently in the formation of axonal
branches. Mol Biol Cell. 19:1485–1498. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen K, Ye Y, Ji Z, Tan M, Li S, Zhang J,
Guo G and Lin H: Katanin p60 promotes neurite growth and collateral
formation in the hippocampus. Int J Clin Exp Med. 7:2463–2470.
2014.PubMed/NCBI
|
|
17
|
Korulu S, Yildiz-Unal A, Yuksel M and
Karabay A: Protein kinase C activation causes neurite retraction
via cyclinD1 and p60-katanin increase in rat hippocampal neurons.
Eur J Neurosci. 37:1610–1619. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ye X, Lee YC, Choueiri M, Chu K, Huang CF,
Tsai WW, Kobayashi R, Logothetis CJ, Yu-Lee LY and Lin SH: Aberrant
expression of katanin p60 in prostate cancer bone metastasis.
Prostate. 72:291–300. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hartman JJ and Vale RD: Microtubule
disassembly by ATP-dependent oligomerization of the AAA enzyme
katanin. Science. 286:782–785. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rasi MQ, Parker JD, Feldman JL, Marshall
WF and Quarmby LM: Katanin knockdown supports a role for
microtubule severing in release of basal bodies before mitosis in
Chlamydomonas. Mol Biol Cell. 20:379–388. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McNally K, Audhya A, Oegema K and McNally
FJ: Katanin controls mitotic and meiotic spindle length. J Cell
Biol. 175:881–891. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Matsuo M, Shimodaira T, Kasama T, Hata Y,
Echigo A, Okabe M, Arai K, Makino Y, Niwa S, Saya H and Kishimoto
T: Katanin p60 contributes to microtubule instability around the
midbody and facilitates cytokinesis in rat cells. PLoS One.
8:e803922013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
McNally KP, Bazirgan OA and McNally FJ:
Two domains of p80 katanin regulate microtubule severing and
spindle pole targeting by p60 katanin. J Cell Sci. 113:1623–1633.
2000.PubMed/NCBI
|
|
25
|
Shankar J, Messenberg A, Chan J, Underhill
TM, Foster LJ and Nabi IR: Pseudopodial actin dynamics control
epithelial-mesenchymal transition in metastatic cancer cells.
Cancer Res. 70:3780–3790. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guirguis R, Margulies I, Taraboletti G,
Schiffmann E and Liotta L: Cytokine-induced pseudopodial protrusion
is coupled to tumour cell migration. Nature (Lond). 329:261–263.
1987. View
Article : Google Scholar
|
|
27
|
Zhang D, Grode KD, Stewman SF,
Diaz-Valencia JD, Liebling E, Rath U, Riera T, Currie JD, Buster
DW, Asenjo AB, et al: Drosophila Katanin is a microtubule
depolymerase that regulates cortical-microtubule plus-end
interactions and cell migration. Nat Cell Biol. 13:361–370. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Toyo-Oka K, Sasaki S, Yano Y, Mori D,
Kobayashi T, Toyoshima YY, Tokuoka SM, Ishii S, Shimizu T,
Muramatsu M, et al: Recruitment of katanin p60 by phosphorylated
NDEL1, an LIS1 interacting protein, is essential for mitotic cell
division and neuronal migration. Hum Mol Genet. 14:3113–3128. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kuo TC, Li LW, Pan SH, Fang JM, Liu JH,
Cheng TJ, Wang CJ, Hung PF, Chen HY, Hong TM, et al: Purine-type
compounds induce microtubule fragmentation and lung cancer cell
death through interaction with Katanin. J Med Chem. 59:8521–8534.
2016. View Article : Google Scholar : PubMed/NCBI
|